Abstract
The three-dimensional structure of Aspergillus niger pectin lyase B (PLB) has been determined by crystallographic techniques at a resolution of 1.7 Å. The model, with all 359 amino acids and 339 water molecules, refines to a final crystallographic R factor of 16.5%. The polypeptide backbone folds into a large right-handed cylinder, termed a parallel β helix. Loops of various sizes and conformations protrude from the central helix and probably confer function. The largest loop of 53 residues folds into a small domain consisting of three antiparallel β strands, one turn of an α helix, and one turn of a 310 helix. By comparison with the structure of Erwinia chrysanthemi pectate lyase C (PelC), the primary sequence alignment between the pectate and pectin lyase subfamilies has been corrected and the active site region for the pectin lyases deduced. The substrate-binding site in PLB is considerably less hydrophilic than the comparable PelC region and consists of an extensive network of highly conserved Trp and His residues. The PLB structure provides an atomic explanation for the lack of a catalytic requirement for Ca2+ in the pectin lyase family, in contrast to that found in the pectate lyase enzymes. Surprisingly, however, the PLB site analogous to the Ca2+ site in PelC is filled with a positive charge provided by a conserved Arg in the pectin lyases. The significance of the finding with regard to the enzymatic mechanism is discussed.
Pectin lyases and Pels (EC 4.2.2.10 and EC 4.2.2.9) are fungal and bacterial proteins that catalyze the degradation of pectin and pectate, respectively, in the middle lamella and cell wall of higher plants (Collmer and Keen, 1986; Barras et al., 1994). Pectin is the methylated form of pectate and carries no net charge. The degradation occurs by a transelimination mechanism, in which the enzymes cleave the α-glycosidic bond between O1 and C4 to generate an unsaturated 4–5 carbon-carbon bond at the nonreducing end of the polygalacturonate (Albersheim et al., 1958). Pectin lyases from Aspergillus spp. are used extensively in the food industry as the primary component of pectinase preparations. Pectin lyases cleave highly esterified pectins without affecting the volatile esters of scent as well as reduce flocculating effects due to fruit Ca2+ complexes if de-esterified pectin derivatives are present (Fogarty and Kelly, 1982; Alana et al., 1989).
PLB belongs to a multigene family of pectin lyases produced by Aspergillus niger (Kuster-van Someren et al., 1992; Kester and Visser, 1994). PLB consists of 359 amino acids with a molecular weight of 37,820 calculated from the amino acid sequence. PLB shares 46 to 65% amino acid sequence identity with other A. niger pectin lyases. Unlike the Pels, pectin lyases do not require Ca2+ for enzymatic activity; nevertheless, Ca2+ and Na+ are known to stimulate enzymatic activity near the pH optimum of 8.5 (Kester and Visser, 1994). PLB is also unusual because the protein can be isolated in two conformational states, one at pH 6.0 and the other at pH 7.5. Unlike the pH 6.0 state, the activity of the pH 7.5 state is dependent upon ionic strength and undergoes a reversible but inactivating change.
The pectin lyases belong to a protein superfamily that includes the Pels as well as plant pollen proteins. A multiple sequence alignment of the superfamily (Henrissat et al., 1995), based upon the structural superposition of members of the Pel subfamily, indicated that pectin lyases share 14 to 20% identity with bacterial Pels. Although the percentage of identical amino acids is relatively low, the presence of key structural regions, such as the Asn ladder in PelC, suggested that all members of the superfamily would share a common structural fold. As predicted, the PLB structure reported here folds into a parallel β helix, a novel topology first observed in the bacterial Pels (Yoder et al., 1993a; Lietzke et al., 1994; Pickersgill et al., 1994). Although the core topology is similar, PLB differs significantly from the Pels in the oligosaccharide-binding loops that protrude from and cover the parallel β helix core. The differences have been exploited in the present research to improve the multiple sequence alignment of the Pel superfamily and to gain further insight into the enzymatic behavior of the pectin lyase subfamily.
MATERIALS AND METHODS
A crystallization kit, Grid Screen A/S, was purchased from Hampton Research (Laguna Hills, CA) and Cryschem plates were obtained from Supper Co. (Natick, MA). A 25% aqueous solution of grade II glutaraldehyde was purchased from ICN. The heavy-atom compounds were purchased from the following companies: PIP from Strem Chemicals (Newburyport, MA); methylmercury acetate from Pfaltz and Bauer, Inc. (Waterbury, CT). All other reagents were obtained from Sigma.
Crystal Preparation
PLB was isolated from an Aspergillus niger multicopy pki-pelB fusion transformant N593(pPK-PLB)6 described by
Kusters-van Someren et al. (1992). The enzyme was purified to homogeneity from the culture filtrate by ammonium sulfate precipitation, dialysis against a 0.01 m Tris-HCl buffer at pH 7.5, and elution from a DEAE-Sepharose column by a 0.0 to 0.3 m sodium chloride gradient, as described by Kester and Visser (1994). The protein was concentrated by ammonium sulfate precipitation, dialyzed against 10 mm N-[2-hydroxyethyl]piperazine-N′-[2-ethane- sulfonic acid], pH 7.5, and stored at −20°C at a concentration of 18.4 A280/mL. The purified recombinant protein had a pI of 5.9 and a specific activity of 280 units/mg at pH 8.5.
For crystal growth, equal volumes of the PLB stock and a solution containing 0.1 m citric acid, pH 5.0, and 1.6 m ammonium sulfate (buffer A) were mixed and centrifuged to remove particulates. Droplets of 10 μL were subsequently placed in a Cryschem plate over 250-μL reservoirs containing buffer A. The largest crystals grew in 3 weeks at 4°C. The diffraction patterns of PLB crystals were consistent with the orthorhombic space group P21212, with unit cell parameters of a = 83.60 Å, b = 88.80 Å, and c = 42.28 Å. Assuming one molecule per asymmetric unit, the Vm and solvent content were estimated to be 2.07 Å3/D and 41%, respectively (Matthews, 1968). To prepare heavy-atom derivatives, crystals were pretreated with a 0.25% glutaraldehyde solution overnight at 4°C and were subsequently transferred to mother liquor at pH 5.0. Each platinum derivative was prepared by adding several grains of PIP to the mother liquor. Crystals of the first platinum derivative, Pt1, were soaked for 2 d and crystals of the second platinum derivative, Pt2, for 7 d at 4°C. For the mercury derivative, the pH of the crystal droplet was raised to pH 7.0 with sodium hydroxide, and subsequently, methylmercury acetate was added to a final concentration of 3 mm. Crystals were removed from the methylmercury acetate solution after 3 d at 4°C.
