Abstract
MYH 3 is a major contractile protein which converts chemical energy into mechanical energy through the ATP hydrolysis. MYH 3 is mainly expressed in the skeletal muscle in different stages especially embryonic period, and it has a role in the development of skeletal muscle and heart. In this study, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) was applied to analyze the genetic variations of the MYH 3 gene and verify the effect on growth and carcass traits in a total of 365 Qinchuan cattles. The PCR product was digested with some restriction enzyme and demonstrated the polymorphism in the population, the single nucleotide polymorphisms (SNPs) at nucleotides g. +1215T>C, g. +3377C>T, and g. +28625C>T were in linkage disequilibrium with each other. The result of haplotype analysis showed that nineteen different haplotypes were identified among the five SNPs. The statistical analyses indicated that the five SNPs were significant association with growth and carcass traits (P < 0.05, N = 365); whereas the five SNPs were no significant association between 18 combined genotypes of MYH 3 gene and growth and carcass traits. Taken together, our results provide the evidence that polymorphisms in MYH 3 are associated with growth and carcass traits in Qinchuan cattle, and may be used as a possible candidate for marker-assisted selection and management in beef cattle breeding program.
Keywords: Qinchuan cattle, Growth and carcass traits, Combined genotypes, Haplotype, MYH3
Introduction
Multiple myosin isoforms are encoded by a multigene family and expressed in different developmental stages and fiber types. There are three skeletal fast myosin heavy chain (MyHC) genes (IIa, IIdx, and IIb), but only the promoter region of the mouse IIb gene has been analyzed to date [1–6]. The MyHCs encoded by a group of genes consisting of IIa, IIb [7], IIx [8], extraocular [9], embryonic [10] and neonatal [11] genes. These genes are located on the chromosome 14 [12], which expressed at different times during development and in different fiber types [13–16].
Myosin proteins consist of both heavy and light chains and are present in skeletal muscle. Heavy chains are associated with the speed of muscular contractions, while the role of light chains is not well defined [17]. Studies have shown that myosin heavy chain 3 (MYH 3) gene may be one of major genes implicated in the differences of muscle fiber property between intact males and castrated Qinchuan cattle [18]. The MYH 3 is a major structural protein of the thick filament of the sarcomere. MYH 3 gene is a member of the MYH family and encodes a protein with an IQ domain [19]. Polymorphism of MHC isoform expression in single myofibers, which points out that single fibers of both developing and adult skeletal muscle exist as hybrids [20–23]. In a word, MYH 3 gene is expressed during the embryonic period [24], at the same time, MYH 3 has a role in skeletal muscle development [13, 25, 26].
In human, the MYH 3 gene is located on chromosome 17, and it is a molecular motor that converts chemical energy into mechanical force [27]. Conventionally, class II myosin is a hexameric protein composed of two MyHC subunits, each with a molecular weight of approximately 220 kDa and two pairs of non-identical myosin light chain subunits. Proteolytic enzymes can cleave the MyHC into two sub-fragments: heavy meromyosin (HMM) and light meromyosin (LMM). The expression of MYH 3 is predominated in myotubes fated to become fast myofibers and is gradually replaced by the expressions of other myosin genes (MYH 1, MYH 2 and MYH 4) [28]. During the development and the adult, α-MYH is predominately expressed in the atrial chamber and β-MYH in the ventricular chamber in both humans and chicks [29–33]. This is further illustrated by the increased expression of β-MHC in the atria of human hearts under pressure overload [34, 35]. MHC analysis of human soleus muscle shows that this muscle expresses an approximately equal mix of type I and IIa isoforms, but the type IIx MHC is not expressed [36]. In contrast, human fast muscles such as the vastus lateralis express a mix of all three types of MHC isoforms at variable proportions [36, 37], depending on the physical fitness and activity of the subject. In addition, the study shows that knockdown in the early embryo leads to abnormal atrial septal development and heart enlargement [38]. These studies all point to the potentially vital function of MYH 3 in regulating gene expression at the skeletal muscle and heart development.
In a rat model, the MYH 3 gene is located on chromosome 10. It is an essential protein with skeletal muscle development. β-MHC has been shown to be down-regulated in the setting of hypoxia, likely as a result of neuroendocrine stimulation [39]. Cytokines and tumor factors such as TNF-α have been demonstrated to selectively target MYH 2 gene and down-regulate transcription in a mouse model [40]. MYH 3 is plastic in the adult rodent heart. In contrast to described human, the adult heart of small animals such as rodents and mice mainly expresses the α-MHC throughout the adult state. These results also support the idea that MYH 3 plays an important role in heart development.
In this study, we used DNA sequencing, PCR-RFLP analysis to investigate allelic variation of MYH 3 gene that encompasses five SNPs. Furthermore, we reported associations among these SNPs of the MYH 3 gene in the Qinchuan cattle breed and the linkage disequilibrium between the variations and we analyzed the relationship between single nucleotide polymorphisms and growth and carcass traits.
Materials and methods
Animal source and preparation of DNA samples
Cattle from the Qinchuan (QC) commercial breeds were randomly selected, for a total of 365 animals. These animals (30 ± 2 months of the age at slaughter) were reared in Shaanxi province, P.R. China. The growth traits (body length, withers height and hip width) and carcass traits (slaughter weight and carcass weight) were measured according to the criteria of GB/T17238-1998 Cutting Standard of Fresh and Chilled Beef in China (China Standard Publishing House). All experimental protocols and animal care were performed according to authorization granted by the Chinese Ministry of Agriculture. Genomic DNA was isolated from 2 % heparin-treated blood samples and stored at −80 °C, following the standard procedures [41].
PCR amplification and DNA sequencing analysis
Primers used to amplify bovine MYH 3 gene were designed from a published gene sequence (GenBank accession number NC_007317). Primers, restriction enzymes selected (ABI, Foster City CA), The primer sequences, location and fragment sizes were listed in Table 1. The detection of allelic variation at the SNPs were based on the electrophoretic pattern of the restriction enzyme-treated PCR products.
Table 1.
