ABSTRACT
Temperature is a critical and ubiquitous environmental signal that governs the development and virulence of diverse microbial species, including viruses, archaea, bacteria, fungi, and parasites. Microbial survival is contingent upon initiating appropriate responses to the cellular stress induced by severe environmental temperature change. In the case of microbial pathogens, development and virulence are often coupled to sensing host physiological temperatures. As such, microbes have developed diverse molecular strategies to sense fluctuations in temperature, and nearly all cellular molecules, including proteins, lipids, RNA, and DNA, can act as thermosensors that detect changes in environmental temperature and initiate relevant cellular responses. The myriad of molecular mechanisms by which microbes sense and respond to temperature reveals an elegant repertoire of strategies to orchestrate cellular signaling, developmental programs, and virulence with spatial and temporal environmental cues.
Introduction
All organisms must contend with environmental perturbations and fluctuations, including exposure to variations in temperature. The capacity to integrate temperature cues and respond appropriately is shared among organisms ranging from bacteria to mammals. For diverse microbial species, including viruses, archaea, bacteria, fungi, and parasites, temperature represents a critical environmental cue that can mediate changes in growth, development, and pathogenesis. These microbes may experience fluctuations in temperature in the form of seasonal changes in environmental temperature, rising global temperatures, interactions with diverse host species, including endothermic species, and upon febrile episodes encountered in the host in response to infection.
The impact of temperature on viral and bacterial pathogens is well established. It has been known for some time that temperature can profoundly influence the replication of viruses, such as the human influenza virus (1), and that viral growth properties differ significantly at different temperatures in diverse host cell types (2). Further, temperature is a critical environmental trigger for many bacterial species. For bacterial pathogens of mammals, including Shigella species, Yersinia species, and other pathogens, elevated temperature can signal successful infection of its host and leads to expression of bacterial virulence genes encoding type III secretion systems, adhesins, and other important virulence regulators (3). Conversely, for bacterial pathogens of plants, such as Agrobacterium tumefaciens, key virulence factors are often repressed in response to elevated temperature (4, 5), suggesting the importance of temperature in the precise coordination of bacterial virulence and pathogenesis.
Fungal pathogens are also profoundly affected by temperature, which can influence developmental programs as well as virulence. This is illustrated by the dimorphic fungal pathogens for which a temperature-induced morphological transition is required for virulence. These dimorphic fungi, including Blastomyces dermatitidis and Histoplasma capsulatum, grow as filamentous molds in the soil at ambient temperature and convert to pathogenic yeast after infectious spores are exposed to elevated temperature upon inhalation into the lungs of a mammalian host (6). In vitro, transitioning these fungi from ambient temperature to an elevated temperature of 37°C is sufficient to induce this morphogenetic switch (6). Temperature also influences developmental transitions in the leading human fungal pathogen, Candida albicans. Temperature controls the C. albicans morphogenetic transition between yeast and filamentous growth (7), phenotypic switching between the white and opaque cellular growth states (8, 9), and resistance to antifungal drugs (10).
Temperature also plays a key role in the virulence and development of diverse species of protozoan parasites. For instance, for the malarial parasite Plasmodium falciparum, the developmental transition from sporozoites, the parasite transmission stage, into early exoerythrocytic forms depends on temperature elevation to mammalian physiological temperature (11). Further, there is evidence to suggest that recurrent febrile episodes in malaria patients can accelerate the intraerythrocytic development of the parasite (12). Similarly, for the parasite Leishmania donovani, the differentiation from the promastigote to the amastigote life cycle stage correlates with the temperature upshift encountered upon transmission from insect to mammal (13, 14), and temperature also controls the differentiation from tachyzoites to bradyzoites in the parasite Toxoplasma gondii (15). Temperature may also influence parasite virulence, as a shift to mammalian physiological temperature contributes to the preservation of sporozoite infectivity of P. falciparum (16), highlighting the important role of temperature sensing for both parasite development and virulence.
As a consequence of the profound impact of temperature on key developmental, virulence, and survival traits in microbial species, an impressive repertoire of temperature-sensing mechanisms has emerged over evolutionary time. In this review, we discuss the diversity of mechanisms by which microbes sense changes in environmental temperature, highlighting findings from bacterial and fungal species, where these mechanisms have been most extensively studied. We explore each of the different biomolecules known to be involved in microbial thermosensing, including proteins, lipids, RNA, and DNA. We expand on the role of diverse classes of proteins, including transcription factors, kinases, two-component systems (TCSs), and chaperones in sensing environmental temperature, the function of lipids and membrane fluidity in response to temperature fluctuations, the role of RNA thermometers, and the importance of DNA structure and topology in mediating microbial thermosensing.
