Abstract
Foxn4, a member of the N-family forkhead transcription factors, controls fate-decision in mouse retina and spinal cord as well as in zebrafish heart. Analysis of Foxn4 amino acid sequence revealed the presence of a region homologous to the activation domain of its close relative Foxn1 in between C-terminal amino acids 402 and 455 of Foxn4 protein. The requirement of Foxn4 putative activation domain remains to be elucidated. Using a gain-of function approach in the rat and chick retinal explants, we report that deletion of Foxn4 putative activation domain results in a complete loss of its activity during retinogenesis. Target promoter transcription assay indicates that this domain is critical for Foxn4 transcriptional regulatory properties in vitro. Accordingly, in chick retinal explants, this domain is required for proper regulation of target retinogenenic factors expression by Foxn4. Thus our study demonstrates that the domain between amino acids 402 and 455 is necessary for Foxn4 transcriptional activity both in vitro and in the retina.
Keywords: Amino Acid Sequence; Animals; Cell Differentiation; Cells, Cultured; Chick Embryo; Eye Proteins; chemistry; genetics; physiology; Forkhead Transcription Factors; chemistry; genetics; physiology; Gene Expression Regulation, Developmental; Genes, Reporter; Mice; Molecular Sequence Data; Neurons; metabolism; Organ Culture Techniques; Protein Structure, Tertiary; RNA, Messenger; biosynthesis; genetics; Rats; Rats, Sprague-Dawley; Recombinant Fusion Proteins; physiology; Retina; embryology; growth & development; metabolism; Sequence Alignment; Sequence Homology, Amino Acid; Transcription, Genetic
Keywords: Foxn4, Foxn1, Forkhead, retinogenesis, activation domain
INTRODUCTION
Forkhead (Fkh) transcription factors form an evolutionarily conserved family of proteins involved in a variety of biological processes (Kaestner, 2000). These transcription factors act either as transactivators or as transrepressors of target gene expression. Their transcriptional properties rely notably on the presence of two specific domains, a transcriptional regulatory domain (either an activation or a repression domain) and a DNA-binding domain (DBD) and are regulated by post-translational modifications (Wijchers, 2006).
Foxn1 and Foxn4 are two closely related forkhead transcription factors belonging to the N-family (Hannenhalli, 2009) involved in cell differentiation during development. Foxn1 drives differentiation in hair follicles and in the thymus (for review (Wijchers, 2006)). Foxn4 is required for the specification of amacrine and horizontal cells during mouse retinogenesis (Li, 2004), the specification of ventral interneurons in mouse spinal cord (Li, 2005) and was more recently shown to be essential for atrio-ventricular canal formation in zebrafish heart (Chi, 2008). Foxn1 protein contains a regulatory domain that acts as an activator of transcription (Schuddekopf, 1996), deleted in almost all reported mutations of Foxn1 leading to T-cell immunodeficiency and congenital alopecia (reviewed in (Boehm, 2003)). With respect to the similarity of sequence with Foxn1, a putative activation domain (AD) was described in the C-terminal end of Foxn4 protein (Fig. 1A) (Gouge, 2001). However, the requirement of this domain and its function as a transcriptional activator or repressor hasn’t been studied so far. To assess the functional relevance of Foxn4 putative AD, we designed a variant of Foxn4 in which the putative AD domain was deleted (Foxn4ΔAD). We analyzed its activity in vivo, using gain-of-function approaches in the embryonic retina, and in vitro and demonstrated that Foxn4 AD is required for Foxn4 activity.
Figure 1. Foxn4 expression in rat retina.
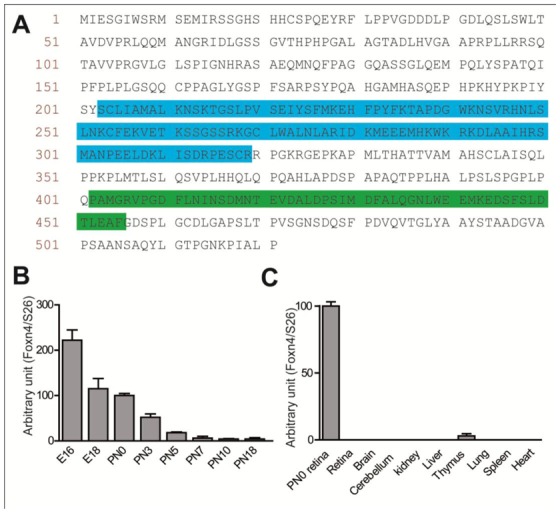
(A) Sequence of mouse Foxn4. DNA binding and putative activation domains (AD) (402–455) are highlighted in blue and green, respectively. (B–C) qPCR analyzes of rat Foxn4 expression during rat retinal development (B) and in adult tissues (C). Foxn4 expression level is normalized by cDNA expression level of S26 ribosomal protein.
