Abstract
Calcium uptake into mitochondria occurs via a recently identified ion channel called the uniporter. Here, we characterize the phylogenomic distribution of the uniporter’s membrane-spanning pore subunit (MCU) and regulatory partner (MICU1). Homologs of both components tend to co-occur in all major branches of eukaryotic life, but both have been lost along certain protozoan and fungal lineages. Several bacterial genomes also contain putative MCU homologs that may represent prokaryotic calcium channels. The analyses indicate that the uniporter may have been an early feature of mitochondria.
Vertebrate mitochondria store large amounts of calcium taken up through a channel called the calcium uniporter. The uniporter, combined with additional intake, buffering, and efflux mechanisms (1), is hypothesized to regulate signaling, energy metabolism, and cell death. Although uniporter activity was documented nearly 50 years ago (2, 3), its molecular identity remained elusive until comparative genomics revealed its regulatory partner, MICU1, and pore-forming subunit, MCU (4–6). MCU represents a fundamentally new class of calcium channels, while MICU1 is a peripheral membrane protein with two EF-hand motifs, resembling known calcium-sensing regulators. Molecular identification of the uniporter machinery, combined with the recent availability of diverse genomes, offers an opportunity to explore this channel’s evolutionary history.
We examined the distribution of MCU and MICU1 across 138 fully sequenced eukaryotic organisms (7). We used sequence similarity followed by inspection of protein domains to annotate organisms that harbor one or more homologs of MCU or of MICU1, and visualized the data on a phylogenomic tree constructed using standard maximum likelihood methods (8).
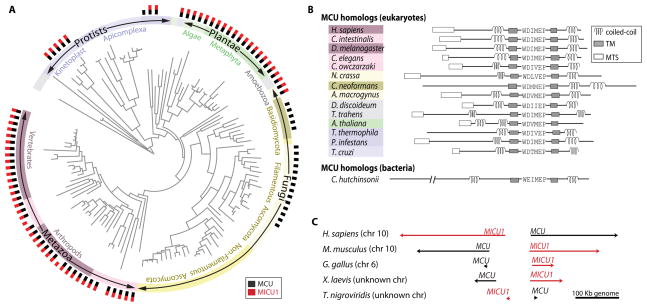
Homologs of MCU are distributed widely across all major branches of eukaryotic life, present in nearly all plants and metazoa, but with apparent loss events in certain protozoan and fungal lineages (Fig. 1A). MCU homologs have highly conserved acidic residues in the vicinity of a “DIME” motif flanked by two transmembrane and coiled coil domains (Fig. 1B). Within protozoa, MCU homologs are found in organisms from diverse clades including kinetoplasts (Trypanosoma cruzi), heterolobosea (Naegleria gruberi), oomycetes (Phytophthora infestans), and ciliates (Tetrahymena thermophila), but they are absent from other major lineages including apicomplexa (Plasmodium falciparum) and mitosome-containing eukaryotes (Encephalitozoon cuniculi, Giardia lamblia). MCU homologs are also present in many fungi, including many basidiomycetes and the early branching Allomyces macrogynus, but they are not found in any of 22 yeast fungi such as Saccharomyces cerevisiae (5). Most MCU-containing species harbor one or two MCU homologs, however Metaphyta tend to have 3–4 homologs and in several interesting cases contain predicted chloroplast targeting signals (7).
Fig. 1.
(A) Distribution of MCU (black) and MICU1 (red) homologs across 138 eukaryotes; a detailed version is available in fig. S1. (B) Protein domain architecture of selected MCU homologs, including mitochondrial targeting sequence (MTS, white rectangle), coiled-coil domain (coil), transmembrane domain (TM, shaded rectangle), and the “DIME” motif. L, Leu; T, Thr; P, Pro; V, Val; W, Trp. (C) Genomic organization of MICU1 and MCU across vertebrate genomes.
Sequenced metazoa, plants, and protozoa typically have homologs of both MCU and MICU1 or lack both proteins (Fig. 1A), but this pattern does not hold in fungi. MCU-containing fungi typically lack MICU1 homologs altogether, suggesting a distinct regulatory mechanism. The two exceptions are deep branching fungi A. macrogynus and Spizellomyces punctatus, which suggests that MICU1 was lost subsequent to the divergence of this branch. Fungi also show greater diversity in MCU domain structure, including the circularly permuted arrangements of coiled-coil and transmembrane domains observed in basidiomycetes (Fig. 1B), which perhaps is coincident with the loss of MICU1.
Mammalian MCU and MICU1 physically interact and are strongly co-expressed (5). Although MICU1 and MCU are not adjacent to each other in most of the analyzed genomes, in all vertebrates they are adjacent and share a potential bi-directional promoter (Fig. 1C), providing a plausible mechanistic basis for their coordinate expression in vertebrates.
Lastly, we asked if any bacteria harbor homologs of this new class of calcium channels. Iterative sequence similarity and protein domain searches revealed three diverse bacteria from the Bacteroidetes/Chlorobi group (Prevotella oris, Chlorobium phaeobacteroides, and Cytophaga hutchinsonii) that contain putative MCU homologs that we now term mitochondrial uniporter homologs(uni). The C. hutchinsoniiuni (CHU_0033) is particularly intriguing given its similar domain organization and conservation of key residues essential for calcium transport (5–6) (Fig. 1B). Functional studies are required to determine if bacterial uni proteins are also selective cationic channels. If shown to transport calcium, they would be among the first identified prokaryotic calcium channels.
Our phylogenomic analysis indicates that the uniporter may have been an early feature of mitochondria. Identification of MCU homologs may help to elucidate the structure of this new class of ion channels and the role of mitochondrial calcium transport in the emergence of eukaryotic life.
Supplementary Material
Acknowledgments
We thank A. Pringle, K. Zimmerman, E. Koonin, and E. Lander for critical reading of the manuscript. Protein sequences were obtained from the KEGG Organisms Database, Release 58, or the Origins of Multicellularity Sequencing Project at the Broad Institute (assemblies generated by the Genome Sequencing Platform and the Genome Sequencing and Analysis Program) as described in the supplementary materials. This work was supported by NIH funds (R01GM077465, R01GM097136 to V.K.M, T32GM007753-33 to A.G.B). Alignments and tree available at TreeBASE (accession S12416).
Footnotes
References and Notes
- 1.Hajnóczky G, Csordás G. Cur Biol. 2010;20:R888. doi: 10.1016/j.cub.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLuca HF, Engstrom GW. Proc Natl Acad Sci. 1961;47:1744. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasington FD, Murphy JV. JBC. 1962;237:2670. [PubMed] [Google Scholar]
- 4.Perocchi F, et al. Nature. 2010;467:291. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baughman JM, et al. Nature. 2011;476:341. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Stefani D, et al. Nature. 2011;476:336. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Materials and methods are available as supporting online material at Science Online.
- 8.Ciccarelli FD, et al. Science. 2006;311:1283. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.