Primer sets for PCR and PCR-RFLP used for genotyping SNPs detected in bovine MYH 3 gene
| SNPS | Amplified region | Primer sequence (5′–3′) | AT(°C) | SAF (bp) | PR | Genotype |
|---|---|---|---|---|---|---|
| SNP1 | Exon5 | AAAGCCCGAGTATCAGAACC TCCAACAGCCTCATCAAACA | 57.4 | 435 | RSaI | CC: 228, 207 |
| CT:228, 207, 150, 78 | ||||||
| TT:207, 150, 78 | ||||||
| SNP2 | Intron5 | AAAGCCCGAGTATCAGAACC TCCAACAGCCTCATCAAACA | 57.4 | 435 | BstXI | TT:435 |
| TC:435, 339, 96 | ||||||
| CC: 339, 96 | ||||||
| SNP3 | Intron7 | TTGCTGCAAATGGCATTATT TGAGATTCCCAACAAAGAGG | 59.8 | 447 | PVUII | GG:447 |
| GC:447, 222, 225 | ||||||
| CC: 222, 225 | ||||||
| SNP4 | Exon14 | TGGAGGATTTTCAGAGGGGT TGATATGGGGTGACAAGTGG | 59.3 | 532 | RSaI | CC:532 |
| CT:532, 364, 168 | ||||||
| TT:364, 168 | ||||||
| SNP5 | Exon14 | TGGAGGATTTTCAGAGGGGT TGATATGGGGTGACAAGTGG | 59.3 | 532 | TaqI | CC:433, 99 |
| CT:433, 243, 190, 99 | ||||||
| TT:243, 190, 99 |
SNPs: single nucleotide polymorphisms; SNP1 = C1878T; SNP2 = T2010C; SNP3 = G3746C; SNP4 = C7294T; SNP5 = C7315T
AT: annealing temperature, SAF: size of amplification fragment, PR: PCR-RFLP
PCR amplifications were performed in a total volume of 15 μL, where the volume mixture contained: 50 ng of genomic DNA as template, 10 mM Tris–HCl buffer (pH 8.8), containing 50 mM KCl, 0.2 μM of each primer, 200 μM dNTP and 0.5 U Taq DNA polymerase (MBI Fermentas, USA). The Mg2+ concentration was optimized for each primer set. PCR conditions were as follows: after an initial denaturation of 5 min at 95 °C, amplicons were generated for 35 cycles of 30 s at 94 °C, 30 s at an optimal annealing temperature, and 45 s at 72 °C, followed by a 10 min final extension at 72 °C.
Restriction fragment length polymorphism analysis was used to identify the genotypes of sequence variants. The PCR products were digested in a total volume of 10 μL containing: 5 μL of PCR product, three units of restriction enzyme (0.3 μL), 1 μL of reaction buffer and 3.7 μL of ddH2O. The mixture was incubated for 10 h at specific temperature (RsaI, BstXI, PVUII and TaqI restriction enzymes were digested at 37, 55, 37 and 65 °C, respectively). Digested PCR products were mixed with 10× loading buffer and subjected to 2.5 % agarose gel electrophoresis in 1× TBE at constant voltage (110 V) for 1.0 h at room temperature. Individuals were then genotyped based on different electrophoresis patterns. Fragments displaying different PCR-RFLP patterns were purified with Qiaquick spin columns (Qiagen) and sequenced with the ABI PRISM 3730 sequencer (ABI, Foster City CA) and sequences were analyzed with BioXM software (Version 2.6). The sequences obtained were named with letters of the alphabet.
Linkage disequilibrium and statistical analysis
Genotypic frequency and allelic frequency were determined for each breed by direct counting. The formulas were as follows:
(ni is the number of i genotypic, “Ni” is the frequency of the i allele, “N” is the number of alleles)
-
(2)
The pattern of pairwise linkage disequilibrium (LD) between the SNPs was measured by LD coefficient (D′) and correlation coefficient (r 2), the measurements were determined using the program Haploview (http://www.broad.mit.edu/mpg/haploview) [42].
-
(3)
The χ2 test was used to determine Hardy–Weinberg equilibrium of the mutation. Population genetic indices, including heterozygosity (He), homozygosity (Ho), polymorphism information content (PIC) and effective allele numbers (Ne) were calculated according to Nei et al. [43]. The formulas were as follows:
(“Pi” is the frequency of the i allele, “n” is the number of alleles)
-
(4)
Haplotypes were obtained for each animal using the PHASE computer program (Ver 2.1) [44].
-
(5)
The SPSS software (Version 16.0) was used to analyze the association between genotypes and traits in Qinchuan cattle.
-
(6)
Combined genotypes of SNPs and growth traits association analyses were carried out to explore the possible interaction between the SNPs. The model was similar to that of single marker association analysis, except that the interaction between the two SVs was included as a fixed effect.
Results
Sequence variants identified of MYH3 gene
The bovine MYH 3 gene located on chromosome 19, and has been revealed that it contained 42 exons and encoded 1249 amino acids. In the present study, Genomic DNA of Qinchuan breeds was successfully amplified using primer pairs for the MYH 3 gene (Table 1).
In total, five variants including three exons and two introns mutation were identified in the study (Table 2). According to the information of cattle MYH 3 (GenBank Accession number: NC_007317), the T2010C and G3746C mutations were in intron 5 and intron 7, respectively, while the C1878T, C7294T and C7315T mutations were in exon 5, exon 14 and exon 14, respectively. According to the sequence mutations, the PCR products could be digested with RSaI, BstXI, PVUII and TaqI restriction enzymes. Distinct banding patterns were analyzed in this study and shown in Table 2. The SNPs of g. +1878C>T and g. +7315C>T resulted in synonymous mutations of Tyr626Tyr and Val2439Val, respectively, while the SNP of g. +7294C>T brought a missense mutation Arg2432Cys. Interestingly, it was firstly found that five novel mutations could be detected by endonucleases restriction site in bovine MYH 3 gene.
Table 2.
Description of the SNPs at the bovine MYH 3 gene
| SNPs name | Alleles | Amino acid change |
|---|---|---|
| SNP1 C1878T | TAC/TAT | Tyr626Tyr |
| SNP2 T2010C | TGT/TGC | – |
| SNP3 G3746C | TGT/TCT | – |
| SNP4 C7294T | CGT/TGT | Arg2432Cys |
| SNP5 C7315T | CGA/TGA | Val2439Val |
Note: SNPs: single nucleotide polymorphisms; SNP1 = C1878T; SNP2 = T2010C; SNP3 = G3746C; SNP4 = C7294T; SNP5 = C7315T
It was a very useful strategy to scan large sample size sequence mutations with the methods of DNA sequencing and PCR-RFLP, which would overcome the inaccuracy, the complicated technical demands, slow speed and unstable reproducibility. Interestingly, in this study, the PCR-RFLP method was successfully carried out to accurately detect the polymorphisms of the MYH 3 gene.
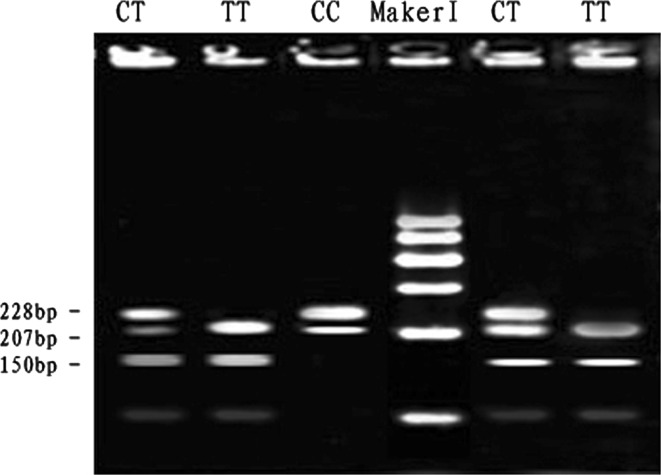
At the SNP1-RSaI locus, digestion of the 435 bp PCR fragment of MYH 3 exon 5 with RSaI resulted in fragment lengths of 228, 207, 150 and 78 bp for genotype CT; 228, 207, 150 and 78 bp for genotype TT and 228 and 207 bp for genotype CC (see Fig. 1). The frequencies of genotype and allele were calculated in the Qinchuan bovine population (Table 2). The frequency of allele T was dominant in the Qinchuan cattles and CT genotype was more frequent than other genotypes. The genotypic frequencies of SNP1-RSaI locus in Qinchuan cattle population agreed with Hardy–Weinberg disequilibrium (0.01 < P < 0.05) (Table 3).