MATERIALS AND METHODS
Animals and Ethics Statement
Gallus gallus white leghorn embryos were obtained from Haas (Kaltenhouse, France). Sprague-Dawley OFA rats were purchased from Charles River. Animals were sacrificed following approval by the French Minister of Agriculture (authorization 75-865-RENEWAL delivered on April 2010) according to the recommendations of our local ethical committee (CNREEA5 Charles Darwin (France)).
Plasmids
The well-characterized mouse Foxn4 cDNA was cloned using specific primers into pDONR221 (pDONR) vector (Invitrogen). Mutation/deletion of activation domain (aa 402 to 455) were performed by PCR-based mutagenesis using specific primers available upon request. pDONR plasmids were recombined with pCIG expression vector (Roger, 2006).
Electroporation, retinal explant culture and dissociation
Rat retinal explants or whole chick retinas were electroporated with pCIG vectors (1 μg/μl) as described previously (Roger, 2006). Rat retinal explants were cultured on polycarbonate filter discs as previously detailled (Roger, 2006). Whole chick retinas were cultured as floating explants in DMEM, 10% FBS, 1% Penicillin/Streptomycin. For cell counting experiments, after the indicated days of in vitro (DIV) culture, retinal cells were dissociated with trypsin, plated on poly-Lysine coated glasses and fixed for 10 min in 4% PFA before immunostaining.
For cell sorting experiments, whole chick retinas were cultured for 36 h, then dissociated and a total of 105 GFP-positive cells (Roger, 2006) were collected for RNA extraction using Vantage Sorter (BD Biosciences).
Immunostaining
The following antibodies were used: anti-rhodopsin (kind gift of R.Molday, Vancouver), anti-syntaxin (Sigma-Aldrich), anti-VC1.1 (Sigma), anti-recoverin (Millipore), anti-visinin (Developmental Studies Hybridoma Bank) antibodies. Secondary Alexa Fluor antibodies were obtained from Molecular Probes (Invitrogen). Cells were immunostained as previously described (Roger, 2006). Fluorescent staining signals were captured with a DM 5500 microscope (Leica) and analyzed with MetaMorph software (Molecular Devices).
Reporter gene assays
A 1 kb genomic DNA fragment upstream of Tbx2 ATG containing Foxn4 responsive sites was cloned into pGL3-basic vector (Promega). Renilla luciferase reporter driven by the herpes simplex virus thymidine kinase promoter (pRL-TK) was used as an internal control for transfection efficiency. 611W cells (Tan, 2004) were cultured in DMEM medium, 10% FBS and 1% Gentamicin, seeded in 24-wells plate and transfected with pCIG expression vectors and the pTbx2-pGL3 reporter plasmid using the calcium-phosphate method (Sambrook 2001). Luciferase activity quantification was performed using Dual-Luciferase® Reporter Assay System (Promega) and a Tristar LB940 luminometer (Berthold technologies). Relative luciferase activity is the ratio of Firefly over Renilla luciferase activities.
RNA isolation and quantification by real-time PCR
Total RNAs were extracted with Nucleospin RNAII Kit (Macherey Nagels). Real-time PCR was performed using 7300 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. Reactions were performed in a 20 μl final volume with Power SYBR® Green PCR Master Mix (Applied Biosystems). Primers are available upon request. For quantification on GFP-positive sorted cells, all samples (pCIG n=5, pCIG-Foxn4 n=5, pCIG-Foxn4-ΔAD n=3) were subjected to two independent retrotranscriptions. For each retrotranscription and each sample, qPCR were run in triplicate and quantification was repeated twice. Data represent the mean ± SEM of all values of each experimental group.
Western blotting and nuclear extract preparation
Western blotting was conducted as described previously (Roger, 2006) using the following primary antibodies : mouse anti-HA (MMS 101R, Covance), goat anti-laminB (sc-6216, SantaCruz), goat anti-GAPDH and mouse anti-actin (Sigma).
For nuclear extract preparation, total protein extracts were collected in hypotonic buffer (20mM HEPES pH7.9, 1mM Na3VO4, 1mM Na Glycerophoshate, 5mM EDTA pH7.5, 1mM EGTA pH7.5, 1mM DTT, Protease inhibitors (IP) (Calbiochem, Merck4Biosciences)) plus 0.2% NP40, incubated on ice 15 minutes and centrifugated 20 secondes at 16000 g. After three washes in hypotonic buffer, pellets were suspended in saline buffer containing IP (120 mM NaCl, 20% glycerol in hypotonic buffer) incubated 30 minutes at 4°C and centrifugated 20 minutes at 16000 g to collect the supernatant (Nuclear Extract, NE).
