ABSTRACT
The distribution, genome location, and evolution of the four paralogous zinc metalloproteases, IgA1 protease, ZmpB, ZmpC, and ZmpD, in Streptococcus pneumoniae and related commensal species were studied by in silico analysis of whole genomes and by activity screening of 154 representatives of 20 species. ZmpB was ubiquitous in the Mitis and Salivarius groups of the genus Streptococcus and in the genera Gemella and Granulicatella, with the exception of a fragmented gene in Streptococcus thermophilus, the only species with a nonhuman habitat. IgA1 protease activity was observed in all members of S. pneumoniae, S. pseudopneumoniae, S. oralis, S. sanguinis, and Gemella haemolysans, was variably present in S. mitis and S. infantis, and absent in S. gordonii, S. parasanguinis, S. cristatus, S. oligofermentans, S. australis, S. peroris, and S. suis. Phylogenetic analysis of 297 zmp sequences and representative housekeeping genes provided evidence for an unprecedented selection for genetic diversification of the iga, zmpB, and zmpD genes in S. pneumoniae and evidence of very frequent intraspecies transfer of entire genes and combination of genes. Presumably due to their adaptation to a commensal lifestyle, largely unaffected by adaptive mucosal immune factors, the corresponding genes in commensal streptococci have remained conserved. The widespread distribution and significant sequence diversity indicate an ancient origin of the zinc metalloproteases predating the emergence of the humanoid species. zmpB, which appears to be the ancestral gene, subsequently duplicated and successfully diversified into distinct functions, is likely to serve an important but yet unknown housekeeping function associated with the human host.
IMPORTANCE
The paralogous zinc metalloproteases IgA1 protease, ZmpB, ZmpC, and ZmpD have been identified as crucial for virulence of the human pathogen Streptococcus pneumoniae. This study maps the presence of the corresponding genes and enzyme activities in S. pneumoniae and in related commensal species of the genera Streptococcus, Gemella, and Granulicatella. The distribution, genome location, and sequence diversification indicate that zmpB is the ancestral gene predating the evolution of today’s humanoid species. The ZmpB protease may play an important but yet unidentified role in the association of streptococci of the Mitis and Salivarius groups with their human host, as it is ubiquitous in these two groups, except for a fragmented gene in Streptococcus thermophilus, the only species not associated with humans. The relative sequence diversification of the IgA1 protease, ZmpB, and ZmpD is striking evidence of differences in selection for diversification of these surface-exposed proteins in the pathogen S. pneumoniae compared to the closely related commensal streptococci.
Introduction
Streptococcus pneumoniae is a leading cause of invasive diseases and respiratory tract infections and one of the most frequent microbial killers worldwide. It is a member of the Mitis group of Streptococcus which currently encompasses 14 species. Apart from S. pneumoniae, all these closely related species (Streptococcus pseudopneumoniae, Streptococcus mitis, Streptococcus oralis, Streptococcus infantis, Streptococcus oligofermentans, Streptococcus sanguinus, Streptococcus gordonii, Streptococcus australis, Streptococcus parasanguinis, Streptococcus cristatus, Streptococcus sinensis, Streptococcus peroris, and Streptococcus lactarius) are commensal colonizers of the upper respiratory tract.
One characteristic feature of S. pneumoniae is the presence of large zinc metalloproteases on the cell surface. The published genome sequences of S. pneumoniae revealed the existence of up to four homologous zinc metalloproteases, the IgA1 protease, ZmpB, ZmpC, and ZmpD (1–3), which all contain the conserved HEXXH…E motif characteristic for zinc metalloproteases (4, 5). Genes encoding the pneumococcal zinc metalloproteases are located at different sites of the chromosome, except for ZmpD, which is next to the IgA1 protease locus (6). Different domains important for enzyme localization have been revealed at the N-terminal region of the IgA1 and ZmpB proteases, so far the only well-characterized proteins of the four proteases. Such domains include a potential signal peptide and a cell wall anchor motif (LPXTG), followed by two hydrophobic domains with the potential of spanning the cytoplasmic membrane and a second putative signal peptidase site (7, 8). In addition, repeat regions that vary in number and sequence have been identified downstream of the anchor motif and may serve immune escape purposes (9–11). A sequence with homology to a G5 domain is located within the repeat region of the IgA1 protease (8). The G5 domain has been suggested to bind N-acetylglucosamine, which is part of the cell wall component peptidoglycan (12), but the exact function of this domain in the IgA1 protease is still unknown. The atypical location of the LPXTG anchor motif, which normally is found at the C terminus in Gram-positive bacterial surface proteins (13), is required for proper enzyme localization of the IgA1 protease at the cell wall (7).