Fig. 1.
The 3 % agarose gels electrophoretic patterns of the MYH 3 gene in bovine. M = DNA molecular weight marker is Marker I; C1878T genotype: CC = 228 + 207 bp, TT = 207 + 150 + 78 bp, CT = 228 + 207 + 150 + 78 bp; It is difficult to see the 78 bp DNA fragment on 3 % agarose gel. Forced PCR-RFLP detection results of MYH 3 gene PCR product
Table 3.
Genotypic and allelic frequencies (%), value of χ2 test and diversity parameter of bovine MYH 3 gene
| SNPs | Genotype | Number | GF | Allele | AF | χ2(HWE)e | He | H O | Ne | PIC |
|---|---|---|---|---|---|---|---|---|---|---|
| SNP1 | CC | 51 | 0.1397 | C | 0.4205 | |||||
| CT | 205 | 0.5616 | T | 0.5795 | 8.4758 | 0.4874 | 0.5126 | 1.9508 | 0.3686 | |
| TT | 109 | 0.2986 | ||||||||
| SNP2 | TT | 56 | 0.1534 | T | 0.4233 | |||||
| TC | 197 | 0.5397 | C | 0.5767 | 4.0605 | 0.4882 | 0.5118 | 1.9539 | 0.3690 | |
| CC | 112 | 0.3068 | ||||||||
| SNP3 | GG | 43 | 0.1178 | G | 0.4384 | |||||
| GC | 234 | 0.6411 | C | 0.5616 | 33.2854 | 0.4924 | 0.5076 | 1.9701 | 0.3712 | |
| CC | 88 | 0.2411 | ||||||||
| SNP4 | CC | 39 | 0.1068 | C | 0.4068 | |||||
| CT | 219 | 0.6000 | T | 0.5932 | 21.5790 | 0.4826 | 0.5174 | 1.9327 | 0.3661 | |
| TT | 107 | 0.2932 | ||||||||
| SNP5 | CC | 46 | 0.1260 | C | 0.4123 | |||||
| CT | 209 | 0.5726 | T | 0.5877 | 12.0281 | 0.4846 | 0.5154 | 1.9402 | 0.3672 | |
| TT | 110 | 0.3014 |
Note: SNPs: single nucleotide polymorphisms; SNP1 = C1878T; SNP2 = T2010C; SNP3 = G3746C; SNP4 = C7294T; SNP5 = C7315T
Number: genotype number
GF: genotypic frequency; AF: allelic frequency; χ 2(HWE): Hardy–Weinberg equilibrium χ2 value; He: gene heterozygosity; Ho: gene homozygosity; Ne: effective allele numbers; PIC: polymorphism information content
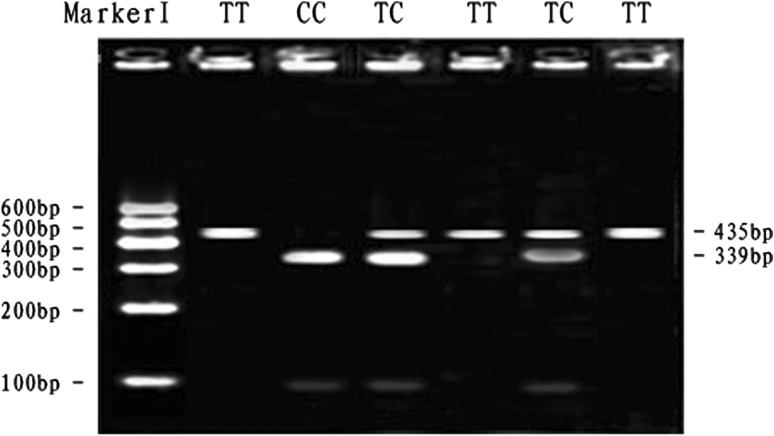
At the SNP2-BstXI locus, digestion of the 435 bp PCR fragment with BstXI resulted in fragment lengths of 435 bp for genotype TT; 435, 339 and 96 bp for genotype TC and 339 and 96 bp for genotype CC (see Fig. 2). The frequency of allele C was dominant in the Qinchuan breeds and TC genotype was more frequent than other genotypes. The genotypic frequencies of SNP2-BstXI locus in Qinchuan cattle population agreed with Hardy–Weinberg equilibrium (P > 0.05) (Table 3).
Fig. 2.
The 3 % agarose gels electrophoretic patterns of the MYH 3 gene in bovine. M = DNA molecular weight marker is Marker I; T2010C genotype: TT = 435 bp, TC = 435 + 339 + 96 bp, CC = 339 + 96 bp; It is difficult to see the 96 bp DNA fragment on 3 % agarose gel
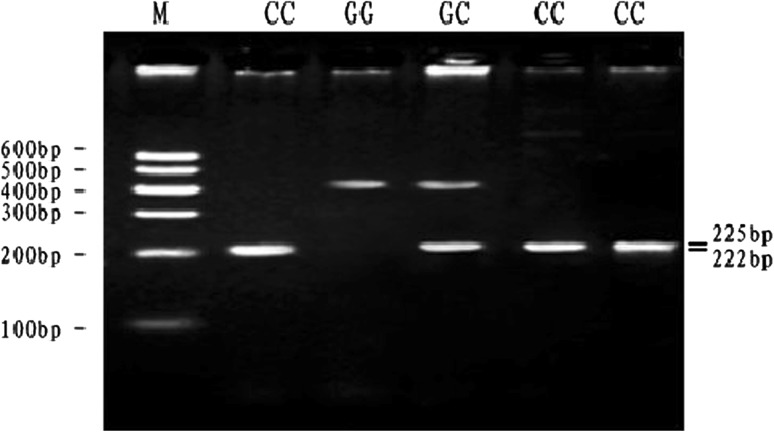
At the SNP3-PVUII locus, digestion of the 447 bp PCR fragment with PVUII resulted in fragment lengths of 447 bp for genotype GG; 447, 225 and 222 bp for genotype GC and 225 and 222 bp for genotype CC (see Fig. 3). The frequency of allele C was dominant in the Qinchuan breeds and GC genotype was more frequent than other genotypes. The genotypic frequencies of SNP3-PVUII locus in Qinchuan cattle population agreed with Hardy–Weinberg disequilibrium (P < 0.01) (Table 3).
Fig. 3.