Statistical analyses
Statistical analyses were conducted using one way analysis of variance followed by Tukey’s multiple comparison tests (Prism 5.0, Graphpad software). (*) p<0.05 (**) p<0.01, (***) p<0.001. If not indicated differently, statistical significances for values compared to pCIG are reported.
RESULTS and DISCUSSION
Foxn4 activity during retinogenesis
The vertebrate retina is composed of six types of neurons. They all derive from multipotent progenitor cells that differentiate in an evolutionarily conserved chronological order (Young 1985; Livesey, 2001). Ganglion cells, amacrine cells (AC), horizontal cells (HC), cone photoreceptors are generated first while rod photoreceptors and bipolar cells are generated last. In mice retina, Foxn4 is expressed from embryonic day 11.5 (E11.5) to postnatal day 6 (P6) in the zone of progenitor cells (Gouge, 2001; Li, 2004). Foxn4 overexpression during development increases the number of syntaxin-positive HC/AC and decreases the number of recoverin-positive photoreceptors (Li, 2004). Here, we show through quantitative PCR analysis that Foxn4 was highly expressed during rat retinal development and became undetectable by P7 (Fig. 1B). Foxn4 was not detected in other adult rat tissues tested except in the thymus (Fig. 1C). The similar temporal expression pattern in mouse and rat retina (Li, 2004) suggests that Foxn4 might have a similar function among rodents. To assay its role on the specification of early and late born cells during rat retinogenesis, Foxn4 was overexpressed in rat retinal explants at E16 and P0 that is just prior the early and late waves of cell differentiation, respectively (Rapaport, 2004). For that purpose we cloned Foxn4 cDNA in the bicistronic pCIG-eGFP (Green Fluorescent Protein) expression vector (Roger, 2006) (Fig. 2A) and control pCIG-eGFP and pCIG-Foxn4-eGFP vectors were electroporated in E16 or P0 rat retinal explants. After 5 or 7 days of in vitro culture (DIV) following E16 electroporation, we stained cell-dissociated explants with anti-GFP and anti-syntaxin to identify AC/HC or anti-recoverin antibodies that label early born cone photoreceptors at these stages (Fig. 2B). After 3 or 6 DIV following P0 electroporation, dissociated cells were stained with anti-GFP and anti-rhodospsin for late born rod photoreceptors. Misexpression of Foxn4 in rat retina at E16 resulted in a 1.9- and 2.3-fold increase in the number of syntaxin-GFP double-positive cells at 5 and 7 DIV, respectively (Fig. 2C), and a 4.9-fold reduction in the number of recoverin-GFP double-positive cone photoreceptors at these two time points (Fig. 2D). Moreover, misexpression at P0 of Foxn4 strongly decreased the number of rhodopsin-GFP double-positive late born rod photoreceptors after 3 or 6 DIV (Fig. 2E). Together these data confirm the phenotype previously observed in the mouse after misexpression of Foxn4 by an in vivo retroviral approach (Li, 2004). Hereafter, we chose Foxn4-induced changes in retinal cell type representation as an appropriate marker of wild type Foxn4 activity.
Figure 2. Effects of misexpression of Foxn4 and FoxnΔAD during rat retinogenesis.