Data Collection and Reduction
X-ray diffraction data were collected at 20°C using a dual-chamber area detector system (San Diego Multiwire Systems, San Diego, CA) (Hamlin, 1985) installed on a rotating anode x-ray generator (Rigaku, Danvers, MA) with a graphite monochromator (Supper) operated at 45 kV × 150 to 175 mA. Each x-ray data set was collected from a single crystal at helium-path distances of 615 and 555 mm between the crystal and each detector. The data were processed with the San Diego Multiwire Systems software package (Howard et al., 1985). Derivative diffraction data sets were scaled, using the Fourier-Bessel program of Terwilliger and Eisenberg (1983), to the native x-ray data set, which had been placed on an absolute scale by Wilson's method (Wilson, 1942). The x-ray diffraction data collection statistics are summarized in Table I.
Table I.
Summary of x-ray diffraction data collection
| Parameter | Native | Pt1a | Pt2b | Hg |
|---|---|---|---|---|
| Resolution (Å) | 1.7 | 1.8 | 1.9 | 1.9 |
| Total observations | 204,035 | 55,844 | 64,912 | 66,919 |
| Unique reflections | 50,124 | 21,076 | 21,900 | 20,372 |
| Total possible (%) | 73.8 | 70.0 | 87.1 | 81.6 |
| Average (I/ς) | 18.9 | 10.1 | 12.2 | 13.8 |
| Rsym (edited)c | 5.4 | 6.5 | 6.6 | 4.0 |
| Rscaled | 9.3 | 6.9 | 5.5 |
First platinum derivative.
Second platinum derivative.
Rsym = 100 × Σ‖Iavg − Iobs‖/ΣIavg, where I is the average (avg) or the observed (obs) intensity of the reflection. Edited Rsym is the value after manual rejection of outlying observations of a reflection.
Rscale = 100 × Σ‖Fnat − Fder‖/Σ(Fnat + Fder).
Phase Determination
The structure was determined by a combination of molecular replacement and MIR techniques. Initial phases were obtained by molecular replacement. A search model was generated from the atomic coordinates of E. chrysanthemi PelE, which shares 20% sequence identity with PLB. Only those amino acids found in the parallel β helix core were included in the model. With the exception of 67 amino acids shared by the 2 proteins, the remaining 140 amino acids in the core were replaced with Ala. The molecular replacement calculations were carried out with the search model in an arbitrary orientation using XPLOR 3.1 (Brunger, 1988, 1990, 1993b), data with F > 4ς in the resolution range of 3.0 to 7.0 Å, and a 5.0 to 24.0 Å shell of integration in the rotation function.
Heavy-atom sites were determined by cross-phasing the difference Fourier of each derivative by the partial model obtained from molecular replacement. Each solution was confirmed by the presence of suitable Harker peaks in difference Patterson maps. The package PHASES (Furey and Swaminathan, 1997) was used for Fourier calculations and the program HEAVY (Terwilliger and Eisenberg, 1983) was used for the refinement of the heavy-atom parameters.
Model Refinement
The model was refined with molecular dynamics techniques (Brünger et al., 1987) using XPLOR 3.1 and later XPLOR 3.8 (Brünger, 1993b). The data, with F > 2ς in the resolution range of 1.7 to 7.0 Å, were randomly divided into two sets, a working set composed of 90% of the data, and a test set composed of the remaining 10% for cross-validation purposes (Brünger, 1993a). The parameters of Engh and Huber (1991) were used. At all except the last stage, the weight was set to one-half the weight of the check stage. The slow cool annealing protocol was used for simulated annealing with a dynamics temperature starting at 4000 K. The refinement alternated between manual building and rebuilding of the model using the program O (Jones et al., 1991). For most of the analysis, partial model phases were combined with heavy-atom derivative phases to generate SIGMAA maps (Read, 1986). The entire model was checked by inspection of annealed omit maps (Brünger, 1993b). At the completion of the protein refinement water molecules were assigned to density using the procedure previously described (Lietzke et al., 1996), with the exception that only peaks greater than 3ς in the Fo − Fc and 1ς in the 2Fo − Fc electron density maps were considered. For the final model and statistics calculations, all reflections with F > 2ς were used and the weight of the crystallographic term was increased in five equal steps to the full value initially suggested by the XPLOR check stage.
Model Analysis
Hydrogen bonds were determined using the program HBPLUS (I.K. McDonald, D.N. Naylor, D.T. Jones, and J.M. Thornton, Department of Biochemistry and Molecular Biology, University College, London) and the criteria of Baker and Hubbard (1984), a donor-acceptor distance of less than 3.9 Å, a hydrogen-acceptor distance of less than 2.5 Å, and associated angles greater than 90o. A hydrophobic contact was assumed to exist between two apolar residues if the carbon-carbon distance was less than 4.0 Å. Amino acids were assigned to secondary structural elements if they exhibited repetitive φ and ψ angles for the structural element and if all backbone amide nitrogens and carbonyl oxygens participated in intra- or interstrand hydrogen bonding. The assignment was initially made with the Kabsch and Sander (1983) algorithm in the PROMOTIF program (Hutchinson and Thornton, 1996), followed by inspection of the main-chain hydrogen bonds.
The structural superposition of PLB and PelC was carried out with the program O. The lsq explicit (a computer command in the program O that performs a least-squares fit between αCs in specified segments) command was initially used to superimpose residues 130 through 150 of PelC with residues 154 through 173 of PLB. The fit was subsequently improved with the lsq improve (a computer command in the program O that improves the least-squares fit previously calculated with lsq explicit) command in O using the αCs of all residues in PLB and PelC, a maximum distance cutoff between paired αCs of 3.0 and 2.0 Å and a minimum fragment length of two. The amino acids with a 3.0-Å or less deviation in the αC position were considered to be structurally related for the sequence alignment of the two proteins. A minimum fragment length of one was ultimately used for aligning residues Val-323 through Tyr-337 of PLB with Thr-310 through Tyr-320 of PelC. The LINEUP routine of the Genetics Computer Group package (Madison, WI) was used for the construction of the sequence alignment table.
The notation of Yoder et al. (1993b) and of Yoder and Jurnak (1995) is used to describe the structural elements of the parallel β helix. The three parallel β sheets are referred to as PB1, PB2, and PB3. The individual β strands are named with a two-part label, PBm,n, where m indicates the β sheet to which the β strand belongs, and n is the sequential number of the strand within the β sheet. Turns of type T1 join PB1 with PB2, turns of type T2 join PB2 with PB3, and turns of type T3 join PB3 with PB1. As with the β strands, turns are named with a two-part label, Tr,s, where r refers to the turn type, and s is the sequential number of the turn within that type.
RESULTS
Structural Determination
The orientation and position of the initial search model, consisting of 207 residues of the parallel β helix, was relatively clear in the molecular replacement searches. The correct solution corresponded to the top peak in all searches. Moreover, the signal-to-noise ratios, defined as the ratios of the correct peak to the top noise peak, were 1.02 for the rotation search, 1.33 after Patterson-correlation refinement, and 1.22 for the translation search. A rigid body refinement of the initial search model resulted in a crystallographic R factor of 0.53.