PROTEIN TEMPERATURE SENSORS
Several classes of proteins, including transcriptional regulators, kinases, and chaperones, have been described as temperature sensors (Fig. 1) (17). In numerous Gram-positive bacteria, including Bacillus subtilis, Lactococcus lactis, and others, the activity of the global transcriptional repressor protein CtsR is regulated by intrinsic heat-sensing capabilities, achieved via a glycine-rich loop, which functions as a protein thermometer (18). When this transcriptional regulator senses temperature stress, it dissociates from DNA associated with heat shock response elements, allowing expression of these transcripts to occur (18). A similar paradigm of temperature-dependent DNA-binding activity has been documented for several other thermosensitive bacterial transcriptional repressors, all of which dissociate from DNA in response to increased temperature (Fig. 1A). Some examples include the TlpA thermosensor in Salmonella (19), RovA in Yersinia (20), RheA in Streptomyces albus (21), and Phr in the archaeal species Pyrococcus furiosus (22). GmaR is a transcription factor and thermosensor protein that controls temperature-dependent transcription of motility genes in Listeria monocytogenes (23). Currently, GmaR is the only example of a bacterial transcription factor thermosensor that functions as an antirepressor of temperature-dependent transcription (23).
FIG 1 .
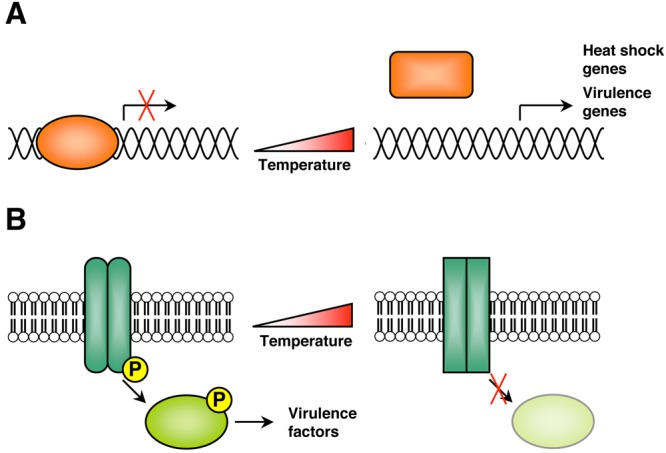
Examples of protein temperature sensors. (A) In several species of bacteria, transcriptional repressors are key temperature sensors. These repressors bind DNA associated with heat shock response genes, and upon temperature stress, changes in protein conformation or oligomerization status cause these transcriptional repressors to dissociate from DNA, allowing transcription to occur. Genes that are expressed upon transition to elevated temperature include heat shock genes and virulence genes. (B) In several species of pathogenic bacteria, the histidine kinases of two-component regulatory systems are involved in temperature sensing. Typically, the temperature-sensing kinase activates its downstream effector via autophosphorylation and effector phosphorylation, allowing the effector protein to activate different signaling pathways and virulence factors in response to temperature. Upon elevated temperature, changes in kinase protein conformation will abolish this signaling.
For bacteria, two-component regulatory systems, comprised of a membrane-associated sensor, typically a histidine kinase, and a cytoplasmic response regulator, are key sensors of environmental fluctuations and frequently influence the expression of virulence genes. The sensor kinases of TCSs are frequently found to have temperature-sensing capabilities and thus regulate bacterial virulence in response to temperature fluctuations (Fig. 1B). For instance, the plant pathogen A. tumefaciens is virulent only at temperatures below 32°C, and a temperature-sensitive TCS comprised of the sensor kinase VirA and the response regulator VirG modulates its virulence. At temperatures above 32°C, VirA undergoes inactivation, as autophosphorylation of VirA and phosphorylation of VirG are both abolished (5). A similar mechanism is observed with the CorS-CorR TCS in Pseudomonas syringae, where temperature-dependent conformational changes in the CorS sensor kinase lead to inactivation at elevated temperatures (24). For the animal pathogen Edwardsiella tarda, the PhoP-PhoQ TCS detects changes in temperature to activate the type III and VI secretion systems that are critical for virulence (25). The PhoQ sensor kinase detects temperature via a conformational change in its secondary structure, to activate protein secretion at optimal temperatures between 35°C and 37°C (25). This differential inactivation or activation of TCSs in bacterial species that are pathogens of plants versus pathogens of animals suggests a mechanism by which bacterial pathogens use temperature as a key environmental signal to govern virulence at the optimal temperature of their host. In Escherichia coli, additional membrane-associated receptors play a role in temperature sensing. The Tsr and Tar proteins, two transmembrane chemoreceptors, function as thermosensors via differential methylation and changes in protein conformation (26, 27).
Although TCSs are ubiquitous in prokaryotes, they are less frequently found in eukaryotic species. However, histidine kinases, which frequently represent the sensor component of TCSs, have been implicated in environmental sensing as well as developmental transitions in diverse fungi (28). Notably, a histidine kinase sensor regulates temperature sensing and development in two species of dimorphic fungal pathogens. These dimorphic fungi, B. dermatitidis and H. capsulatum, grow as filamentous molds at ambient temperature and convert to pathogenic yeast after exposure to elevated temperature upon inhalation into the lungs of a mammalian host (6). In B. dermatitidis and H. capsulatum, the histidine kinase Drk1 functions as an environmental sensor of temperature and controls morphogenesis, as well as adaptation to environmental stress within the mammalian host (29). Drk1 is further required for the expression of virulence genes as well as fungal pathogenicity in vivo, reinforcing the relationship between fungal thermotolerance and virulence (29).