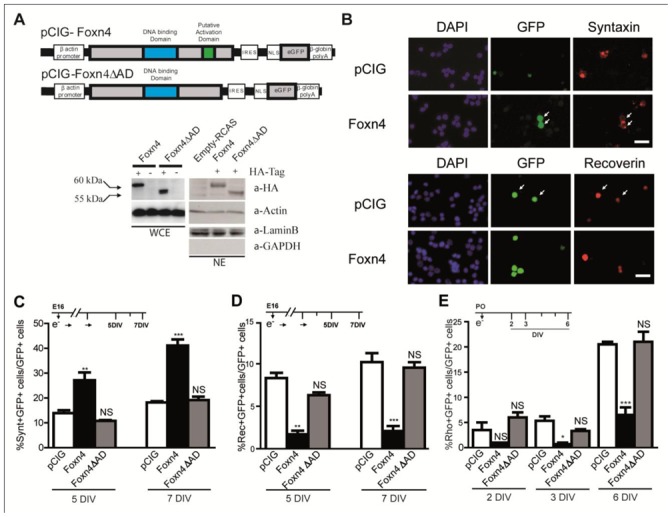
(A) Schematic structure of pCIG-Foxn4 and pCIG-Foxn4ΔAD GFP-encoding plasmids. IRES, Internal Ribosomal Entry Site, NLS, Nuclear Localisation Signal. Blue box: DNA Binding Domain, green box: putative activation domain. Foxn4 and Foxn4ΔAD were also recombined into pRCAS plasmid without HA-tag or with a HA-tag in-frame. Whole cell extract (WCE) and nuclear extract (NE) from cells transfected with RCAS vectors were subjected to Western Blotting using anti-HA antibody. Actin was used as a loading control and anti-LaminB and anti-GAPDH as markers of nuclear and cytosolic fractions, respectively as well as loading control for anti-LaminB. (B) Rat retinal explants were electroporated at E16 with pCIG or pCIG-Foxn4. After 7 days in vitro (DIV), cells were dissociated and immunostained with anti-GFP and anti-syntaxin (upper panel) or anti-recoverin (lower panel) antibodies. White arrows point at double-positive cells. Scale bar: 10 μm. (C–E) Retinal explants were electroporated at E16 (C–D) or P0 (E) with pCIG, pCIG-Foxn4 or pCIG-Foxn4ΔAD plasmids. The number of syntaxin (C), recoverin (D) or rhodopsin (E) and GFP double-positive cells were scored at the indicated DIV. Data represent the mean ± SEM of at least three dissociated explants counts and are representative of two independent experiments.
Activity of Foxn4ΔAD variant in the retina
Transcription activity of the Fox family member is thought to be dependent of the presence of a regulatory domain in addition to their DBD. Foxn1 is the only member of the N-family in which the activity of an trans-Activation Domain (AD) has been demonstrated. In mouse, it is located between amino acid (aa) 509 and 562 (Schuddekopf, 1996). Gouge et al. have initially described a putative activation domain located in the C-terminal part of the mouse Foxn4 protein (Gouge, 2001). Since that, C-terminal putative activation domains with various boundaries have been hypothesized in chick, xenopus, human and zebrafish Foxn4 proteins (Schuddekopf, 1996; Schlake, 2000; Gouge, 2001; Boije, 2008; Chi, 2008). Here we have tested the requirement of the region between aa402 and aa455 for Foxn4 activity (reference sequence NP_683737) (Fig. 1A). This region is rich in acidic amino acids and has been delimited to encompass the highly conserved amino acids of previously described activation domains (Boije, 2008; Chi, 2008). A Foxn4ΔAD cDNA missing aa402 to aa455 was generated and recombined within the pCIG vector (Fig. 2A) to allow for misexpression in the retina and, concomitantly, recombined in frame with the HA-tag of pRCAS plasmid to ensure the proper synthesis and the stability of the Foxn4ΔAD tagged-protein in chick fibroblast cells (DF1). Foxn4ΔAD protein was expressed to a nearly equal level to the wild type Foxn4 protein (Fig. 2A, WCE). Moreover, purification of nuclear extracts indicated that Foxn4ΔAD was still present in the nucleus (Fig. 2A, NE). Overexpression at E16 of Foxn4ΔAD failed to increase the percentage of syntaxin-GFP double-positive early-born AC/HC cells and to decrease the percentage of recoverin-GFP double-positive early born cone cells (Fig. 2C,D). Furthermore, as opposed to Foxn4, Foxn4ΔAD did not modify the number of rhodopsin-GFP double-positive late born rod photoreceptors (Fig. 2E) when misexpressed at P0. Thus these results indicate that Foxn4 AD is required for Foxn4 effects on retinal cell type representation in vivo. A dominant negative effect of the AD defective form of Foxn4 could have been expected. However none of the characteristics of Foxn4 knock-out mouse phenotype (Li, 2004), that is a decrease of syntaxin-positive amacrine/horizontal cells and an increase of recoverin-positive photoreceptor cells, was observed following forced expression of Foxn4ΔAD.
Transcriptional properties of Foxn4 variants in vitro
Loss of Foxn4ΔAD activity during retinal development strongly suggests that this putative AD domain is necessary for the transcriptional regulatory properties of Foxn4. Thus we tested the transcriptional activity of Foxn4 and this variant with an in vitro reporter assay using a Tbx2 promoter, recently described as a direct target of Foxn4 in the zebrafish heart, driving the expression of the luciferase gene (Chi, 2008). For this purpose, we co-transfected 661W mouse retinal cells with Foxn4 or Foxn4ΔAD and the pTbx2-pGL3 reporter plasmid. Foxn4 transfection reduced by 2.7-fold the basal level of Tbx2 promoter transcription (Fig. 3A). On the contrary, deletion of Foxn4 AD increased by 1.9-fold Tbx2 transcription level when compared to pCIG vector (Fig. 3A) showing that this domain might be indeed a trans-regulatory domain of Foxn4 transcriptional activity. Transactivation properties of Foxn1 AD rely mostly on the presence of four aspartic acid residues (Schuddekopf, 1996). Three of them are conserved within the Foxn4 AD and thus might be involved in Foxn4 AD regulatory activity (Fig. 3B). Mutation of these aspartic acid residues, separately, did not significantly change the reporter transcription level when compared to Foxn4 suggesting that none of these residues are involved in Foxn4 trans-repression ability (Fig. 3A). The effect of the combination of the three mutations should be further studied. However, in genetic screen tests of affected individuals, it is unlikely that all three mutations would be identified in a single patient.