Efforts to complete the polypeptide tracing with the available model phases were unsuccessful. Therefore, additional phasing information was derived from heavy-atom derivatives. Four unique heavy-atom positions were deduced from Fourier syntheses cross-phased with partial model phases and confirmed by difference Patterson techniques. The refined heavy-atom parameters are listed in Table II and the MIR phasing statistics are presented in Table III. The heavy atoms substitute at chemically reasonable sites. In 2 d, Pt binds at pH 5.0 to Met-352, and in 7 d to Met-352 as well as to Cys-206, which forms a disulfide bond with Cys-72. At pH 7.0, Hg binds to His-79 and to His-259. The overall figure of merit for the heavy-atom phases is 0.44 for 9028 reflections in the resolution range of 2.7 to 50.0 Å. The inclusion of the heavy atoms in the phasing process occurred in steps, as the data were measured and/or the need for them in map interpretation arose. Continued model building and refinement were interspersed with the addition of derivative phasing information.
Table II.
Refined heavy-atom derivative parameters for PLB in the resolution range of 50.0 to 2.7 Å
| Derivative | Occupancy | Atomic Coordinates
|
B(Å2) | ||
|---|---|---|---|---|---|
| X | Y | Z | |||
| Pt1a | 0.323 | 0.400 | 0.112 | 0.868 | 32.6 |
| Pt2b | 0.319 | 0.400 | 0.111 | 0.866 | 37.1 |
| 0.283 | 0.045 | 0.171 | 0.707 | 32.3 | |
| Hg | 0.178 | 0.794 | 0.157 | 0.458 | 33.3 |
| 0.133 | 0.502 | 0.187 | 0.367 | 44.4 | |
First platinum derivative.
Second platinum derivative.
Table III.
PLB MIR phasing statistics
Fh/E, Ratio of heavy atom structure factor and the residual lack of closure. Rcullis, Cullis R for centric reflections = Σ‖‖FpH ± Fp‖ − FH‖/Σ‖FpH ± Fp‖ where Fp, FpH, and FH are the structure factors of the protein, derivative, and heavy atom, respectively.
| Resolution Range (Å) | Pt1a
|
Pt2b
|
Hg
|
Figure of Merit | |||
|---|---|---|---|---|---|---|---|
| Fh/E | Rcullis | Fh/E | Rcullis | Fh/E | Rcullis | ||
| 9.74 | 0.77 | 0.67 | 1.20 | 0.59 | 0.77 | 0.68 | 0.54 |
| 6.14 | 0.87 | 0.69 | 1.50 | 0.54 | 0.87 | 0.67 | 0.60 |
| 4.80 | 0.69 | 0.77 | 1.29 | 0.66 | 0.63 | 0.70 | 0.47 |
| 4.07 | 0.55 | 0.70 | 1.12 | 0.68 | 0.55 | 0.68 | 0.45 |
| 3.59 | 0.48 | 0.75 | 0.92 | 0.73 | 0.51 | 0.73 | 0.39 |
| 3.25 | 0.41 | 0.73 | 0.83 | 0.73 | 0.50 | 0.73 | 0.43 |
| 2.99 | 0.44 | 0.76 | 0.75 | 0.69 | 0.44 | 0.77 | 0.39 |
| 2.79 | 0.42 | 0.77 | 0.73 | 0.66 | 0.40 | 0.76 | 0.44 |
| Total | 0.55 | 0.73 | 1.03 | 0.65 | 0.58 | 0.71 | 0.44 |
First platinum derivative.
Second platinum derivative.
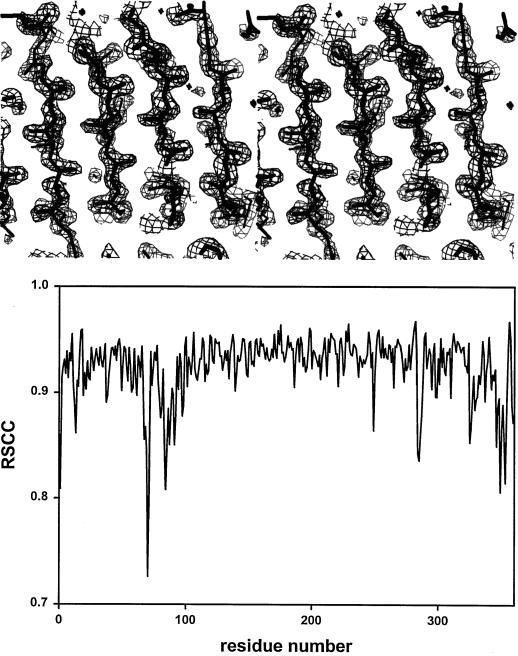
The PLB model presented here consists of 2662 nonhydrogen atoms in 359 amino acids and 339 water molecules. The final crystallographic R factor, including all reflections with F > 2ς in the 1.7- to 7.0-Å shell, is 16.5%. A summary of the model refinement statistics is presented in Table IV. A Ramachandran plot of the backbone dihedral angles in Figure 1 illustrates that 82.7% of non-Gly residues lie in the most favored region and that 15.9% lie in allowed regions. Four residues are within the generously allowed regions, with φ and ψ angles of 38.8o and 62.5o for Asn-146; 84.5o and 10.8o for Trp-151; 75.3o and 84.8o for Asp-160; and 48.9o and 121.8o for Asn-187. However, the latter residues all fall in well-defined electron density in the annealed omit maps. A representative section of the final annealed omit map is shown in Figure 2, top. The final map contoured at 0.9 ς shows continuous density for all main-chain atoms and most side chains, with the exception of some surface groups, mostly Lys's. The electron density also indicates a few errors in the amino acid sequence: Arg-47 is a Gln, Ile-281 is a Val, and Asn-346 is an Ala. The real space correlation coefficient of the main-chain atoms of PLB as a function of residue number is shown in Figure 2, bottom.
Table IV.
Final refinement statistics for PLB
| Resolution range (Å) | 1.7–7.0 |
| No. of reflections (F > 2ς) | 32,007 |
| Nonhydrogen protein atoms/asymmetric unit | 2,662 |
| Waters/asymmetric unit | 339 |
| Rfree for 10% of data | 0.198 |
| Rwork for 90% of data | 0.162 |
| Combined R-factor for last cycle | 0.165 |
| rms deviations from ideal geometry | |
| Bond length (Å) | 0.010 |
| Bond angle (°) | 2.10 |
| Impropers (°) | 1.44 |
| Average thermal factors | |
| Main chain (Å2) | 13.4 |
| Side chain (Å2) | 16.8 |
| All protein atoms (Å2) | 15.0 |
| Waters (Å2) | 37.7 |
| All atoms (Å2) | 17.5 |
Figure 1.