The 2.5 % agarose gels electrophoretic patterns of the MYH 3 gene in bovine. M = DNA molecular weight marker is Marker I; G3746C genotype: GG = 447 bp, GC = 447 + 225 + 222 bp, CC = 225 + 222 bp; It is difficult to see the 222 bp and 225 bp DNA fragment on 2.5 % agarose gel
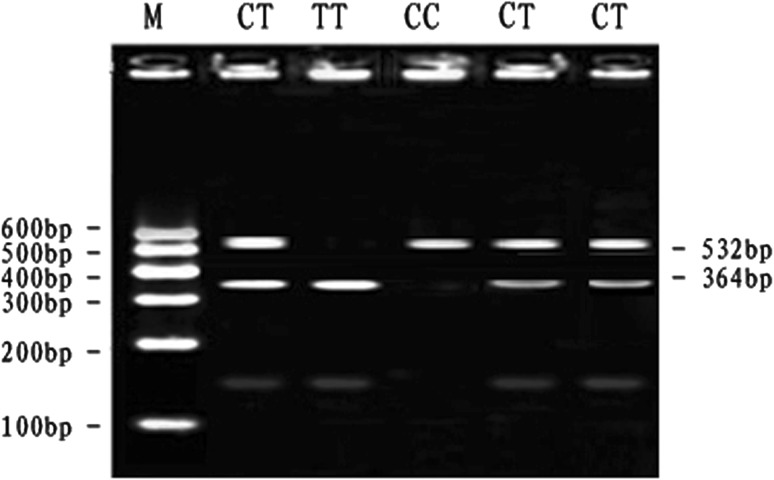
At the SNP4-RSaI locus, digestion of the 532 bp PCR fragment with RSaI resulted in fragment lengths of 532 bp for genotype CC; 532, 364 and 168 bp for genotype CT and 364 and 168 bp for genotype TT (see Fig. 4). The frequency of allele T was dominant in the Qinchuan breeds and CT genotype was more frequent than other genotypes. The genotypic frequencies of SNP4-RSaI locus in Qinchuan cattle population agreed with Hardy–Weinberg disequilibrium (P < 0.01) (Table 3).
Fig. 4.
The 2.5 % agarose gels electrophoretic patterns of the MYH 3 gene in bovine. M = DNA molecular weight marker is Marker I; C7294T genotype: CC = 532 bp, CT = 532 + 364 + 168 bp, TT = 364 + 168 bp; It is difficult to see the 168 bp DNA fragment on 2.5 % agarose gel
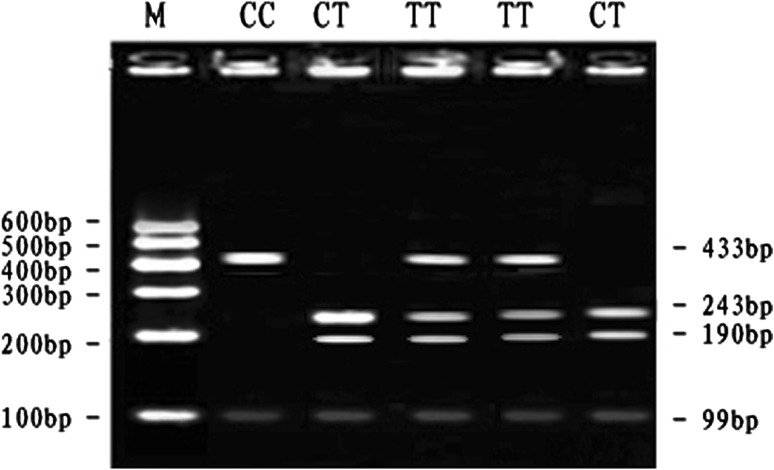
At the SNP5-TaqI locus, digestion of the 433 bp PCR fragment with TaqI resulted in fragment lengths of 433 bp for genotype CC; 433, 243 and 190 bp for genotype CT and 243 and 190 bp for genotype TT (see Fig. 5). The frequency of allele T was dominant in the Qinchuan and CT genotype was more frequent than other genotypes. The genotypic frequencies of SNP5-TaqI locus in Qinchuan cattle population agreed with Hardy–Weinberg disequilibrium (P < 0.01) (Table 3).
Fig. 5.
The 3 % agarose gels electrophoretic patterns of the MYH 3 gene in bovine. M = DNA molecular weight marker is MarkerI; C7315T genotype: CC = 433 + 99 bp, CT = 243 + 190 + 99 bp, TT = 433 + 243 + 190 + 99 bp
Analysis of polymorphism of the MYH3 in Qinchuan cattle population
The means and standard deviations (SD) for traits were analyzed in this study shown in Table 4. The allelic and genotypic frequencies, genetic diversity parameters (Ho, He, Ne and PIC) of the five SNPs were shown in Table 3. According to Table 3, only one loci of Qinchuan cattle was in Hardy–Weinberg equilibrium (P > 0.05). It showed that the Qinchuan cattle breed was in a dynamic disequilibrium in artificial selection, migration and genetic drift function. The maximum and minimum PIC values were 0.3661 and 0.3690. According to the genetic diversity classification of PIC (PIC value <0.25, low polymorphism; 0.25 < PIC value < 0.5, intermediate polymorphism and PIC value >0.5, high polymorphism), at the five SNPs, the result reflected that an intermediate genetic diversity of Qinchuan bovine MYH 3 gene in the population analyzed.
Table 4.
Means and standard deviations (SD) for traits
| Traits | Mean | SD |
|---|---|---|
| Body height (cm) | 140.22 | 4.26 |
| Withers length (cm) | 150.71 | 6.14 |
| Hip width (cm) | 48.00 | 3.66 |
| Slaughter weight (kg) | 497.96 | 63.67 |
| Carcass weight (kg) | 268.87 | 36.99 |
Linkage disequilibrium and haplotype analysis of the MYH3 in Qinchuan cattle population
Linkage disequilibrium and haplotype analysis of the MYH 3 gene in Qinchuan cattle population were shown in Tables 5 and 6. The linkage disequilibrium between the five SNPs in the population was estimated, which indicated that the D′ values ranged from 0.011 to 0.58; the r 2 values were from 0.000 to 0.292. Moreover, SNP1 and SNP2–SNP5 had little linkage equilibrium (D′ < 0.07 and r 2 < 0.003). SNP2 and SNP3–SNP5 had little linkage equilibrium (D′ < 0.08 and r 2 < 0.003).
Table 5.
The estimated values of linkage equilibrium analysis between 5 mutation sites within MYH 3 gene of studied population
| SNPs | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | D′ = 0.077 | D′ = 0.080 | D′ = 0.026 | D′ = 0.011 | |
| 2 | r 2 = 0.003 | D′ = 0.087 | D′ = 0.080 | D′ = 0.110 | |
| 3 | r 2 = 0.002 | r 2 = 0.003 | D′ = 0.439 | D′ = 0.580 | |
| 4 | r 2 = 0.001 | r 2 = 0.004 | r 2 = 0.110 | D′ = 0.422 | |
| 5 | r 2 = 0.000 | r 2 = 0.005 | r 2 = 0.292 | r 2 = 0.117 |
SNPs single nucleotide polymorphisms; 1 = C1878T; 2 = T2010C; 3 = G3746C; 4 = C7294T; 5 = C7315T
D′ and r 2 above and below the diagonal, respectively
Table 6.
Haplotype and haplotype frequency within studied population of 3 SNPs in bovine MYH 3 gene
| Haplotype | SNPs | Frequency in population | Cumulative frequency | ||
|---|---|---|---|---|---|
| G3746C | C7294T | C7315T | |||
| Hap 1 | C | C | C | 0.080 | 0.080 |
| Hap 2 | C | C | T | 0.149 | 0.229 |
| Hap 3 | C | T | C | 0.046 | 0.275 |
| Hap 4 | C | T | T | 0.393 | 0.668 |
| Hap 5 | G | C | C | 0.210 | 0.878 |
| Hap 6 | G | C | T | 0.038 | 0.916 |
| Hap 7 | G | T | C | 0.034 | 0.950 |
| Hap 8 | G | T | T | 0.050 | 1.000 |
The haplotype analysis showed that eight different haplotypes were identified among the three SNPs. Three major haplotypes accounting for 75.2 % of the alleles were obtained as follows, haplotype 2: –CCT– (14.9 %), haplotype 4, –CTT– (39.3 %) and haplotype 5, –GCC– (21.0 %).