IgA1 protease activity is a characteristic of all three principal causes of bacterial meningitis, S. pneumoniae, Haemophilus influenzae, and Neisseria meningitidis (14–16). The IgA1 proteases share the unique ability to cleave human immunoglobulin A1, the primary mediator of specific humoral immunity of the upper respiratory tract, in the extended and heavily glycosylated hinge region but do so by different catalytic mechanisms (17). Comparison of gene sequences and enzymatic properties have shown that the IgA1 protease activity evolved along at least five independent evolutionary lineages in bacteria (17). Cleavage of IgA1 separates Fc-mediated secondary effector mechanisms from the antigen-binding part (Fab) of the antibody (18) and results in Fab-mediated promotion of adherence (19, 20). Among the pneumococcal zinc metalloproteases, only the IgA1 protease has the ability to cleave IgA1. However, yet unidentified substrates and functions are implied for the pneumococcal IgA1 protease, as it contributes to virulence in mice, although it has no activity on murine IgA (21, 22). So far, no alternative substrates have been identified, which is in contrast to the finding of alternative substrates for the serine IgA1 proteases of Neisseria gonorrhoeae and N. menigitidis that are able to cleave lysosome-associated membrane protein (LAMP-1) (23), the tumor necrosis factor alpha receptor II (TNF-α-RII) (24), gonadotropin (25), and vesicle-associated membrane protein 2 (Vamp-2) (26). Pneumococcal ZmpC has been shown to activate human matrix metalloprotease 9 (MMP-9), an enzyme involved in cell migration by remodeling of the extracellular matrix and in opening of the blood-brain barrier during inflammation (6). In addition, ZmpC causes shedding of syndecan-1, a general host response to tissue injury and inflammation and a broadly used pathogenic strategy to enhance microbial virulence (27). Recently, ZmpC was shown to induce ectodomain shedding of mucin 16 (MUC16), a crucial defense component of the epithelial microcolony-associated meshwork (MAM) glycocalyx barrier (28). The function and substrate specificity of ZmpB and ZmpD remain unknown, although ZmpB has been shown to contribute to inflammation by increasing the proinflammatory cytokine TNF-α in the lower respiratory tract (29). However, the mechanism by which ZmpB alters the levels of TNF-α remains unknown.
The existence of the four zinc metalloproteases is assumed to be a result of gene duplications, considered a major source of new protein functions in bacteria. However, limited knowledge exists regarding the evolution and exact functions of these proteases, although all four zinc metalloproteases previously have been detected in large-scale identification studies of pneumococcal virulence factors (22, 30). In addition, all proteases were shown to be crucial in pneumococcal pathogenicity in an experimental mouse model, although the main impact on virulence was assigned to the IgA1 protease and ZmpB (21).
The distribution of the individual zinc metalloproteases has primarily been investigated in the Mitis group of Streptococcus. IgA1 protease activity has been used as a character of taxonomic significance for classification of viridians streptococci (31), but reclassifications and introduction of several new species render the overall picture blurred. IgA1 protease activity is characteristic of S. pneumoniae (32). Accordingly, the iga gene (zmpA) encoding the IgA1 protease has been identified in all pneumococcal strains examined so far, together with the zmpB gene encoding ZmpB (33). In contrast, ZmpC and ZmpD have been found to be variably present among pneumococci. The presence of IgA1 protease activity varies among the closely related commensal species S. mitis, S. oralis, S. infantis, S. sanguinis, and S. gordonii (34). More recently, a homologous IgA1 protease in Gemella haemolysans was reported (8), and a putative IgA1 protease has been identified and characterized in the major swine pathogen and emerging zoonotic agent, Streptococcus suis, suggested to contribute to the pathogenesis of this species (35, 36).
In the present study, we examined the phylogenetic relationship and distribution of the paralogous genes iga, zmpB, zmpC, and zmpD among bacterial species in an attempt to better understand the origin and evolution of the zinc metalloproteases. Such analyses may contribute to an improved understanding of the functions played by the proteases and their role in pathogenesis.
RESULTS
To ensure correct species assignment of strains and genomes from which zmp sequences were retrieved, all genomes were subjected to multilocus sequence analysis (MLSA) using the seven housekeeping loci previously used for online identification of viridans streptococci (37). According to our results, some of the genomes were mislabeled; for example, strains ATCC 6249 and ATCC 49296 were assigned to Streptococcus mitis and S. sanguinis, respectively, in the ATCC catalogue and in the NCBI database. Our findings show that ATCC 6249 is not an S. mitis strain but is closer to the S. oralis/S. oligofermentans cluster and that ATCC 49296 is an S. oralis strain (Fig. 1). Throughout this paper, these strains will be referred as S. oralis strains. Most genomes of strains published as unidentified Streptococcus strains could be unequivocally assigned to recognized species (Fig. 1). On the basis of the MLSA identification results, the extracted sequences represented strains of S. pneumoniae (n = 67, including seven single gene sequences), S. pseudopneumoniae (n = 1), S. mitis (n = 10, including one single gene sequence), “S. mitis biovar 2” (n = 3), S. oralis (n = 7, including two single gene sequences), S. oligofermentans (n = 1), S. infantis (n = 3), S. sanguinis (n = 27, including seven single gene sequences), S. gordonii (n = 2), S. salivarius (n = 3), S. parasanguinis (n = 4), S. vestibularis (n = 2), S. australis (n = 1), S. cristatus (n = 1), S. suis (n = 7), unclassified Streptococcus sp. (n = 2), Gemella haemolysans (n = 3, including one single gene sequence), Gemella sanguinis (n = 1), Gemella morbillorum (n = 1), Granulicatella adiacens (n = 1), and Granulicatella elegans (n = 1) (see Table S1 in the supplemental material).