Ramachandran plot of the φ and ψ dihedral angles of the PLB polypeptide backbone. Gly residues are shown as triangles; all other residues are shown as squares. The plot was prepared with the program PROCHECK (Laskowski et al., 1993).
Figure 2.
Top, Stereo view of an annealed omit electron density map. Several parallel β strands are superimposed in thick black lines upon an electron density map in which a region of 8 Å around Trp-212 had been omitted and the remaining model refined. The map is contoured at 1.0 ς. The figure was generated with the program O. Bottom, The real space correlation coefficients of the main chain atoms of PLB are plotted as a function of residue number. The correlation coefficients were calculated with the program O.
Structural Results
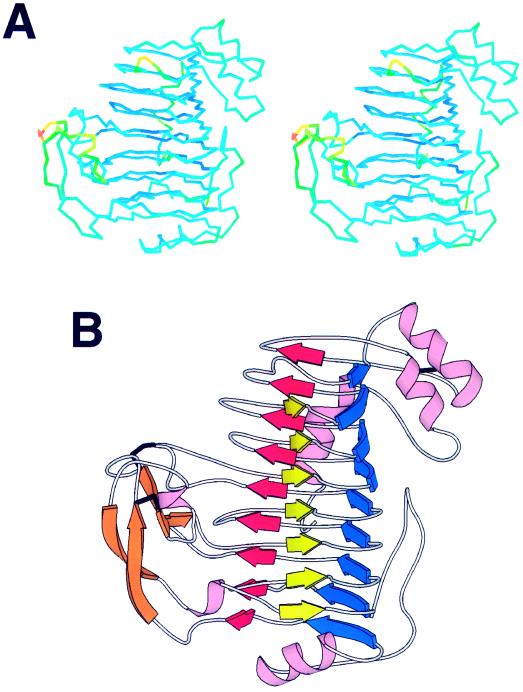
The PLB molecule is a single structural domain with approximate dimensions of 34 × 44 × 56 Å. Table V lists the amino acids that comprise the secondary structural elements. Of the 359 amino acids, 105 or 29.2% form β structures, 25 or 7% form α helices, and 20 or 5.6% form 310 helices. As illustrated in Figure 3, the core of the protein consists of three parallel β sheets that have individual parallel β strands that coil into a large right-handed cylinder. The folding topology is similar to that first observed for PelC (Yoder et al., 1993a) and is termed a parallel β helix. Within PLB, there are 7 complete turns in the parallel β helix and a minimum of 22 residues per turn. A schematic of the PLB polypeptide backbone, highlighting the β structural features is shown in Figure 3. The three parallel β sheets consist of seven, nine, and nine strands, respectively. Within each β sheet, the β strands are relatively short, ranging from two to six residues in length and generating only a small right-handed twist. As a consequence of the packing arrangement of the parallel β sheets, the cross-section of the parallel β helix is not circular, but L shaped. Two of the β sheets form a parallel β sandwich with the third parallel β sheet oriented approximately 110o to the parallel β sandwich. In addition to the β structure, there are four α helices and four 310 helices, all of which are peripheral to the core structure.
Table V.
Secondary structural elements of PLB
| β Strand | Residues | β Strand | Residues | β Strand | Residues |
|---|---|---|---|---|---|
| PB2,1 | 41 –45 | PB3,1 | 48 –50 | PB1,1 | 104 –105 |
| PB2,2 | 110 –114 | PB3,2 | 120 –122 | PB1,2 | 126 –128 |
| PB2,3 | 135 –138 | PB3,3 | 141 –143 | PB1,3 | 156 –158 |
| PB2,4 | 164 –167 | PB3,4 | 170 –171 | PB1,4 | 179 –181 |
| PB2,5 | 189 –192 | PB3,5 | 195 –197 | PB1,5 | 214 –216 |
| PB2,6 | 221 –226 | PB3,6 | 229 –231 | PB1,6 | 239 –240 |
| PB2,7 | 244 –249 | PB3,7 | 252 –254 | PB1,7 | 262 –263 |
| PB2,8 | 267 –272 | PB3,8 | 275 –277 | ||
| PB2,9 | 290 –292 | PB3,9 | 315 –317 | ||
| α Helices | Residues | 310 Helices | Residues | Antiparallel β | |
| A | 28 –36 | 57–63 | |||
| B | 81 –84 | 74–76 | |||
| C | 98 –101 | 91–96 | |||
| D′ | 304 –307 | D | 298 –303 | ||
| E | 327 –333 | Other parallel β | |||
| F′ | 347 –354 | F | 344 –346 | 149–150 |
Parallel β strands within the parallel β helix are named with a two-part label, as described in Methods. Helices are named with an uppercase letter. In the two examples in which α helices immediately follow 310 helices, the same letter is used for both but with a prime for the α helix.
Figure 3.
A, Stereo view of the αC backbone of PLB. The color represents the temperature factor and varies smoothly from blue to red as the temperature factor increases. B, Schematic ribbon diagram of the PLB backbone, illustrating the secondary structure. Helices are represented by pink coils and β structure is shown by arrows. The parallel β sheet, PB1, is yellow; PB2 is blue; and PB3 is red. The antiparallel β structure within the first T3 turn and a short β strand within the third T3 loop are indicated in orange. Disulfide bonds are designated by thick, black lines. The image in A was generated with the program O and the image in B was done with MOLSCRIPT (Kraulis, 1991).
The interior of the parallel β helix of PLB is filled with ordered arrays of side chains. The majority of the interior residues are aliphatic, either Val or Ile, and to a lesser extent, Leu or Ala. As described in Table VI, there are four aliphatic stacks within the PB1-PB2 β sandwich, with the longest running eight side chains in length. The Val and Ile side chains have similar orientations but, with an αC-to-αC distance of 4.5 to 5.5 Å, the adjacent groups in the stacks are not sufficiently close for a strong hydrophobic interaction. The side chains of the two stacks of PB1 are interdigitated with the side chains of the two stacks of PB2. PB3 also contributes to an interior mixed stack of aromatic and aliphatic side chains as well as to an exterior stack of mostly aliphatic groups.
Table VI.
Aliphatic side-chain stacks within the parallel β structure of PLB
Solid boxes enclose the stacked aliphatic side chains. Boxes in broken lines enclose stacked aromatic residues in mixed aliphatic/aromatic stacks. The letter i on the top of a column indicates that the side chains of the residues of the column point toward the interior of the parallel β helix.