Association analysis of single markers and combined genotypes
The results of the association analyses between five mutations in MYH 3 and growth and carcass traits were shown in Table 7. Growth and carcass traits were associated by the analysis of Qinchuan cattle at 30 ± 2 months old. According to Table 7, at locus C1878T, the animals with the CT genotype had greater withers height and heavier slaughter weight than those with the TT genotype (P < 0.05). At locus T2010C, the TT genotype was better than the TC and CC genotypes, and the animals with the TT genotype had greater withers height than those with the CC genotype (P < 0.05), the animals with the TT genotype had heavier slaughter weight than those with the TC and CC genotypes (P < 0.05). At locus G3746C, the animals with the GG genotype had greater body length and withers height than those with the GC and CC genotypes (P < 0.05), and GG genotype had greater hip width than those with GC genotype (P < 0.05). At locus C7315T, the animals with the CC genotype had heavier slaughter weight than those with CT genotype (P < 0.05). The rest of the records of growth and carcass traits had no significant association (P > 0.05).S
Table 7.
Association of SNPs with growth and carcass traits
| Locus | Genotype | N | Body length (cm) (mean ± SE) | Withers height (cm) (mean ± SE) | Hip width (cm) (mean ± SE) | Slaughter weight (kg) (mean ± SE) | Carcass weight (kg) (mean ± SE) |
|---|---|---|---|---|---|---|---|
| C1878T | CC | 51 | 140.681 ± 0.882 | 151.306 ± 0.991ab | 47.292 ± 0.622 | 497.056 ± 10.171ab | 265.425 ± 5.656 |
| CT | 205 | 140.382 ± 0.524 | 150.382 ± 0.589b | 46.750 ± 0.370 | 495.824 ± 6.043b | 266.104 ± 3.360 | |
| TT | 109 | 140.753 ± 0.603 | 152.506 ± 0.678a | 47.299 ± 0.425 | 505.623 ± 6.955a | 273.209 ± 3.868 | |
| P | 0.889 | 0.063 | 0.564 | 0.049 | 0.321 | ||
| T2010C | TT | 56 | 140.568 ± 0.777 | 149.676 ± 0.947b | 46.676 ± 0.573 | 487.243 ± 9.266 | 258.881 ± 5.260b |
| TC | 197 | 140.055 ± 0.637 | 150.855 ± 0.777ab | 47.491 ± 0.470 | 497.036 ± 7.600 | 268.102 ± 4.314a | |
| CC | 112 | 141.206 ± 0.811 | 152.735 ± 0.988a | 46.941 ± 0.598 | 509.706 ± 9.666 | 265.806 ± 5.487a | |
| P | 0.536 | 0.043 | 0.521 | 0.248 | 0.032 | ||
| G3746C | GG | 43 | 138.629 ± 0.940b | 148.742 ± 1.065b | 45.758 ± 0.666b | 484.290 ± 10.908 | 264.461 ± 6.115 |
| GC | 234 | 140.964 ± 0.575a | 151.892 ± 0.651a | 47.301 ± 0.407a | 507.000 ± 6.667 | 271.022 ± 3.737 | |
| CC | 88 | 140.832 ± 0.521a | 151.594 ± 0.590a | 47.213 ± 0.369ab | 498.089 ± 6.043 | 267.742 ± 3.388 | |
| P | 0.085 | 0.035 | 0.117 | 0.198 | 0.625 | ||
| C7294T | CC | 39 | 140.353 ± 0.680 | 151.138 ± 0.782 | 46.914 ± 0.494 | 492.552 ± 7.986 | 266.414 ± 4.518 |
| CT | 219 | 140.146 ± 0.572 | 150.524 ± 0.658 | 47.067 ± 0.416 | 500.061 ± 6.716 | 271.487 ± 3.800 | |
| TT | 107 | 141.308 ± 0.643 | 152.123 ± 0.739 | 47.146 ± 0.467 | 501.539 ± 7.544 | 267.275 ± 4.268 | |
| P | 0.376 | 0.272 | 0.942 | 0.681 | 0.637 | ||
| C7315T | CC | 46 | 140.283 ± 0.964 | 149.700 ± 1.094 | 46.467 ± 0.681 | 485.100 ± 11.141b | 268.123 ± 6.243 |
| CT | 209 | 140.112 ± 0.533 | 151.837 ± 0.605 | 47.026 ± 0.377 | 504.102 ± 6.164a | 268.125 ± 3.454 | |
| TT | 110 | 141.14 ± 0.569 | 151.198 ± 0.646 | 47.192 ± 0.402 | 499.372 ± 6.580ab | 269.176 ± 3.687 | |
| P | 0.404 | 0.231 | 0.657 | 0.030 | 0.976 |
Note: Values with different superscripts within the same column differ significantly at P < 0.05 (a, b)
SE standard error
The combined genotypes association results for SNP3, SNP4 and SNP5 were shown in Table 8. The associated analysis suggested that no significant differences were detected between the combined genotypes of three SNPs and growth and carcass traits in Qinchuan cattle (P > 0.05).
Table 8.