Figure 3. In vitro transcriptional activity of Foxn4 and its variants.
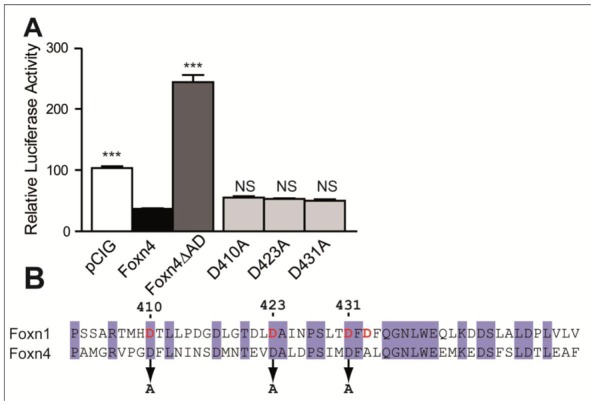
(A) Luciferase assays in 611W cells transfected with pTbx2-pGL3 containing Foxn4-responsive enhancer. Cells were co-transfected with empty control vector (pCIG) or pCIG expression vectors for Foxn4, Foxn4ΔAD or Foxn4 variants in which aspartic residues within activation domain were mutated (D410A, D420A and D431A). Data represent the mean ± SEM (n=6). Statistical significances for values compared to Foxn4 are reported. (B) Alignment of the sequences of the mouse Foxn1 and Foxn4 activation domains. Conserved aspartic acid residues D410, D423, D431 were bolded and printed in red. They were replaced by alanine in D410A, D423A and D431A Foxn4 variants.
Foxn4 AD regulatory properties in vivo
We next monitored Foxn4 AD functional relevance in the regulation of Foxn4 candidate target genes involved in retinogenesis using the chick retina that offers a reliable model to study transcriptional regulation following the overexpression of a transcription factor (Tetreault, 2009).
To validate this model, we first analyzed whether Foxn4 misexpression in the cone-dominated chick retina biased cell specification. Whole E5 chick retinas were electroporated with pCIG, pCIG-Foxn4 or pCIG-Foxn4ΔAD GFP-encoding plasmids and collected after 3 DIV corresponding approximately to E8 in vivo. At this stage most amacrine/horizontal cells and photoreceptors were differentiated in the chick retina (Prada, 1991). Dissociated cells were stained with anti-GFP and anti-syntaxin or anti-VC1.1 antibodies to identify AC/HC (Alexiades, 1997). Anti-visinin antibody was used to label all photoreceptors (cones and rods) as both types of photoreceptors are born concomitantly in chick retina (Prada, 1991). Misexpression of Foxn4 in chick retina increased by 2-fold the number of syntaxin-GFP double-positive cells (Fig. 4A) and VC1.1-GFP double-positive cells (Fig. 4B) at 3 DIV. On the contrary, no significant increase of syntaxin-GFP or VC1.1-GFP cells was observed following pCIG-Foxn4ΔAD electroporation (Fig. 4A–B). As for photoreceptors, the number of visinin-GFP double-positive cells was decreased by three fold in Foxn4 misexpressing retinas and this effect was abolished following deletion of Foxn4 AD (Fig. 4C). Together results in mouse, chick and rat retinas suggest that the function of Foxn4 may be conserved among vertebrates at least concerning amacrine/horizontal and photoreceptor cell specification.
Figure 4. Effects on cell type specification and transcriptional activity of Foxn4 and FoxnΔAD in chick retina.