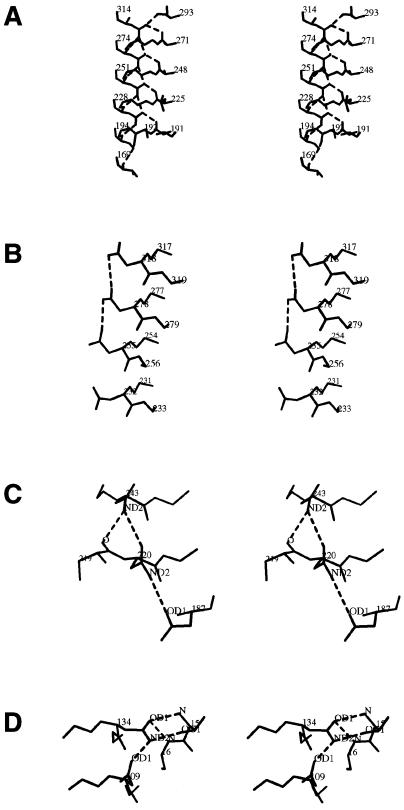
A second type of vertical stack involves the polar residues Ser and Asn. One prominent stack is the Asn ladder located at the second position of the T2 turn between PB2 and PB3. As shown in Figure 4A, the ladder in PLB contains four Asns: Asn-228, Asn-251, Asn-274, and Asn-314, and is preceded by Ser-194. As in the Pels, the Asn side chains are oriented so that the maximum number of hydrogen bonds are formed. Unlike the Pels, there are additional short Asn stacks that are formed on the exterior of the parallel β helix of PLB, as illustrated in Figure 4, B through D. The first stack consists of a central aspartate flanked by Asn-232, Asn-255, Asp-278, and Asn-318. The second unusual grouping involves a hydrogen bonding ladder formed by a triplet of Asns, two from adjacent rungs of the parallel β helix, Asn-220 and Asn-243, and the third, Asn-187, from a neighboring loop. The last noteworthy grouping is a hydrogen bond ladder between Asn-109 and Asn-134, and the neighboring Thr-16.
Figure 4.
Stereo views of stacked Asn interactions on the interior and exterior of the parallel β helix in PLB. A, Hydrogen bonding pattern of the Asn ladder in PLB. With the exception of the first Asn, Asn-228, and the last in the ladder, Asn-314, each Asn forms five hydrogen bonds. Not shown is the short aliphatic stack, Ile-140 and Val-169, which precedes Ser-194 in the Asn ladder. B, Mixed stack, illustrating the hydrogen bonding between the carboxyl oxygens of Asp-278 and the flanking ND2 nitrogens of Asn-255 and Asn-318. Because the side chain of Asn-232 is not parallel to the others in the stack, there is no interaction with its neighbor, Asn-255. C, Triplet Asn interactions involving hydrogen bonds among a neighboring Asn-187 and the stacked Asn-220 and Asn-243 residues. The ND2 nitrogens of Asn-220 and Asn-243 form hydrogen bonds with the carboxyl groups of Asn-187 and Asn-220, respectively. ND2 of Asn-243 also forms a hydrogen bond with the carbonyl oxygen of residue 219. D, Triplet interactions, involving hydrogen bonds among a neighboring Thr-16 and the stacked Asn-109 and Asn-134 residues. The figure was generated with the program O.
A third notable side-chain grouping involves aromatic residues. In PLB the aromatic residues form either vertical stacks, in which the aromatic planar rings lie parallel to one another, or clusters, in which the rings are at an angle to neighboring rings. There are two vertical stacks. One is composed of four phenylalanines, Phe-230, Phe-253, Phe-276, and Phe-316, with the side chains oriented toward the interior from PB3. The other, a mixed stack composed of His-186, Tyr-193, and Trp-350, lies on the exterior of the parallel β helix. In both examples, the inter-ring distance is approximately 3.6 Å and the nearly parallel rings are tilted slightly with respect to the stacking axis. There are two prominent aromatic clusters in which the adjacent rings have the typical edge-on appearance (Burley and Petsko, 1985). The first is found in all Pels and in PLB it consists of Phe-10 in the N-terminal loop, and His-247, Phe-307, and Phe-333 in the C-terminal loop. The second, shown in Figure 5, is a very extensive network found in the putative oligosaccharide-binding region of PLB, consisting of Trp-66, Trp-81, Trp-151, His-178, His-210, Tyr-211, Trp-212, and His-259. A smaller cluster of four residues is found in the comparable location in Bacillus subtilus Pel, but not in PelC or PelE. The increased hydrophobicity conferred by the large aromatic cluster in a PLB region analogous to the oligosaccharide-binding site in PelC (R. Scavetta and F. Jurnak, personal observations) may be related to the fact that the pectin substrate is not charged in contrast to pectate, which is recognized by the Pels.
Figure 5.
Stereo view of the extensive network of Trp and His residues in the putative oligosaccharide binding of PLB. The αC backbone is drawn in gray, the side chains of the Trp and His residues are drawn in black, and each residue is labeled. The figure was generated with the program O.
As found in the Pel structures, one of three peptide segments connecting the β strands in each helical coil, the T2 turn, forms a regular secondary structural element consisting of two amino acids. In PLB there are six examples of the bend that resembles a distorted γβE turn. The first residue has an αL conformation with average φ and ψ angles of 52.3o and 35.5o, respectively, and the second, a βE conformation with average φ and ψ angles of −102.5o and 158.8o, respectively. The remaining T2 loops in PLB vary considerably in geometry and cannot be readily classified as a distinct nonrepetitive secondary structural element. The rare αL conformation is also found for three of the seven amino acids initiating the T3 loops, Asn-109, Asn-134, and Leu-163, and three of the seven amino acids terminating the T1 loops, Asp-144, Asn-232, and Asn-255. The peptide segments comprising the T1 turns are limited in length, varying from three to seven residues. The peptide segments comprising the T3 turns are the longest and most varied of all external loops. The first T3 loop is the longest, 53 residues in length, and folds into a small domain consisting of three antiparallel β strands, one turn of an α helix, and one turn of a 310 helix. The tertiary fold of the T3 turn is stabilized by a disulfide bond between Cys-63 in the middle strand of the antiparallel β structure, and Cys-82 in the second helical segment. The unit is further stabilized by interaction with two other T3 turns. The third T3 turn forms a β strand that extends the β sheet of the first T3 turn. The fifth T3 turn is connected to the first T3 turn by a disulfide bond between Cys-72 and Cys-206. Several of the residues in the T1 and T3 loops participate in extensive backbone hydrogen bonding between adjacent coils of the parallel β helix.
PLB has six Cys residues, all of which participate in disulfide bonds. In addition to the two disulfide bonds involving T3 loops, the third disulfide bond between Cys-303 and Cys-311 is found at the C-terminal end of the parallel β helix. Other amino acids of note include three residues with the unusual cis conformation. The first is Pro-238, analogous to the invariant Pro found at position 220 in the PelC sequence (Henrissat et al., 1995) and observed to be a cis-Pro in all known Pels structures. The second amino acid is also a cis-Pro, Pro-286, but has no counterpart in other Pel structures. The third cis-amino acid is Thr-183. Although cis-residues other than Pro are unusual, they are occasionally observed. For example, lectin IV of Griffonia simplicifolia contains a cis-Pro, a cis-Gly, and a cis-Asp (Delbaere et al., 1993).