Chaperone proteins can also act as temperature sensors to control thermal regulation in diverse microbial species. In Saccharomyces cerevisiae, the small heat shock protein Hsp26, which functions as part of an oligomer complex, directly senses temperature and shifts from a low- to a high-affinity chaperone state with increased temperature (30). Evidence suggests that the small heat shock protein Hsp21 may play a similar role in temperature sensing in the fungal pathogen C. albicans, though the precise mechanism involved remains to be elucidated (31). For the bacteria E. coli and Thermus thermophilus, the Hsp70 family chaperone DnaK, its cochaperone DnaJ, and the nucleotide exchange factor GrpE function as a critical thermosensor that detects temperature fluctuations via thermally controlled conformational changes in GrpE (32–35). The chaperone DegP (HtrA) also functions as a unique bacterial thermosensor in both E. coli and Thermotoga maritima, where the protein transitions between activities as a chaperone and a protease in a temperature-dependent manner (36, 37).
The environmentally responsive chaperone protein HSP90 has long been known to function as a temperature sensor in mammalian cells (38, 39), where global problems in protein folding at elevated temperature titrate HSP90 away from client proteins, such as the heat shock factor HSF1, due to the increase in denatured cellular targets requiring HSP90’s chaperone function (40). Although this mechanism of temperature sensing regulated by overwhelming HSP90 function has not been directly validated in microbial species, there is evidence to suggest that Hsp90 may play a similar role as a temperature sensor in the fungal pathogen C. albicans (7) and that Hsp90’s interaction with Hsf1 may be instrumental to regulating this process (41). Further, in protozoan parasites, including P. falciparum and L. donovani, evidence suggests that Hsp90 may act as a key thermosensor in mediating temperature-dependent developmental transitions (42).
LIPIDS AND MEMBRANE FLUIDITY AS TEMPERATURE SENSORS
For many microbes, the cellular membrane is among the first to sense fluctuations in environmental temperatures. Exposure to extreme temperatures, hot or cold, can dramatically alter membrane properties, allowing membranes themselves to act as a kind of thermosensor (43). Membrane fluidity, which decreases at lower temperature, is regulated in part by the ratio of unsaturated to saturated fatty acids present in membrane lipids (43). Changes in environmental temperature have been demonstrated to alter both the fatty acid composition and the degree of unsaturation of membrane lipids in diverse bacterial and archaeal species (44, 45, 46). For the bacterium B. subtilis, decreased membrane fluidity caused by lower temperature favors a kinase-dominant state of the TCS sensor kinase DesK, which is then able to activate the downstream response regulator DesR. DesR subsequently activates a transcriptional program that is able to restore bacterial membrane fluidity (47, 48). In Francisella bacteria, the acyltransferases LpxD1 and LpxD2 control temperature-dependent remodeling of membrane lipid A under different environmental temperature conditions. LpxD2 adds shorter acyl chains to lipid A, allowing growth in colder environments, whereas LpxD1 adds longer acyl chains upon transitioning to higher temperatures experienced in the mammalian host (49). The cyanobacterium Synechocystis responds to decreased temperature by upregulating expression of acyl-lipid desaturases, which increase the cis-unsaturation of membrane-bound lipid fatty acids, thus restoring membrane fluidity (50, 51).
RNA THERMOMETERS
Temperature-responsive RNAs, or RNA thermometers, are RNA control elements located in the 5′ untranslated region (UTR) of bacterial genes involved in virulence, heat shock, or cold shock (17, 52). The basic principle of RNA thermometers is that the Shine-Dalgarno (SD) sequence (located upstream of the AUG start codon) is base paired to form a hairpin structure with the AUG start codon in the 5′ UTR of an mRNA transcript at low temperature (Fig. 2). Increasing temperature destabilizes the structure, allowing the ribosome-binding site to become accessible and facilitating the initiation of translation (52). By this mechanism, temperature fluctuations can have an instantaneous effect on the expression of RNA thermometer transcripts involved in mediating heat shock response, cold shock response, or virulence, and gene expression correlates with the severity of the temperature change (Fig. 2) (53, 54).
FIG 2 .