Associations between combined genotypes of three SNPs and growth and carcass traits in Qinchuan cattle (N = 365): Mean ± SE
| Genotype of combination (19) | Number of combination | Body length (cm) (mean ± SE) | Withers height (cm) (mean ± SE) | Hip width (cm) (mean ± SE) | Slaughter weight (kg) (mean ± SE) | Carcass weight (kg) (mean ± SE) |
|---|---|---|---|---|---|---|
| 1 CCCCCC | 7 | 139.33 ± 3.09 | 150.67 ± 3.49 | 48.33 ± 2.18 | 467.33 ± 36.02 | 283.73 ± 19.78 |
| 2 CCCCCT | 15 | 140.29 ± 2.03 | 152.29 ± 2.29 | 46.43 ± 1.43 | 515.14 ± 23.58 | 259.40 ± 12.95 |
| 3 CCCCTT | 8 | 137.50 ± 3.79 | 153.00 ± 4.28 | 45.50 ± 2.68 | 496.00 ± 44.11 | 264.10 ± 24.22 |
| 4 CCCTCC | 8 | 145.00 ± 3.79 | 152.50 ± 4.28 | 47.00 ± 2.68 | 524.00 ± 44.11 | 255.90 ± 24.22 |
| 5 CCCTCT | 25 | 140.13 ± 1.38 | 151.40 ± 1.56 | 47.57 ± 0.98 | 490.80 ± 16.11 | 263.33 ± 8.85 |
| 6 CCCTTT | 40 | 141.00 ± 1.05 | 150.81 ± 1.19 | 46.73 ± 0.74 | 498.31 ± 12.23 | 271.83 ± 6.72 |
| 7 CCTTCC | 6 | 142.00 ± 3.79 | 152.00 ± 4.28 | 47.50 ± 2.68 | 503.50 ± 44.11 | 285.10 ± 24.22 |
| 8 CCTTCT | 16 | 139.67 ± 1.79 | 153.33 ± 2.02 | 46.90 ± 1.26 | 490.11 ± 20.79 | 270.44 ± 11.42 |
| 9 CCTTTT | 50 | 141.30 ± 0.88 | 151.68 ± 0.99 | 47.65 ± 0.66 | 501.70 ± 10.26 | 266.39 ± 5.63 |
| 10 GCCCCC | 10 | 140.50 ± 2.68 | 147.75 ± 3.02 | 45.75 ± 1.89 | 463.50 ± 31.19 | 264.75 ± 17.13 |
| 11 GCCCCT | 28 | 141.80 ± 1.20 | 153.95 ± 1.35 | 48.45 ± 0.85 | 503.25 ± 13.95 | 264.54 ± 7.66 |
| 12 GCCTCT | 40 | 140.07 ± 1.03 | 151.78 ± 1.16 | 47.37 ± 0.73 | 520.41 ± 12.01 | 279.10 ± 6.59 |
| 13 GCCTTT | 22 | 140.73 ± 1.62 | 150.09 ± 1.82 | 47.00 ± 1.14 | 485.82 ± 18.81 | 265.65 ± 10.33 |
| 14 GCTTCT | 15 | 141.40 ± 1.70 | 152.40 ± 1.91 | 45.80 ± 1.20 | 512.60 ± 19.73 | 263.54 ± 10.83 |
| 15 GCTTTT | 11 | 141.88 ± 1.90 | 150.75 ± 2.14 | 47.38 ± 1.34 | 511.25 ± 22.06 | 266.58 ± 12.11 |
| 16 GGCCCC | 25 | 139.79 ± 1.30 | 149.18 ± 1.47 | 46.53 ± 0.91 | 483.76 ± 15.13 | 265.93 ± 8.31 |
| 17 GGCCCT | 9 | 136.40 ± 2.40 | 148.20 ± 2.71 | 44.00 ± 1.69 | 479.20 ± 27.90 | 258.28 ± 15.32 |
| 18 GGCTCT | 10 | 136.67 ± 2.19 | 146.00 ± 2.47 | 45.67 ± 1.54 | 480.83 ± 25.47 | 264.87 ± 13.99 |
| 19 GGTTCC | 5 | 139.00 ± 3.79 | 151.50 ± 4.28 | 43.00 ± 2.68 | 509.00 ± 44.11 | 265.35 ± 24.22 |
| Significance (P value) | 0.83 | 0.59 | 0.75 | 0.93 | 0.99 |
SE standard error
Discussion
In previous studies, the MYH 3 gene is the majority myosin isoform in embryonic and neonatal muscle fibers and its expression declines after birth to become undetectable around 3 weeks postnatal [45]. MYH 3 expression demonstrates in the early looping heart, and subsequently throughout the myocardium of the outflow tract, and at lower levels to the ventricular chamber [38]. Knockdown of MYH 3 in the chick resulted in abnormal atrial septal development, similar to that seen upon α-MYH knockdown [46, 47]. These results support the idea that MYH 3 is a fundamental process in skeletal muscle and heart development. Another study finds that myosin myopathies have evolved as a new group of muscle diseases caused by mutations in skeletal muscle myosin heavy-chain genes [48].
Up to now, there are few polymorphisms detected in the MYH 3 gene, it may be related to meat quality traits. We choosed it as a candidate gene to identify its SNPs and analyze the associations between the polymorphism and growth and carcass traits in the Qinchuan cattle population, which would lay the foundation for the study on the function of MYH 3 gene in the cattle breed.
The allelic and genotypic frequencies, PIC, He, Ne and χ2 values for SNPs showed considerable variability in Qinchuan cattle, the allelic frequency of the T alleles in five loci were up to 0.42 and C alleles in five loci were up to 0.40. PIC, He and Ne of SNP3 were higher than other four SNPs, showed the high polymorphism, high genetic variability, high ability to maintain allelic stability during selection or mutation, so SNP3 was likely to be useful in breeding programs for Qinchuan cattle. Chi-squared tests of SNP1, SNP3, SNP4 and SNP5 did not agreed with Hardy–Weinberg equilibrium (0.01 < P < 0.05, P < 0.01, P < 0.01 and P < 0.01, respectively), while SNP2 met with the Hardy–Weinberg equilibrium (P > 0.05).
In this study, we first reported the novel five SNPs in bovine MYH 3 gene, and its association analysis with the growth and carcass traits in bovine. The genotypes at locus C1878T had a significant effect on slaughter weight. The genotypes at locus T2010C had a significant effect on withers height and carcass weight. The genotypes at locus G3746C had a significant effect on withers height. The genotypes at locus C7315T had a significant effect on slaughter weight. Based on these study, the individuals with better performance with the CT genotype at locus C1878T and TT genotypes at locus T2010C and GG genotypes at locus G3746C and CC genotypes at locus C7315T could be used for the development of new breeds of beef cattle in China, and these four locus could be used as remarkable molecular markers for better performance in the bovine industry.
The result of haplotype analysis of three major loci showed that eight different haplotypes were identified which get nineteen combined haplotypes, and all nineteen combined genotypes were found in this study animal DNA samples, four major combined haplotypes accounting for 51.9 % of the haplotypes were obtained as follows, CCCTTT: (12.3 %), CCTTTT: (17.5 %), GCCTCT: (9.4 %), GCCCCT: (12.7 %). But the associated analysis suggested that no significant differences were detected between the combined genotypes of three SNPs and growth and carcass traits in Qinchuan cattle (P > 0.05). So this study indicated that combined haplotypes did not increase growth or carcass traits of Qinchuan cattle.
In our study, three mutations in the bovine MYH 3 gene were found in the exon region, polymorphisms in these regions could alter gene expression. The SNP g. +7294C>T was a missense mutation Arg2432Cys. It is possible to change the important physiological function in secondary and tertiary configuration of MYH 3 protein as well as affect the biological function of protein, and it needs to study further. Our study was the first report of five novel SNPs in the bovine MYH 3 gene, and we analyzed their association with growth and carcass traits in Qinchuan cattle. The results showed that some genotypes had a significant effect on the growth and carcass traits. Therefore, these mutations of MYH 3 gene might positively influence the growth and carcass traits in Qinchuan population. In conclusion, this study will be contributed to geneticists and breeders as a molecular marker for better performance in the bovine industry. While the further research is still needed to clarify the role on the genetic variants of the MYH 3 gene, and to analyze the mRNA expression levels of the MYH 3 gene. Hence, would be a candidate gene worthy of further investigation.