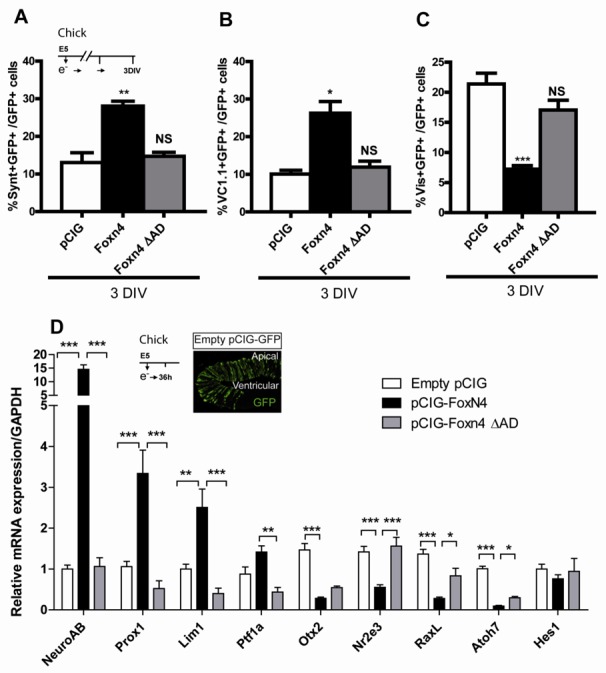
E5 chick retinal cells were electroporated with pCIG-GFP (Empty pCIG), pCIG-Foxn4(-GFP) or pCIG-Foxn4-ΔAD(-GFP). (A–C) After 3 DIV, the number of syntaxin (A), VC1.1 (B) or visinin (C) and GFP double-positive cells were scored. Data represent the mean ± SEM of at least three dissociated retina counts. D) Expression of Foxn4 candidate downstream target genes was analyzed 36h after electroporation by quantitative PCR on RNA from GFP-positive sorted cells. Candidate cDNA level was normalized by GAPDH cDNA level. Scale bar: 50μm.
We next assessed gene regulation in pCIG, pCIG-Foxn4 or pCIG-Foxn4ΔAD transfected E5 chick retinas. To study early changes in candidate gene expression, electroporated cells were harvested as shortly as 36 h after electroporation. This condition, soon after the onset of GFP expression, was chosen as the earliest time point where a sufficient number of GFP-expressing cells can be collected. We then isolated GFP-positive electroporated cells by Fluorescence Activated Cell Sorting and quantified by quantitative Polymerase Chain reaction (qPCR) the expression level of candidate genes selected according to their known function during retinogenesis (Fig. 4D).
We first analyzed genes that are representative of the AC or/and HC pathways. NeuroAB is expressed in GABAergic AC during chick retinogenesis (Ohkawara, 2004) and Prox1 and Lim1 are HC specific genes (Dyer, 2003; Poche, 2007; Poche, 2009; Suga, 2009). Loss of Foxn4 AD abolished the Foxn4-mediated NeuroAB upregulation. Foxn4 misexpression induced a 3.5- and 2.5-fold increase in Prox1 and Lim1 mRNA expression levels, respectively. However, Lim1 and Prox1 were not upregulated when Foxn4ΔAD was overexpressed (Fig. 4D). Ptf1a is a bHLH transcription factor required for AC and HC specification that acts as a downstream target of Foxn4 in the mouse retina (Fujitani, 2006). Deletion of Foxn4 AD abolished the Ptf1a upregulation observed following Foxn4 misexpression. We next studied genes involved in specifying photoreceptors and their subtypes. The homeodomain transcription factors Otx2 and RaxL are essential for photoreceptor cell development in the retina (Chen, 2002; Nishida, 2003) and are expressed in photoreceptor cells in chick retina (Bovolenta, 1997; Chen, 2002; Plouhinec, 2005). Nr2e3 is detected in the photoreceptor cell layer in chick retina (Kobayashi, 2008) and is critical for rod specification (Cheng, 2004). Foxn4 strongly downregulated Otx2, Nr2e3 and RaxL in chick electroporated retinal cells (Fig. 4D). Loss of its AD decreased the repressive activity of Foxn4 on Otx2 albeit not significantly and abolished Foxn4 repression of Nr2e3 (Fig. 4D). Foxn4ΔAD repression of RaxL was significantly lower compared to wild type Foxn4 (Fig. 4D). Furthermore, we found that Atoh7 involved in ganglion (Liu, 2001) and photoreceptor cell (Ma, 2004) genesis in chick was inhibited following Foxn4 overexpression while deletion of Foxn4 AD decreased significantly this repression (Fig. 4D). Therefore, in vivo Foxn4 transcriptional regulation of its target genes during retinogenesis is dependent on its putative AD. Loss of this AD only affects Foxn4 regulation of its target genes since AD deletion did not change the expression level of Hes1, a marker of proliferating cells which is not modified following Foxn4 misexpression (Fig. 4D).
Together our findings support that Foxn4 Activation Domain is indeed involved in Foxn4 transcriptional regulation properties both in vivo and in vitro.