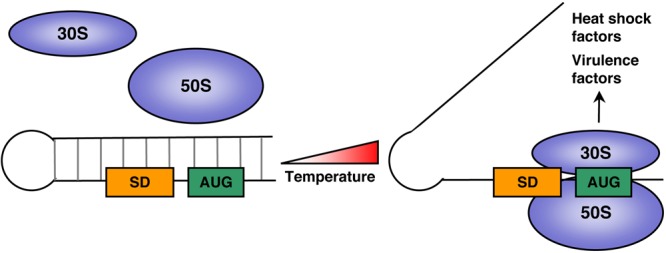
The basic principle of RNA thermometers. For temperature-responsive RNA thermometers, the Shine-Dalgarno (SD) sequence and the AUG start codon are base paired to form a hairpin structure in the 5′ UTR of an mRNA transcript at low temperature. Increasing temperature destabilizes the structure, allowing the ribosome-binding site to become accessible, allowing the 30S and 50S ribosomal subunits to bind, and facilitating the initiation of translation. Many transcripts encoding heat shock factors and virulence factors are regulated this way.
Many RNA thermometers govern the expression of heat shock genes in response to elevated temperature. The most abundant class of bacterial RNA thermometers is the repression of the heat shock gene expression (ROSE) family elements (55, 56), which are found in numerous alpha- and gamma-proteobacteria, including E. coli and Salmonella species (57). ROSE elements are 60 to 100 nucleotides long, located in the 5′ UTR of small heat shock genes, and regulate the translation of these heat shock transcripts in response to temperature change (52). Another class of RNA thermometers that controls the expression of heat shock genes is the fourU class of elements, containing a stretch of four uridines that pair with the SD sequence (58). One of the best examples of a fourU element is upstream of the agsA small heat shock gene in Salmonella enterica, which forms a translation initiation complex at high but not low temperatures (58). An RNA thermometer regulating expression of heat shock genes has also been identified in the cyanobacterium Synechocystis, which controls translation of the hsp17 heat shock gene in response to elevated temperature stress (59).
In response to low temperature, the expression of cold shock genes can also be controlled via RNA thermometers, although this phenomenon has not been explored as extensively as RNA thermometers controlling heat shock (60, 61). In E. coli, a sequence upstream of the cold shock protein CspA has been postulated to function as an RNA thermometer, governing translation efficiency of this transcript (62). The mRNA of cspA forms different structures at 37°C compared to cold shock temperatures, and the structure produced under cold shock conditions is translated more efficiently, suggesting a mechanism by which this bacterium responds to cold shock conditions (63). Other factors involved in bacterial response to cold shock are postulated to be regulated by RNA thermometers, including the cold shock factor CspE and the small RNA involved in low-temperature growth, DsrA (64–66), although the precise mechanisms remain to be elucidated.
RNA thermometers can also control the expression of virulence genes, when an increase in temperature to 37°C signals that the bacterium has successfully invaded its host. For the plague bacterium Yersinia pestis, LcrF is a crucial virulence factor that regulates the expression of adhesin proteins at 37°C but not at ambient temperature (67). The 5′ UTR of LcrF contains a fourU RNA thermometer, which enables translation of this virulence factor specifically at host temperature (68, 58). A similar fourU RNA thermometer controls key virulence factors in Yersinia pseudotuberculosis in response to host temperature (69), as Y. pseudotuberculosis expressing a stabilized RNA thermometer variant is avirulent in a murine infection model (69). In L. monocytogenes, an RNA thermometer controls translation of the virulence factor PrfA, which activates several virulence genes, including adhesins, phagosome escape factors, and immune-modulating factors (70). The 5′ UTR of the prfA transcript is folded such that the SD sequence is poorly accessible at 30°C but is destabilized at 37°C, enabling translation of this virulence factor in response to host temperature (70). An RNA thermometer-like mechanism also controls the life cycle of phage lambda, as temperature controls translation of the cIII mRNA, involved in the lysis-lysogeny decision (71). High concentrations of cIII protein at optimal growth temperatures (37°C) favor lysogeny, while under heat stress conditions (45°C), cIII is low and the lytic life cycle is favored (71). Regulation of this temperature-dependent transition is accomplished by alternative RNA structures in the 5′ UTR of cIII, which change in response to temperature fluctuations (71). Thus, temperature-sensitive RNA thermometers can have a profound impact on virulence and development in response to temperature fluctuations.
DNA STRUCTURE AND TOPOLOGY AS A THERMOSENSOR
Conditions that influence the integrity and topology of DNA, including changes in temperature, ultimately impact on gene expression (Fig. 3). In this way, DNA can act as a thermosensor of environmental temperature change. DNA supercoiling of bacterial and archaeal plasmids is one of the primary parameters of DNA topology that is affected by temperature, as has been documented for numerous species that grow at moderate temperatures (mesophiles) or at high temperatures (hyperthermophiles) (Fig. 3A) (72). In these species, changes in DNA supercoiling can function as a sensor of temperature stress under both heat shock and cold shock conditions, and DNA transcription efficiency is highly sensitive to changes in DNA supercoiling (72). For mesophiles, such as bacterial species E. coli and Salmonella, DNA is negatively supercoiled, and heat stress induces a transient increase in positive supercoiling, leading to plasmid relaxation, while cold shock leads to a transient decrease in supercoiling (Fig. 3A) (73, 74). For hyperthermophiles, such as the archaeal species of Sulfolobus and Thermococcus, DNA is positively supercoiled, and heat stress induces increased positive supercoiling, while colder temperatures induce negative supercoiling (72, 75). Transcription efficiency is extremely sensitive to changes in DNA supercoiling, and temperature changes can profoundly influence gene expression, including the expression of key virulence determinants in these bacterial and archaeal species (Fig. 3A) (76, 77).