Comparison of PLB and PelC
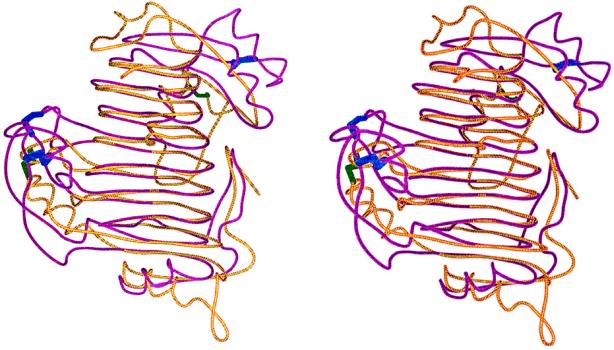
The three-dimensional structure of PLB was superimposed upon the PelC structure by first aligning the αC of the parallel β strands within the parallel β helix core and then minimizing the rms distances between αC in both models. The results of the superposition, illustrating the alignment of secondary structural elements and αC backbones, is shown in Figure 6. The core parallel β helix aligns very well, within an rms deviation between corresponding αC atoms of 1.1 Å for 197 pairs with a 2.0-Å distance cutoff. In both structures the parallel β helix core consists of seven complete turns, with the number of amino acids per β strand and parallel β strands per β sheet varying only slightly between the two enzymes.
Figure 6.
Stereo view of the superposition of the ribbon diagrams of PLB and PelC. PLB is illustrated in purple; PelC is in yellow. The disulfide bonds of PLB are shown in blue and PelC are green. The figure was drawn using MOLSCRIPT.
As illustrated in Figure 6, the loops that protrude from and cover the central parallel β helix occur at analogous positions in PLB and PelC. A larger distance cutoff allows for a more accurate comparison of the peptide protrusions that are similar in conformation but tilted or rotated somewhat between the two proteins. With a 3.0-Å cutoff, 221 pairs of αCs have a deviation of 1.3 Å. Three of the peptide protrusions are structurally conserved in all known Pel structures. These include the N terminal branch from residues 7 to 20 in PLB; the helix covering the amino end of the parallel β helix, residues 28 to 36 in PLB; and the C-terminal helix, from residues 344 to 354 in PLB. A fourth region, residues 325 to 343 in PLB, is similar to a large omega loop found in all Pel structures, with the exception that one side is composed of a 310 helix instead of the typical β element. Most interesting is the similarity in the cap structure at the C-terminal end of the parallel β helix of PLB and PelC, in distinct contrast to the varied C-terminal caps of other known parallel β helices. In PLB and in PelC, the parallel β helices terminate in a helix followed by a β ribbon before entering the omega loop. However, the location of the helical segment is displaced in PLB relative to PelC. As yet, no functional significance can be attributed to the C-terminal cap structure.
There are pronounced differences in the T3 loops protruding from the first, third, and fifth rungs of the parallel β helix. In the Pel structures the loops form part of the walls of the oligosaccharide-binding groove (R. Scavetta and F. Jurnak, personal observation) and are rich in hydrophilic residues. In PLB the same loops are longer in length and less hydrophilic in character. In particular, the first T3 loop of PLB, with 53 residues, is longer than the analogous loops in PelC or PelE. It is interesting that the same PLB loop is comparable in length, but totally different in secondary structure to the analogous loop in Bacillus subtilus Pel. The three T3 loops in the Pel structures also form the binding site for Ca2+, an essential cation for pectinolytic activity in the Pels. In contrast, Ca2+ is not required for pectinolytic activity in the pectin lyases, but is observed to stimulate activity in the high pH range.
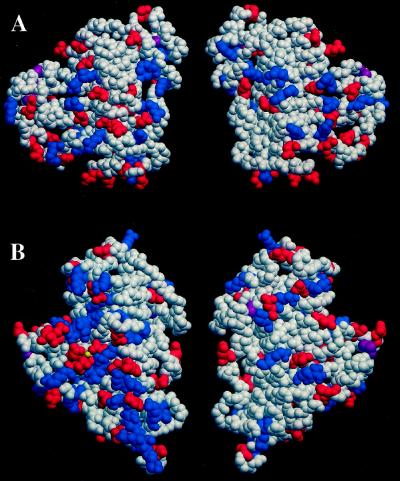
As shown in Figure 7, two of the three acidic amino acids that coordinate directly with Ca2+ in the Pels, Glu-166 and Asp-170 in PelC, are replaced with Arg-176 and Val-180 in PLB. Only the third acidic residue, Asp-131 in PelC, has a counterpart in PLB, Asp-154. With amino acid substitutions in two of the three acidic groups that coordinate Ca2+ in the Pels, it is not surprising that PLB or other pectin lyases do not bind Ca2+ strongly or require the cation for pectinolytic activity. The decreased hydrophilicity conferred by the T3 loops in PLB is best represented by a comparative view of the surface charges of PLB and PelC in the putative oligosaccharide-binding groove shown in Figure 8.
Figure 7.
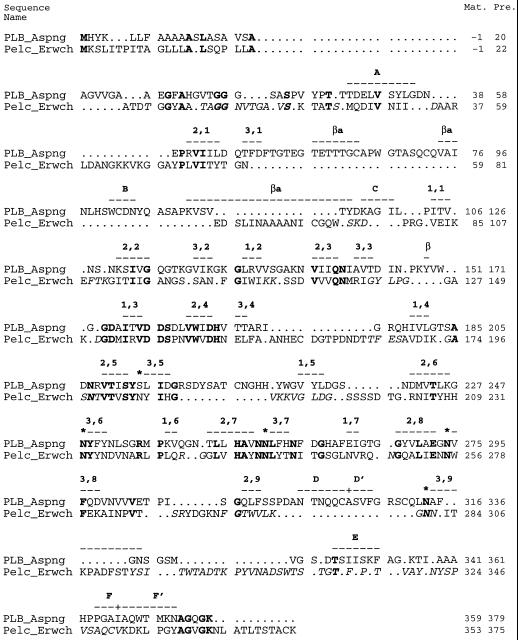
Sequence alignment of PLB and PelC based upon their structural superposition. Identical residues bewteen PLB and PelC are given in bold letters. Italic letters in the PelC sequence indicate the residues that have been aligned incorrectly with PLB in Henrissat et al. (1995). In addition, the sequence numbering for PLB has been changed and differs by −3 in the sequence number before Tyr-96 and by −2 after Tyr-96 from that previously published in Henrissat et al. (1995). Broken lines represent the residue range of a particular structural element. Structural assignments are given in bold type. The parallel β strands within the parallel β helix are designated by their number, antiparallel β strands are designated as βa, and parallel β strands outside the parallel β helix are designated as β. Helix lettering for PLB is given in Table V. The + sign within a broken line indicates the switch of a helix from 310 to α. Residue numbers are given on the right for mature protein (Mat.) and for the precursor form (Pre.). Asterisks (*) indicate the Asp ladder for PLB. The initial part of the figure includes the N-terminal signal peptide.