We here show that Foxn4ΔAD was detected in the nuclear cell extracts as reported for the AD-deleted form of Foxn1StL (Schorpp, 2000). Thus, the loss of trans-activation and/or trans-repression activity of Foxn4ΔAD could possibly result from an alteration of its DNA binding property or by a loss of interaction with its cofactors, as previously stated for Foxn family members (Schuddekopf, 1996; Schorpp, 2000; Gouge, 2001). We show that the AD-deleted form of Foxn4 was able to activate the Tbx2 promoter. The deleted domain is enriched in negatively charged acidic amino acids, a feature of transcription domains, that favor the interaction with the general transcription machinery (Mitchell, 1989). It is therefore likely that this domain is involved in the regulation of transcription initiation by recruiting co-factors. It is also possible that Foxn4 AD is required for the binding of Foxn4 to DNA. However, other Fkh members such as Foxn1, Foxa1 (HFN3α), Foxa2 (HFN3β) were still capable of binding DNA regardless the lengh of deletions of their C terminal and/or N-terminal domains if their DBD sequences are preserved (Lai, 1990; Lai, 1991; Pani, 1992; Qian, 1995; Schlake, 2000).
Using various experimental conditions, we were not able to detect a specific binding of mouse Foxn4 proteins or their mutant forms to zebrafish Tbx2b or mouse Tbx2 promoter specific probes by electromobility shift assay (EMSA). The identification of a consensus sequence of mouse Foxn4 binding sites will be required to perform EMSA experiments to determine if Foxn4 AD is involved in Foxn4 binding to DNA.
Selected genes in the AC/HC pathway were systematically upregulated by Foxn4. Whether this transcriptional regulation is direct or not remains to be elucidated. Foxn4 repressed Tbx2 in the 661W cell line suggesting that Foxn4 is a transcriptional repressor contrary to Foxn1, and acts indirectly on the transcription AC/HC genes in vivo. However, its transcriptional regulatory properties might be cellular context- and gene-dependent. Notably, Foxn4 downregulated Tbx2 in 611W cell line while it positively regulates it in the zebrafish heart but has no effect on it in zebrafish retina (Chi, 2008). As already described for other Fkh transcription factors (Wijchers, 2006), Foxn4 might therefore act either as a repressor or as an activator depending on the recruitment of co-repressors or co-activators by its AD. In light of our study, the putative trans-activation domain of Foxn4 should thus be referred to as a transcriptional “trans-regulatory domain”.
So far most of the mutations of the Fox family associated with pathologies have been found in the DBD (Lehmann, 2003). However, the deletion of the trans-activation domain of Foxn1 in the nude mice has been proposed as a molecular explanation for the majority of known loss-of-function alleles of whn genes (Schuddekopf, 1996; Boehm, 2003). Taken together our results and others strongly suggest that the regulatory domain is critical for Foxn protein family activity and may be an additional target of mutation resulting in a loss-of-function phenotype. Identification of cofactors interacting with Foxn4 regulatory domain within Foxn4-expressing structures in vivo are now required to fully understand tissue-dependent Foxn4 transcriptional regulatory properties and may also help to decipher the genetic of human retinal, heart or spinal cord defects.
Acknowledgments
We thank J. Dreux for critical reading of the manuscript. This work was financed by INSERM, Retina France, EU (LSHG-CT-2005-512036, ERC-StG-210345), French ANR (ANR-Geno-031-03, ANR-08-MNPS-003).