FIG 3 .
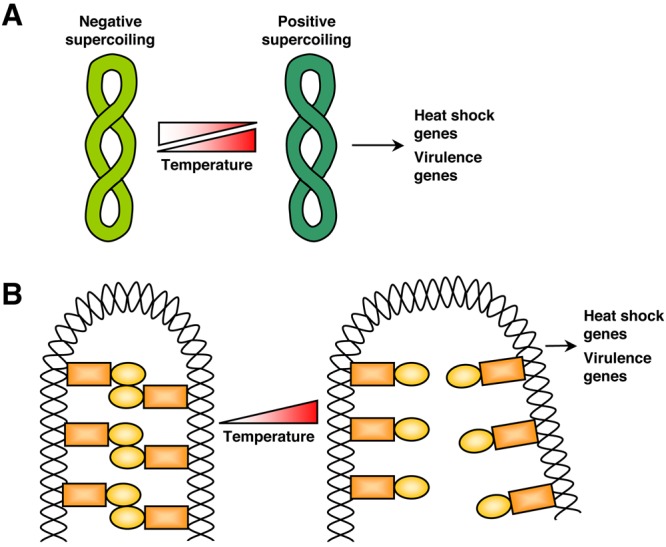
DNA structure and topology as a thermosensor. (A) For mesophilic bacterial species, plasmid DNA is negatively supercoiled and heat stress induces a transient increase in positive supercoiling, leading to plasmid relaxation, while cold shock leads to a transient decrease in supercoiling. Transcription efficiency is sensitive to changes in DNA supercoiling, and temperature changes can influence gene expression in these species, including the expression of virulence determinants and heat shock factors. (B) The histone-like nucleoid structuring protein (H-NS) binds DNA and represses transcription in numerous bacterial species. H-NS regulation of DNA is sensitive to fluctuations in temperature, and the formation of higher-order oligomers and overall DNA binding capacity are reduced in response to elevated host temperature. Expression of many virulence genes and heat shock genes are regulated in response to host temperature in this manner.
Another mechanism by which DNA topology is influenced by temperature is by local DNA structures. The promoters of bacterial species such as E. coli contain intrinsically curved DNA regions due to AT-rich sequences (78, 79). Temperature fluctuations induce topological changes in these regions, affecting RNA polymerase binding and gene expression. In the cyanobacterium Synechocystis, DNA curvature found in certain promoter regions is influenced by temperature changes and regulates the expression of genes important for membrane fluidity (80). DNA curvature also influences the expression of phospholipase genes in response to temperature fluctuations in the bacterium Clostridium perfringens (81), suggesting a mechanism by which these species rapidly adapt to environmental temperature perturbations.
Finally, DNA-mediated temperature sensing can be achieved through the binding of silencer proteins, such as the histone-like nucleoid structuring protein (H-NS), which binds DNA and represses transcription in numerous bacterial species. H-NS regulation of DNA is exquisitely sensitive to fluctuations in temperature, as the formation of higher-order oligomers and overall DNA binding capacity are reduced in response to sensing host temperature (37°C) (Fig. 3B) (82). This temperature-dependent accessibility of promoter regions occupied by H-NS is a highly conserved mechanism of ensuring that virulence genes are expressed in response to host temperature. For example, in S. enterica, expression of a type III secretion system that is critical for virulence is repressed by H-NS at temperatures below 30°C (83). Similarly, in Shigella flexneri, the virF gene, encoding an invasion regulator, is expressed in a host temperature-dependent manner, controlled by the binding of H-NS (84). Overall, approximately 69% and 77% of temperature-regulated genes are controlled by H-NS in E. coli and Salmonella, respectively (82, 85), demonstrating the fundamental importance of H-NS in coordinating gene expression with environmental temperature.
CONCLUSION
Temperature is a universal environmental stimulus that influences all microbes, and fluctuations in temperature may be experienced by environmental change or upon infection of a host. Accordingly, these changes in temperature may activate heat shock or cold shock stress response pathways to cope with severe changes in environmental temperature or may inform microbial pathogens of successful host infection and initiate virulence programs. As demonstrated here, diverse and disparate microbes, including bacteria, archaea, and fungi, have all evolved molecular strategies to sense temperature and activate appropriate response pathways. Many of the canonical mechanisms of temperature sensing have been elucidated in bacterial species, and future work may inform how these mechanisms, or perhaps novel mechanisms of thermosensing, operate in microbes for which temperature is an important environmental cue, including fungi and parasites. Temperature-dependent control of microbial development, virulence, and survival may have even broader implications for the origin of mammalian endothermy, which may have evolved to optimally restrict pathogens such as fungi, many of which lose growth capacity above ambient temperatures (86, 87). The stunning complexity of molecular mechanisms used to sense and control temperature fluctuations reflects on the pervasive impact of temperature as one of the most powerful selective forces in nature.