Figure 8.
A, Space-filling models of PLB. B, Space-filling model of PelC. Neutral residues are shown in white, positively charged residues are in blue, and negatively charged residues are in red. The location of the disulfide bonds are shown in purple and the Ca2+ ion in PelC is in yellow. In the left view, the putative oligosaccharide-binding site is located on the left. The right view is rotated 180o around the vertical axis of the left view. The figure was generated with the program O.
The structural superposition of PLB and PelC has been used to generate an accurate sequence alignment between the two proteins. The new sequence alignment improves an earlier multiple sequence alignment between the Pel and the pectin lyase subfamilies (Henrissat et al., 1995). The previous alignment had been based upon a combination of structural and evolutionary relationships among protein sequences in the Pel superfamily. The new structural alignment primarily corrects the subfamily alignments in loop regions that differ in length or structure. The most pronounced corrections occur in three regions. Within the N-terminal branch, Val-14 through Ser-21 of PLB structurally aligns with Thr-10 through Ser-20 of PelC. Thr-10 was previously designated as an invariant residue in the pectate and pectin lyase subfamilies but the corrected alignment indicates that Thr-10 is not conserved. The second region involves a region of insertions from Arg-199 through Asn-220 in PLB or the corresponding sequence in PelC, Val-188 through Gly-202. The third correction in the sequence alignment is a consequence of a structural difference in the large omega loop near the C terminus. In PLB, a 310 helix replaces one of the β strands found in the omega loop of PelC. Consequently, Gly-317 through Ile-347 of PLB is realigned with Tyr-292 through Val-330 in PelC. None of the present corrections to the multiple sequence alignment alter the invariant residues previously identified in the Pel superfamily or the conclusions, suggesting two active site regions (Henrissat et al., 1995).
DISCUSSION
The early biochemical studies in the 1960s were stymied by the findings that many pectate and pectin lyases existed as isoenzymes. With the introduction of recombinant DNA techniques in the 1980s, individual lyase genes were cloned and overexpressed, providing a source of homogeneous product for further characterization. The coincidental advances in crystallographic techniques quickly led to an atomic description of three bacterial Pels, PelC and PelE from E. chrysanthemi (Yoder et al., 1993a; Lietzke et al., 1994) and Pel from B. subtilus (Pickersgill et al., 1994). The present structure of PLB confirms the predictions that the pectin lyases, like the Pels, fold into the unusual parallel β helix topology. PLB is the fourth example of a parallel β helix in the Pel superfamily and the sixth example of a right-handed parallel β helix. The other examples include the tailspike protein from P22 phage (Steinbacher et al., 1994) and Bordetella pertussis P.69 pertactin (Emsley et al., 1996). Although the parallel β helix structure was anticipated for PLB, both the structural similarities and the differences are instructive. The similarities contribute to a general understanding of which amino acids are compatible with the parallel β helix and consequently improve algorithms to predict the fold in other primary sequences. The structural differences are particularly informative for further insight into the mechanistic differences between the pectate and the pectin subfamilies at the atomic level.
Pectin lyases are believed to cleave the α-1,4-glycosidic bond between neutral, methylated forms of galacturonate moieties. Like the Pel, no single amino acid has been implicated in the active site region of a pectin lyase by classical methods. To compensate for the dearth of biochemical data, the active site regions have been deduced by a comparative structural and sequence analysis of the Pel superfamily (Henrissat et al., 1995). The clustering of 10 amino acids, invariant in the superfamily, into two distinct structural regions has led to the postulate of two active sites for all members. Subsequently, the pectinolytic region of the Pels has been confirmed by site-specific mutational techniques (Kita et al., 1996). The structural similarity of PLB and PelC is further confirmation that the pectinolytic region, found in a long, narrow groove, is probably the same in both lyase classes. Not surprisingly, given the charge difference in the substrates, the putative oligosaccharide-binding groove in PLB is lined with an extended network of Trp and His residues, whereas the comparable groove in PelC is lined with Lys's and Args. Most significantly, the three invariant residues in the active site region of the Pels have the same orientation and conformation in PLB. The residues include Asp-154, Arg-236, and Pro-238 in PLB, which are analogous to Asp-131, Arg-218, and Pro-220 in PelC. As in all Pel structures, Pro-238 in PLB folds into the less common cis conformation to properly orient the invariant Arg-236 for a key role in catalysis. Because the Arg is present in pectin lyases as well as in Pels, its catalytic role is believed to be shared as well. The most likely role is that of providing a positive charge to neutralize the negatively charged intermediate generated by the initial proton abstraction during the β-elimination pathway.
The role of the invariant Asp remains an enigma. It is tempting to speculate that the carboxyl group participates in the proton abstraction step. However, neither carboxyl oxygen of the invariant Asp is free to abstract a proton, in either the presence or absence of the substrate (R. Scavetta and F. Jurnak, personal observation). In the Pel structures, both oxygens of the carboxyl group are coordinated to Ca2+ and in PLB, both oxygens form salt bridges with Arg-176, an invariant amino acid within the A. niger pectin lyase isozymes. It is interesting that the positive charge residing on Arg-176 in PLB is positioned at the same relative location as the Ca2+ site in the Pels. This finding suggests that a positive charge, approximately 7.0 Å from the invariant, catalytically important Arg-236 in PLB is critical and that the invariant Asp-154 in PLB aids in its proper orientation. The catalytic necessity of a second positively charged residue, particularly one that is completely neutralized by an Asp, is not at all clear from the structural data. It is more likely that the invariant electrostatic interaction serves as an active or passive marker that aids in the correct positioning of the scissle glycosidic bond of the oligosaccharide substrate in the catalytic site. Attempts to test the hypothesis with structural studies of Pel-oligosaccharide complexes are underway.
Unfortunately, the PLB structure is static at a single pH of 5.0 and does not provide any clues as to the reversible conformational change triggered by changing the pH from 6.0 to 7.5. Nor does the PLB structure provide any clues as to the function of the second putative active site region involving the invariant WiDH sequence. The answers must await further investigations, both structural and otherwise.
Note Added in Proof
Since the initial submission of the PLB manuscript, the three-dimensional structure of a related protein, A. niger pectin lyase A (PLA), was independently determined and reported (Mayans et al., 1997). PLA shares 65% sequence identity with PLB and, in general, the two structures are similar. Thus, the conclusions regarding the catalytic residues and the aromatic cluster in the putative oligosac-charide-binding site are the same. PLB and PLA primarily differ in the pH and resolution of the structure determination, in the number and type of Asn stacks, and in the number of residues with a cis conformation.