References
- Alexiades MR, Cepko CL. Subsets of retinal progenitors display temporally regulated and distinct biases in the fates of their progeny. Development. 1997;124:1119–1131. doi: 10.1242/dev.124.6.1119. [DOI] [PubMed] [Google Scholar]
- Boehm T, Bleul CC, Schorpp M. Genetic dissection of thymus development in mouse and zebrafish. Immunol Rev. 2003;195:15–27. doi: 10.1034/j.1600-065x.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- Boije H, Edqvist PH, Hallbook F. Temporal and spatial expression of transcription factors FoxN4, Ptf1a, Prox1, Isl1 and Lim1 mRNA in the developing chick retina. Gene Expr Patterns. 2008;8:117–123. doi: 10.1016/j.modgep.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Bovolenta P, Mallamaci A, Briata P, Corte G, Boncinelli E. Implication of OTX2 in pigment epithelium determination and neural retina differentiation. J Neurosci. 1997;17:4243–4252. doi: 10.1523/JNEUROSCI.17-11-04243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CM, Cepko CL. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development. 2002;129:5363–5375. doi: 10.1242/dev.00114. [DOI] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Chi NC, Shaw RM, De Val S, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Gouge A, Holt J, Hardy AP, Sowden JC, Smith HK. Foxn4--a new member of the forkhead gene family is expressed in the retina. Mech Dev. 2001;107:203–206. doi: 10.1016/s0925-4773(01)00465-8. [DOI] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- Kobayashi M, Hara K, Yu RT, Yasuda K. Expression and functional analysis of Nr2e3, a photoreceptor-specific nuclear receptor, suggest common mechanisms in retinal development between avians and mammals. Dev Genes Evol. 2008;218:439–444. doi: 10.1007/s00427-008-0232-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE., Jr HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Tao WF, Chen WS, Darnell JE., Jr Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 1991;5:416–427. doi: 10.1101/gad.5.3.416. [DOI] [PubMed] [Google Scholar]
- Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- Li S, Misra K, Matise MP, Xiang M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc Natl Acad Sci U S A. 2005;102:10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mo Z, Yang X, Price SM, Shen MM, Xiang M. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43:795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci U S A. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Ma W, Yan RT, Xie W, Wang SZ. A role of ath5 in inducing neuroD and the photoreceptor pathway. J Neurosci. 2004;24:7150–7158. doi: 10.1523/JNEUROSCI.2266-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, et al. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Ohkawara T, Shintani T, Saegusa C, Yuasa-Kawada J, Takahashi M, Noda M. A novel basic helix-loop-helix (bHLH) transcriptional repressor, NeuroAB, expressed in bipolar and amacrine cells in the chick retina. Brain Res Mol Brain Res. 2004;128:58–74. doi: 10.1016/j.molbrainres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Pani L, Overdier DG, Porcella A, Qian X, Lai E, Costa RH. Hepatocyte nuclear factor 3 beta contains two transcriptional activation domains, one of which is novel and conserved with the Drosophila fork head protein. Mol Cell Biol. 1992;12:3723–3732. doi: 10.1128/mcb.12.9.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouhinec JL, Leconte L, Sauka-Spengler T, Bovolenta P, Mazan S, Saule S. Comparative analysis of gnathostome Otx gene expression patterns in the developing eye: implications for the functional evolution of the multigene family. Dev Biol. 2005;278:560–575. doi: 10.1016/j.ydbio.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE, Behringer RR. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007;27:14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Reese BE. Retinal horizontal cells: challenging paradigms of neural development and cancer biology. Development. 2009;136:2141–2151. doi: 10.1242/dev.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada C, Puga J, Perez-Mendez L, Lopez, Ramirez G. Spatial and Temporal Patterns of Neurogenesis in the Chick Retina. Eur J Neurosci. 1991;3:1187. doi: 10.1111/j.1460-9568.1991.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Qian X, Costa RH. Analysis of hepatocyte nuclear factor-3 beta protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res. 1995;23:1184–1191. doi: 10.1093/nar/23.7.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Roger J, Brajeul V, Thomasseau S, et al. Involvement of Pleiotrophin in CNTF-mediated differentiation of the late retinal progenitor cells. Dev Biol. 2006;298:527–539. doi: 10.1016/j.ydbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Sambrook S. Molecular Cloning : A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Schlake T, Schorpp M, Maul-Pavicic A, Malashenko AM, Boehm T. Forkhead/winged-helix transcription factor Whn regulates hair keratin gene expression: molecular analysis of the nude skin phenotype. Dev Dyn. 2000;217:368–376. doi: 10.1002/(SICI)1097-0177(200004)217:4<368::AID-DVDY4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Schorpp M, Schlake T, Kreamalmeyer D, Allen PM, Boehm T. Genetically separable determinants of hair keratin gene expression. Dev Dyn. 2000;218:537–543. doi: 10.1002/1097-0177(200007)218:3<537::AID-DVDY1007>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Schuddekopf K, Schorpp M, Boehm T. The whn transcription factor encoded by the nude locus contains an evolutionarily conserved and functionally indispensable activation domain. Proc Natl Acad Sci U S A. 1996;93:9661–9664. doi: 10.1073/pnas.93.18.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga A, Taira M, Nakagawa S. LIM family transcription factors regulate the subtype-specific morphogenesis of retinal horizontal cells at post-migratory stages. Dev Biol. 2009;330:318–328. doi: 10.1016/j.ydbio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- Tan E, Ding XQ, Saadi A, Agarwal N, Naash MI, Al-Ubaidi MR. Expression of cone-photoreceptor-specific antigens in a cell line derived from retinal tumors in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:764–768. doi: 10.1167/iovs.03-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault N, Champagne MP, Bernier G. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol. 2009;327:541–550. doi: 10.1016/j.ydbio.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Burbach JP, Smidt MP. In control of biology: of mice, men and Foxes. Biochem J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]