ACKNOWLEDGMENTS
We thank Cowen lab members for helpful discussions.
R.S.S. is supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) CGS-D award, and L.E.C. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, by a Canada Research Chair in Microbial Genomics and Infectious Disease, by a Ministry of Research and Innovation Early Researcher Award, by NSERC Discovery grant number 355965, and by Canadian Institutes of Health Research (CIHR) grants MOP-86452 and MOP-119520.
Footnotes
Citation Shapiro RS, Cowen LE. 2012. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. mBio 3(5):e00238-12. doi:10.1128/mBio.00238-12.
REFERENCES
- 1. Scholtissek C, Rott R. 1969. Effect of temperature on the multiplication of an influenza virus. J. Gen. Virol. 5:283–290 [DOI] [PubMed] [Google Scholar]
- 2. Hatta M, et al. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 3:1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Konkel ME, Tilly K. 2000. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. 2:157–166 [DOI] [PubMed] [Google Scholar]
- 4. Banta LM, Bohne J, Lovejoy SD, Dostal K. 1998. Stability of the Agrobacterium tumefaciens VirB10 protein is modulated by growth temperature and periplasmic osmoadaption. J. Bacteriol. 180:6597–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin S, Song YN, Deng WY, Gordon MP, Nester EW. 1993. The regulatory VirA protein of Agrobacterium tumefaciens does not function at elevated temperatures. J. Bacteriol. 175:6830–6835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein BS, Tebbets B. 2007. Dimorphism and virulence in fungi. Curr. Opin. Microbiol. 10:314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shapiro RS, et al. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soll DR. 2004. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. Bioessays 26:10–20 [DOI] [PubMed] [Google Scholar]
- 9. Srikantha T, Soll DR. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53–60 [DOI] [PubMed] [Google Scholar]
- 10. Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189 [DOI] [PubMed] [Google Scholar]
- 11. Kaiser K, Camargo N, Kappe SH. 2003. Transformation of sporozoites into early exoerythrocytic malaria parasites does not require host cells. J. Exp. Med. 197:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavithra SR, Banumathy G, Joy O, Singh V, Tatu U. 2004. Recurrent fever promotes Plasmodium falciparum development in human erythrocytes. J. Biol. Chem. 279:46692–46699 [DOI] [PubMed] [Google Scholar]
- 13. Saar Y, et al. 1998. Characterization of developmentally regulated activities in axenic amastigotes of Leishmania donovani. Mol. Biochem. Parasitol. 95:9–20 [DOI] [PubMed] [Google Scholar]
- 14. Wiesgigl M, Clos J. 2001. Heat shock protein 90 homeostasis controls stage differentiation in Leishmania donovani. Mol. Biol. Cell 12:3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soête M, Camus D, Dubremetz JF. 1994. Experimental induction of bradyzoite-specific antigen expression and cyst formation by the RH strain of Toxoplasma gondii in vitro. Exp. Parasitol. 78:361–370 [DOI] [PubMed] [Google Scholar]
- 16. Siau A, et al. 2008. Temperature shift and host cell contact up-regulate sporozoite expression of Plasmodium falciparum genes involved in hepatocyte infection. PLoS Pathog. 4:e1000121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klinkert B, Narberhaus F. 2009. Microbial thermosensors. Cell. Mol. Life Sci. 66:2661–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elsholz AK, Michalik S, Zühlke D, Hecker M, Gerth U. 2010. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 29:3621–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hurme R, Berndt KD, Normark SJ, Rhen M. 1997. A proteinaceous gene regulatory thermometer in Salmonella. Cell 90:55–64 [DOI] [PubMed] [Google Scholar]
- 20. Herbst K, et al. 2009. Intrinsic thermal sensing controls proteolysis of Yersinia virulence regulator RovA. PLoS Pathog. 5:e1000435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Servant P, Grandvalet C, Mazodier P. 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc. Natl. Acad. Sci. U. S. A. 97:3538–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu W, Vierke G, Wenke AK, Thomm M, Ladenstein R. 2007. Crystal structure of the archaeal heat shock regulator from Pyrococcus furiosus: a molecular chimera representing eukaryal and bacterial features. J. Mol. Biol. 369:474–488 [DOI] [PubMed] [Google Scholar]
- 23. Kamp HD, Higgins DE. 2011. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 7:e1002153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Braun Y, Smirnova AV, Weingart H, Schenk A, Ullrich MS. 2007. A temperature-sensing histidine kinase: function, genetics, and membrane topology. Methods Enzymol. 423:222–249 [DOI] [PubMed] [Google Scholar]
- 25. Chakraborty S, et al. 2010. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J. Biol. Chem. 285:38876–38888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee L, Mizuno T, Imae Y. 1988. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J. Bacteriol. 170:4769–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizuno T, Imae Y. 1984. Conditional inversion of the thermoresponse in Escherichia coli. J. Bacteriol. 159:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santos JL, Shiozaki K. 2001. Fungal histidine kinases. Sci. STKE 2001:re1. [DOI] [PubMed] [Google Scholar]