In addition to PLA, another example of right-handed parallel β helix was observed in the structure of Aspergillus aculeatus rhamnogalacturonase A (Peterson et al., 1997).
Abbreviations:
- αC
alpha carbon
- crystallographic R factor
agreement factor between observed structure factor amplitudes Fo, and calculated structure factor amplitudes Fc, based on atomic model
- Hg
methylmercury acetate derivative
- MIR
multiple isomorphous replacement
- Pel
pectate lyase
- PelC
Erwinia chrysanthemi pectate lyase C
- PelE
Erwinia chrysanthemi pectate lyase E
- φ
rotation angle about the N-αC bond in the peptide backbone
- PIP
di-μ-iodobis(ethylenediamino) diplatinum (II) nitrate
- PLB
Aspergillus niger pectin lyase B
- ψ
rotation angle about the αC-C bond in the peptide backbone
- rms
root-mean-square
- ς
standard deviation
Footnotes
F.J. was supported by the U.S. Department of Agriculture (award 96-02966), Academic Computing Graphics and Visual Imaging Lab, University of California, Riverside, and the San Diego Supercomputer Center.
LITERATURE CITED
- Alana A, Gabilondo A, Hernando F, Moragues M, Dominguez J, Llama M, Serra J. Pectin lyase production by a Penicillium italicum strain. Appl Environ Microbiol. 1989;5:1612–1616. doi: 10.1128/aem.55.6.1612-1616.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim P, Neukom H, Stutz E. Pectic substances and pectic enzymes. Adv Enzymol. 1958;20:341–382. doi: 10.1002/9780470122655.ch11. [DOI] [PubMed] [Google Scholar]
- Baker EN, Hubbard RE. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol. 1994;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Barras F, van Gijsegem F, Chatterjee AK. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol. 1994;32:201–234. [Google Scholar]
- Brünger A. Crystallographic refinement by simulated annealing: application to a 2.8 Å resolution structure of aspartate aminotransferase. J Mol Biol. 1988;208:803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- Brünger A. Extension of molecular replacement: a new search strategy based on Patterson correlation refinement. Acta Crystallogr. 1990;A46:46–57. [Google Scholar]
- Brünger AT. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr. 1993a;D49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- Brünger AT (1993b) XPLOR Manual, Version 3.1. Yale University, New Haven, CT
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Collmer A, Keen NT. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- Delbaere LT, Vandonselaar M, Prasad L, Quail JW, Wilson KS, Dauter Z. Structures of the lectin IV of Griffonia simplicifolia and its complex with the Lewis B human blood group determinant at 2.0 Å resolution. J Mol Biol. 1993;230:950–965. doi: 10.1006/jmbi.1993.1212. [DOI] [PubMed] [Google Scholar]
- Emsley P, Charks IG, Fairweather NF, Isaacs NW. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- Engh RA, Huber R. Accurate bond and angle parameters for x-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- Fogarty W, Kelly C (eds) (1982) Microbial Enzymes and Biotechnology. Applied Science Publisher, London, pp 131–182
- Furey W, Swaminathan S (1997) PHASES-95: a program package for processing and analyzing of diffraction data from macromolecules. In CW Carter, Jr, RM Sweet, eds, Methods in Enzymology, Vol 277. Academic Press, New York, pp 590–620 [DOI] [PubMed]
- Hamlin R. Multiwire area x-ray diffractometers. Methods Enzymol. 1985;114:416–452. doi: 10.1016/0076-6879(85)14029-2. [DOI] [PubMed] [Google Scholar]
- Henrissat B, Heffron SE, Yoder MD, Lietzke SE, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard AJ, Nielson C, Xoung N-H. Software for a diffractometer with multiwire area detector. Methods Enzymol. 1985;114:452–472. doi: 10.1016/0076-6879(85)14030-9. [DOI] [PubMed] [Google Scholar]
- Hutchinson EG, Thornton JM. PROMOTIF: a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kester HCM, Visser J. Purification and characterization of pectin lyase B, a novel pectinolytic enzyme from Aspergillus niger. FEMS Microbiol. 1994;120:63–68. [Google Scholar]
- Kita N, Boyd CM, Garrett MR, Jurnak F, Keen NT. Construction of site-directed mutations in pelC and their differential effect on pectate lyase activity, plant tissue maceration and elicitior activity. J Biol Chem. 1996;43:26529–26535. doi: 10.1074/jbc.271.43.26529. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- Kusters-van Someren M, Flipphi M, de Graaff L, van den Broeck H, Kester HCM, Hinnen A, Visser J. Characterization of the Aspergillus niger pelB gene: structure and regulation of expression. Mol Gen Genet. 1992;234:113–120. doi: 10.1007/BF00272352. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Lietzke SE, Scavetta RD, Yoder MD, Jurnak F. The refined three-dimensional structure of pectate lyase E from Erwinia chrysanthemi. Plant Physiol. 1996;111:73–92. doi: 10.1104/pp.111.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Yoder MD, Keen NT, Jurnak F. The three dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant Physiol. 1994;106:849–862. doi: 10.1104/pp.106.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Mayans O, Scott M, Connerton I, Gravesen T, Benen J, Visser J, Pickersgill R, Jenkins J. Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergences in the substrate-binding clefts of pectin and pectate lyases. Structure. 1997;5:677–689. doi: 10.1016/s0969-2126(97)00222-0. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Kauppinen S, Larsen S. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed β helix. Structure. 1997;5:533–544. doi: 10.1016/s0969-2126(97)00209-8. [DOI] [PubMed] [Google Scholar]
- Pickersgill R, Jenkins J, Harris G, Nasser W, Robert-Baudouy J. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat Struct Biol. 1994;1:717–723. doi: 10.1038/nsb1094-717. [DOI] [PubMed] [Google Scholar]
- Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
- Steinbacher S, Seckler R, Miller S, Steipe B, Huber R, Reinemer P. Crystal structure of P22 tailspike-protein: interdigitated subunits in a thermostable trimer. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Eisenberg DE. Unbiased three dimensional refinement of heavy atom parameters by correlation of origin removed Patterson functions. Acta Crystallogr. 1983;A39:813–817. [Google Scholar]
- Wilson AJC. Determination of absolute from relative x-ray intensity data. Nature. 1942;150:90–112. [Google Scholar]
- Yoder MD, Jurnak F. The refined three-dimensional structure of pectate lyase C from Erwinia chrysanthemi at 2.2 Å resolution. Plant Physiol. 1995;107:349–364. doi: 10.1104/pp.107.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MD, Keen NT, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993a;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- Yoder MD, Lietzke SE, Jurnak F. Unusual structural features in the parallel β-helix in pectate lyases. Structure. 1993b;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]