Acknowledgments
Research supported by the 12th “Five-Year” National Science and Technology Key Project (No. 2011AA100307), the National “863” Program of China (No. 2008AA101010), “13115” Sci-Tech Innovation Program of Shaanxi Province (No. 2008ZDKG-11).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Abbreviations
- PCR-RFLP
Polymerase chain reaction-restriction fragment length polymorphism
- SNPs
Single nucleotide polymorphisms
- MyHC
Myosin heavy chain
- MYH3
Myosin heavy chain 3
- HMM
Heavy meromyosin
- LMM
Light meromyosin
- LD
Linkage disequilibrium
- He
Heterozygosity
- Ho
Homozygosity
- PIC
Polymorphism information content
References
- 1.Diagana TT, North DL, Jabet C, Fiszman MY, Takeda S, Whalen RG. The transcriptional activity of a muscle-specific promoter depends critically on the structure of the TATA element and its binding protein. J Mol Biol. 1997;265(5):480–493. doi: 10.1006/jmbi.1996.0752. [DOI] [PubMed] [Google Scholar]
- 2.Lakich MM, Diagana TT, North DL, Whalen RG. MEF-2 and Oct-1 bind to two homologous promoter sequence elements and participate in the expression of a skeletal muscle-specific gene. J Biol Chem. 1998;273(24):15217–15226. doi: 10.1074/jbc.273.24.15217. [DOI] [PubMed] [Google Scholar]
- 3.Swoap SJ. In vivo analysis of the myosin heavy chain IIB promoter region. Am J Physiol. 1998;274(3Pt1):C681–C687. doi: 10.1152/ajpcell.1998.274.3.C681. [DOI] [PubMed] [Google Scholar]
- 4.Takeda S, North DL, Lakich MM, Russell SD, Whalen RG. A possible regulatory role for conserved promoter motifs in an adult-specific muscle myosin gene from mouse. J Biol Chem. 1992;267(24):16957–16967. [PubMed] [Google Scholar]
- 5.Takeda S, North DL, Diagana T, Miyagoe Y, Lakich MM, Whalen RG. Myogenic regulatory factors can activate TATA-containing promoter elements via an E-box independent mechanism. J Biol Chem. 1995;270(26):15664–15670. doi: 10.1074/jbc.270.26.15664. [DOI] [PubMed] [Google Scholar]
- 6.Wheeler MT, Snyder EC, Patterson MN, Swoap SJ. An E-box within the MHC IIB gene is bound by MyoD and is required for gene expression in fast muscle. Am J Physiol. 1999;276(5Pt1):C1069–C1078. doi: 10.1152/ajpcell.1999.276.5.C1069. [DOI] [PubMed] [Google Scholar]
- 7.Lieber RL, Bodine SC, Burkholder TJ, Pierotti DJ, Ryan AF. Cloning and in situ hybridization of type 2A and 2B rat skeletal muscle myosin tail region: implications for filament assembly. Biochem Biophys Res Commun. 1993;197(3):1312–1318. doi: 10.1006/bbrc.1993.2620. [DOI] [PubMed] [Google Scholar]
- 8.DeNardi C, Ausoni S, Moretti P, Gorza L, Velleca M, Buckingham M, Schiaffino S. Type 2X-myosin heavy chain is coded by a muscle fiber type-specific and developmentally regulated gene. J Cell Biol. 1993;123(4):823–835. doi: 10.1083/jcb.123.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieczorek DF, Periasamy M, Butler-Browne GS, Whalen RG, Nadal-Ginard B. Co-expression of multiple myosin heavy chain genes, in addition to a tissue-specific one, in extraocular musculature. J Cell Biol. 1985;101(2):618–629. doi: 10.1083/jcb.101.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strehler EE, Strehler-Page MA, Perriard JC, Periasamy M, Nadal-Ginard B. Complete nucleotide and encoded amino acid sequence of a mammalian myosin heavy chain gene. Evidence against intron-dependent evolution of the rod. J Mol Biol. 1986;190(3):291–317. doi: 10.1016/0022-2836(86)90003-3. [DOI] [PubMed] [Google Scholar]
- 11.Periasamy M, Wieczorek DF, Nadal-Ginard B. Characterization of a developmentally regulated perinatal myosin heavy-chain gene expressed in skeletal muscle. J Biol Chem. 1984;259(21):13573–13578. [PubMed] [Google Scholar]
- 12.Weiss A, McDonough D, Wertman B, Acakpo-Satchivi L, Montgomery K, Kucherlapati R, Leinwand L, Krauter K. Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proc Natl Acad Sci USA. 1999;96(6):2958–2963. doi: 10.1073/pnas.96.6.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyons GE, Ontell M, Cox R, Sassoon D, Buckingham M. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol. 1990;111(4):1465–1476. doi: 10.1083/jcb.111.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weydert A, Daubas P, Caravatti M, Minty A, Bugaisky G, Cohen A, Robert B, Buckingham M. Sequential accumulation of mRNAs encoding different myosin heavy chain isoforms during skeletal muscle development in vivo detected with a recombinant plasmid identified as coding for an adult fast myosin heavy chain from mouse skeletal muscle. J Biol Chem. 1983;258(22):13867–13874. [PubMed] [Google Scholar]
- 15.Whalen RG, Sell SM, Butler-Browne GS, Schwartz K, Bouveret P, Pinset-Harstom I. Three myosin heavy-chain isozymes appear sequentially in rat muscle development. Nature. 1981;292(5826):805–809. doi: 10.1038/292805a0. [DOI] [PubMed] [Google Scholar]
- 16.Rubinstein NA, Kelly AM. Development of muscle fiber specialization in the rat hindlimb. J Cell Biol. 1981;90(1):128–144. doi: 10.1083/jcb.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hundley AF, Yuan L, Visco AG. Skeletal muscle heavy-chain polypeptide 3 and myosin binding protein H in the pubococcygeus muscle in patients with and without pelvic organ prolapse. Am J Obstet Gynecol. 2006;194(5):1404–1410. doi: 10.1016/j.ajog.2006.01.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YY, Zan LS, Wang HB, Qing L, Wu KX, Quan SA, Li CQ, Zhong X, Wang CJ. Differentially expressed genes in skeletal muscle tissues from castrated Qinchuan cattle males compared with those from intact males. Livest Sci. 2011;135(1):76–83. doi: 10.1016/j.livsci.2010.06.070. [DOI] [Google Scholar]
- 19.Bober E, Buchberger-Seidl A, Braun T, Singh S, Goedde HW, Arnold HH. Identification of three developmentally controlled isoforms of human myosin heavy chains. Eur J Biochem. 1990;189(1):55–65. doi: 10.1111/j.1432-1033.1990.tb15459.x. [DOI] [PubMed] [Google Scholar]
- 20.di Maso NA, Caiozzo VJ, Baldwin KM. Single-fiber myosin heavy chain polymorphism during postnatal development: modulation by hypothyroidism. Am J Physiol Regul Integr Comp Physiol. 2000;278(4):R1099–R1106. doi: 10.1152/ajpregu.2000.278.4.R1099. [DOI] [PubMed] [Google Scholar]
- 21.Staron RS, Pette D. The multiplicity of combinations of myosin light chains and heavy chains in histochemically typed single fibres. Rabbit tibialis anterior muscle. Biochem J. 1987;243(3):695–699. doi: 10.1042/bj2430695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Termin A, Staron RS, Pette D. Changes in myosin heavy chain isoforms during chronic low-frequency stimulation of rat fast hindlimb muscles. A single-fiber study. Eur J Biochem. 1989;186(3):749–754. doi: 10.1111/j.1432-1033.1989.tb15269.x. [DOI] [PubMed] [Google Scholar]