- 29. Nemecek JC, Wüthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588 [DOI] [PubMed] [Google Scholar]
- 30. Franzmann TM, Menhorn P, Walter S, Buchner J. 2008. Activation of the chaperone Hsp26 is controlled by the rearrangement of its thermosensor domain. Mol. Cell 29:207–216 [DOI] [PubMed] [Google Scholar]
- 31. Mayer FL, et al. 2012. Small but crucial: the novel small heat shock protein Hsp21 mediates stress adaptation and virulence in Candida albicans. PLoS One 7:e38584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Groemping Y, et al. 2001. Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE. J. Mol. Biol. 305:1173–1183 [DOI] [PubMed] [Google Scholar]
- 33. Groemping Y, Reinstein J. 2001. Folding properties of the nucleotide exchange factor GrpE from Thermus thermophilus: GrpE is a thermosensor that mediates heat shock response. J. Mol. Biol. 314:167–178 [DOI] [PubMed] [Google Scholar]
- 34. McCarty JS, Walker GC. 1991. DnaK as a thermometer: threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc. Natl. Acad. Sci. U. S. A. 88:9513–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siegenthaler RK, Christen P. 2006. Tuning of DnaK chaperone action by nonnative protein sensor DnaJ and thermosensor GrpE. J. Biol. Chem. 281:34448–34456 [DOI] [PubMed] [Google Scholar]
- 36. Kim DY, Kwon E, Shin YK, Kweon DH, Kim KK. 2008. The mechanism of temperature-induced bacterial HtrA activation. J. Mol. Biol. 377:410–420 [DOI] [PubMed] [Google Scholar]
- 37. Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455–459 [DOI] [PubMed] [Google Scholar]
- 38. Nair SC, et al. 1996. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor HSF1, and the aryl hydrocarbon receptor. Cell Stress Chaperones 1:237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. 1998. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94:471–480 [DOI] [PubMed] [Google Scholar]
- 40. Sangster TA, Lindquist S, Queitsch C. 2004. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays 26:348–362 [DOI] [PubMed] [Google Scholar]
- 41. Leach MD, Tyc KM, Brown AJ, Klipp E. 2012. Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS One 7:e32467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Acharya P, Kumar R, Tatu U. 2007. Chaperoning a cellular upheaval in malaria: heat shock proteins in Plasmodium falciparum. Mol. Biochem. Parasitol. 153:85–94 [DOI] [PubMed] [Google Scholar]
- 43. Digel I. 2011. Primary thermosensory events in cells. Adv. Exp. Med. Biol. 704:451–468 [DOI] [PubMed] [Google Scholar]
- 44. Matsuno Y, et al. 2009. Effect of growth temperature and growth phase on the lipid composition of the archaeal membrane from Thermococcus kodakaraensis. Biosci. Biotechnol. Biochem. 73:104–108 [DOI] [PubMed] [Google Scholar]
- 45. Mykytczuk NC, Trevors JT, Twine SM, Ferroni GD, Leduc LG. 2010. Membrane fluidity and fatty acid comparisons in psychrotrophic and mesophilic strains of Acidithiobacillus ferrooxidans under cold growth temperatures. Arch. Microbiol. 192:1005–1018 [DOI] [PubMed] [Google Scholar]
- 46. Paulucci NS, Medeot DB, Dardanelli MS, de Lema MG. 2011. Growth temperature and salinity impact fatty acid composition and degree of unsaturation in peanut-nodulating rhizobia. Lipids 46:435–441 [DOI] [PubMed] [Google Scholar]
- 47. Albanesi D, Mansilla MC, de Mendoza D. 2004. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J. Bacteriol. 186:2655–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Albanesi D, et al. 2009. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 106:16185–16190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li Y, et al. 2012. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:8716–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mikami K, Kanesaki Y, Suzuki I, Murata N. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp PCC 6803. Mol. Microbiol. 46:905–915 [DOI] [PubMed] [Google Scholar]
- 51. Suzuki I, Los DA, Murata N. 2000. Perception and transduction of low-temperature signals to induce desaturation of fatty acids. Biochem. Soc. Trans. 28:628–630 [PubMed] [Google Scholar]
- 52. Kortmann J, Narberhaus F. 2012. Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10:255–265 [DOI] [PubMed] [Google Scholar]
- 53. Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. 2010. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 38:3834–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H. 2011. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 39:8258–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Narberhaus F, Käser R, Nocker A, Hennecke H. 1998. A novel DNA element that controls bacterial heat shock gene expression. Mol. Microbiol. 28:315–323 [DOI] [PubMed] [Google Scholar]
- 56. Nocker A, Krstulovic NP, Perret X, Narberhaus F. 2001. ROSE elements occur in disparate rhizobia and are functionally interchangeable between species. Arch. Microbiol. 176:44–51 [DOI] [PubMed] [Google Scholar]
- 57. Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. 2005. RNA thermometers are common in alpha- and gamma-proteobacteria. Biol. Chem. 386:1279–1286 [DOI] [PubMed] [Google Scholar]
- 58. Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. 2007. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65:413–424 [DOI] [PubMed] [Google Scholar]
- 59. Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. 2011. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 39:2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Phadtare S. 2011. Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 8:394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Phadtare S, Severinov K. 2010. RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 7:788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamanaka K, Mitta M, Inouye M. 1999. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181:6284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Giuliodori AM, et al. 2010. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 37:21–33 [DOI] [PubMed] [Google Scholar]
- 64. Lybecker MC, Samuels DS. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64:1075–1089 [DOI] [PubMed] [Google Scholar]
- 65. Repoila F, Gottesman S. 2003. Temperature sensing by the dsrA promoter. J. Bacteriol. 185:6609–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Uppal S, Akkipeddi VS, Jawali N. 2008. Posttranscriptional regulation of cspE in Escherichia coli: involvement of the short 5′-untranslated region. FEMS Microbiol. Lett. 279:83–91 [DOI] [PubMed] [Google Scholar]
- 67. Skurnik M, Toivanen P. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 174:2047–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoe NP, Goguen JD. 1993. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J. Bacteriol. 175:7901–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Böhme K, et al. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 8:e1002518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Johansson J, et al. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551–561 [DOI] [PubMed] [Google Scholar]
- 71. Altuvia S, Kornitzer D, Teff D, Oppenheim AB. 1989. Alternative mRNA structures the cIII gene of bacteriophage lambda determine the rate of its translation initiation. J. Mol. Biol. 210:265–280 [DOI] [PubMed] [Google Scholar]
- 72. López-García P, Forterre P. 2000. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays 22:738–746 [DOI] [PubMed] [Google Scholar]
- 73. Kataoka K, Mizushima T, Ogata Y, Miki T, Sekimizu K. 1996. Heat shock-induced DNA relaxation in vitro by DNA gyrase of Escherichia coli in the presence of ATP. J. Biol. Chem. 271:24806–24810 [DOI] [PubMed] [Google Scholar]
- 74. Mizushima T, Kataoka K, Ogata Y, Inoue R, Sekimizu K. 1997. Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol. Microbiol. 23:381–386 [DOI] [PubMed] [Google Scholar]
- 75. López-García P, Forterre P. 1997. DNA topology in hyperthermophilic archaea: reference states and their variation with growth phase, growth temperature, and temperature stresses. Mol. Microbiol. 23:1267–1279 [DOI] [PubMed] [Google Scholar]
- 76. Dorman CJ, Corcoran CP. 2009. Bacterial DNA topology and infectious disease. Nucleic Acids Res. 37:672–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dorman CJ, Ní Bhriain N. 1993. DNA topology and bacterial virulence gene regulation. Trends Microbiol. 1:92–99 [DOI] [PubMed] [Google Scholar]
- 78. Mizuno T. 1987. Random cloning of bent DNA segments from Escherichia coli chromosome and primary characterization of their structures. Nucleic Acids Res. 15:6827–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nickerson CA, Achberger EC. 1995. Role of curved DNA in binding of Escherichia coli RNA polymerase to promoters. J. Bacteriol. 177:5756–5761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Los DA. 2004. The effect of low-temperature-induced DNA supercoiling on the expression of the desaturase genes in Synechocystis. Cell. Mol. Biol. 50:605–612 [PubMed] [Google Scholar]
- 81. Katayama S, Matsushita O, Jung CM, Minami J, Okabe A. 1999. Promoter upstream bent DNA activates the transcription of the Clostridium perfringens phospholipase C gene in a low temperature-dependent manner. EMBO J. 18:3442–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ono S, et al. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Duong N, et al. 2007. Thermosensing coordinates a cis-regulatory module for transcriptional activation of the intracellular virulence system in Salmonella enterica serovar Typhimurium. J. Biol. Chem. 282:34077–34084 [DOI] [PubMed] [Google Scholar]
- 84. Prosseda G, et al. 2004. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51:523–537 [DOI] [PubMed] [Google Scholar]
- 85. White-Ziegler CA, Davis TR. 2009. Genome-wide identification of H-NS-controlled, temperature-regulated genes in Escherichia coli K-12. J. Bacteriol. 191:1106–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bergman A, Casadevall A. 2010. Mammalian endothermy optimally restricts fungi and metabolic costs. mBio 1(5):e00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Robert VA, Casadevall A. 2009. Vertebrate endothermy restricts most fungi as potential pathogens. J. Infect. Dis. 200:1623–1626 [DOI] [PubMed] [Google Scholar]