- 23.Termin A, Staron RS, Pette D. Myosin heavy chain isoforms in histochemically defined fiber types of rat muscle. Histochemistry. 1989;92(6):453–457. doi: 10.1007/BF00524756. [DOI] [PubMed] [Google Scholar]
- 24.Eller M, Stedman HH, Sylvester JE, Fertels SH, Wu QL, Raychowdhury MK, Rubinstein NA, Kelly AM, Sarkar S. Human embryonic myosin heavy chain cDNA. Interspecies sequence conservation of the myosin rod, chromosomal locus and isoform specific transcription of the gene. FEBS Lett. 1989;256(1–2):21–28. doi: 10.1016/0014-5793(89)81710-7. [DOI] [PubMed] [Google Scholar]
- 25.Tajsharghi H, Kimber E, Kroksmark AK, Jerre R, Tulinius M, Oldfors A. Embryonic myosin heavy-chain mutations cause distal arthrogryposis and developmental myosin myopathy that persists postnatally. Arch Neurol. 2008;65(8):1083–1090. doi: 10.1001/archneur.65.8.1083. [DOI] [PubMed] [Google Scholar]
- 26.Lagrutta AA, McCarthy JG, Scherczinger CA, Heywood SM. Identification and developmental expression of a novel embryonic myosin heavy-chain gene in chicken. DNA. 1989;8(1):39–50. doi: 10.1089/dna.1.1989.8.39. [DOI] [PubMed] [Google Scholar]
- 27.Ruppel KM, Spudich JA. Structure–function analysis of the motor domain of myosin. Annu Rev Cell Dev Biol. 1996;12:543–573. doi: 10.1146/annurev.cellbio.12.1.543. [DOI] [PubMed] [Google Scholar]
- 28.Toydemir RM, Rutherford A, Whitby FG, Jorde LB, Carey JC, Bamshad MJ. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat Genet. 2006;38(5):561–565. doi: 10.1038/ng1775. [DOI] [PubMed] [Google Scholar]
- 29.Oana S, Machida S, Hiratsuka E, Furutani Y, Momma K, Takao A, Matsuoka R. The complete sequence and expression patterns of the atrial myosin heavy chain in the developing chick*. Biol Cell. 1998;90(9):605–613. [PubMed] [Google Scholar]
- 30.Reiser PJ, Portman MA, Ning XH, Moravec CS. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol. 2001;280(4):H1814–H1820. doi: 10.1152/ajpheart.2001.280.4.H1814. [DOI] [PubMed] [Google Scholar]
- 31.Somi S, Klein ATJ, Houweling AC, Ruijter JM, Buffing AAM, Moorman AFM, van den Hoff MJB. Atrial and ventricular myosin heavy-chain expression in the developing chicken heart: strengths and limitations of non-radioactive in situ hybridization. J Histochem Cytochem. 2006;54(6):649. doi: 10.1369/jhc.5A6846.2006. [DOI] [PubMed] [Google Scholar]
- 32.Wessels A, Anderson RH, Markwald RR, Webb S, Brown NA, Viragh SZ, Moorman AFM, Lamers WH. Atrial development in the human heart: an immunohistochemical study with emphasis on the role of mesenchymal tissues. Anat Rec. 2000;259(3):288–300. doi: 10.1002/1097-0185(20000701)259:3<288::AID-AR60>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 33.Wessels, Vermeulen JLM, Vir A, Gh S, Kalman F, Lamers WH, Moorman AFMA. Spatial distribution of “Tissue-Specific” antigens in the developing human heart and skeletal muscle. II. An immunohistochemical analysis of myosin heavy chain isoform expression patterns in the embryonic heart. Anat Rec. 1991;229(3):355–368. doi: 10.1002/ar.1092290309. [DOI] [PubMed] [Google Scholar]
- 34.Buttrick PM, Malhotra A, Brodman R, McDermott L, Lam L. Myosin isoenzyme distribution in overloaded human atrial tissue. Circulation. 1986;74(3):477. doi: 10.1161/01.CIR.74.3.477. [DOI] [PubMed] [Google Scholar]
- 35.Schaub MC, Hefti MA, Zuellig RA, Morano I. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc Res. 1998;37(2):381–404. doi: 10.1016/S0008-6363(97)00258-7. [DOI] [PubMed] [Google Scholar]
- 36.Harridge SDR, Bottinelli R, Canepari M, Pellegrino M, Reggiani C, Esbj O, Rnsson M, Balsom PD, Saltin B. Sprint training, in vitro and in vivo muscle function, and myosin heavy chain expression. J Appl Physiol. 1998;84(2):442–449. doi: 10.1152/jappl.1998.84.2.442. [DOI] [PubMed] [Google Scholar]
- 37.Larsson L, Moss RL. Maximum velocity of shortening in relation to myosin isoform composition in single fibres from human skeletal muscles. J Physiol. 1993;472(1):595–614. doi: 10.1113/jphysiol.1993.sp019964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutland CS, Polo-Parada L, Ehler E, Alibhai A, Thorpe A, Suren S, Emes RD, Patel B, Loughna S. Knockdown of embryonic myosin heavy chain reveals an essential role in the morphology and function of the developing heart. Development. 2011;138(18):3955–3966. doi: 10.1242/dev.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, Taegtmeyer H, Adrogue J, Razeghi P, Sen S, Ngumbela K, Essop MF. Dynamic changes of gene expression in hypoxia-induced right ventricular hypertrophy. Am J Physiol Heart Circ Physiol. 2004;286(3):1185–1192. doi: 10.1152/ajpheart.00916.2003. [DOI] [PubMed] [Google Scholar]
- 40.Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. Cancer cachexia is regulated by selective targeting of skeletal muscle gene products. J Clin Invest. 2004;114(3):370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook JG, Russell R, Umrania Y, Edwards YJ, Campbell RD, Elgar G, Clark MS. Fugu orthologues of human major histocompatibility complex genes: a genome survey. Immunogenetics. 2002;54(6):367–380. doi: 10.1007/s00251-002-0478-0. [DOI] [PubMed] [Google Scholar]
- 42.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 43.Nei M, Roychoudhury AK. Sampling variances of heterozygosity and genetic distance. Genetics. 1974;76(2):379–390. doi: 10.1093/genetics/76.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen DL, Leinwand LA. Postnatal myosin heavy chain isoform expression in normal mice and mice null for IIb or IId myosin heavy chains. Dev Biol. 2001;229(2):383–395. doi: 10.1006/dbio.2000.9974. [DOI] [PubMed] [Google Scholar]
- 46.Rutland C, Warner L, Thorpe A, Alibhai A, Robinson T, Shaw B, Layfield R, Brook JD, Loughna S. Knockdown of alpha myosin heavy chain disrupts the cytoskeleton and leads to multiple defects during chick cardiogenesis. J Anat. 2009;214(6):905–915. doi: 10.1111/j.1469-7580.2009.01079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, Robinson TE, Dearlove AM, Ribas G, Bonser AJ, Thomas NR, Scotter AJ, Caves LS, Tyrrell GP, Newbury-Ecob RA, Munnich A, Bonnet D, Brook JD. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet. 2005;37(4):423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 48.Oldfors A. Hereditary myosin myopathies. Neuromuscul Disord. 2007;17(5):355–367. doi: 10.1016/j.nmd.2007.02.008. [DOI] [PubMed] [Google Scholar]