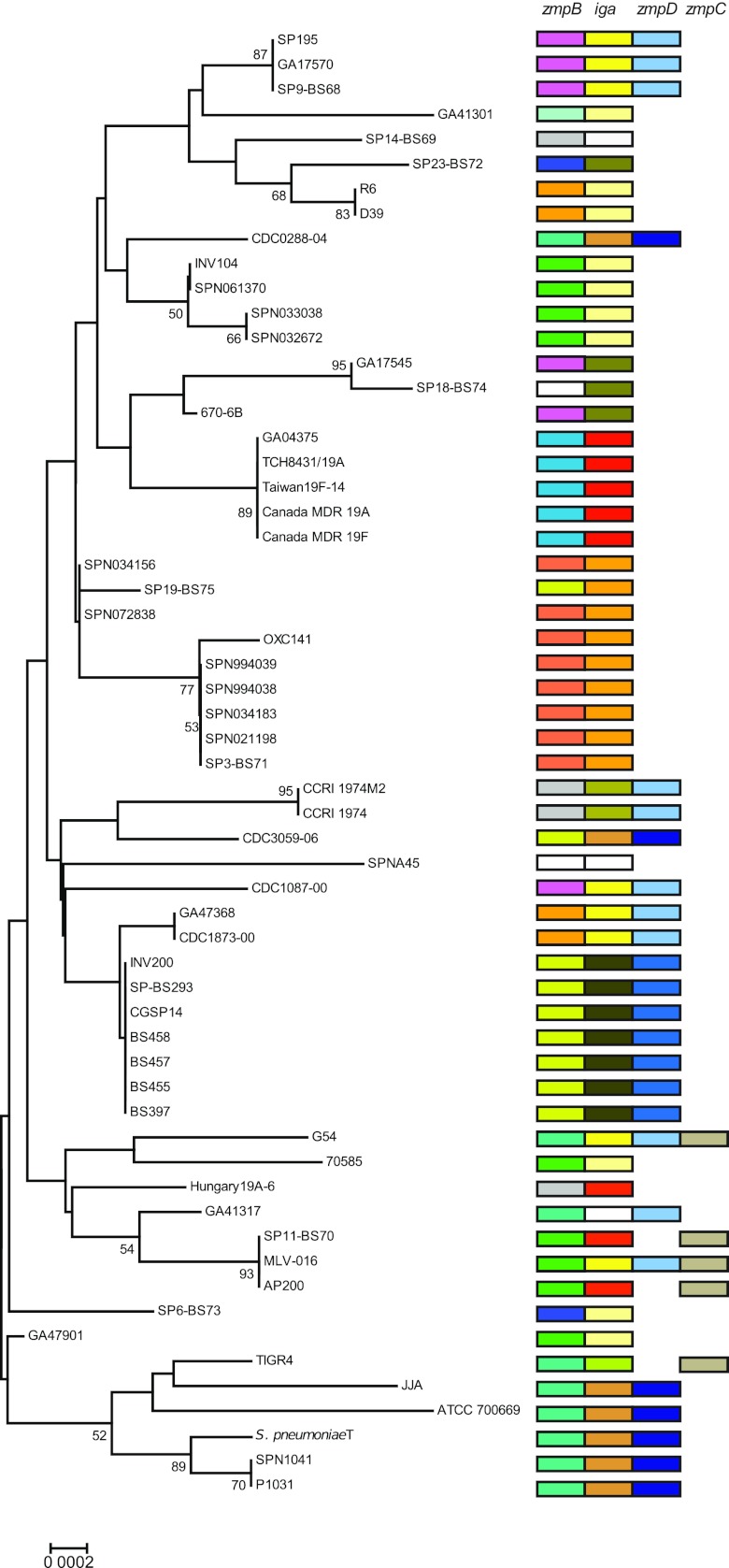
FIG 1 .
Phylogenetic tree constructed with the minimum evolution algorithm and based on partial sequences of the housekeeping genes map, pfl, ppaC, pyk, rpoB, sodA, and tuf. Type strains of individual species are shown with the species name. The presence of Zmp genes iga (A), zmpB (B), zmpC (C), and zmpD (D) is indicated by the letter. Red uppercase letters indicate the presence of the Zmp genes, and black lowercase letters indicate the absence of the Zmp genes. Bootstrap values (shown as percentages) at the nodes of the tree are based on 500 replications. str., strain. The bar indicates the genetic distance (number of substitutions per nucleotide).
Nomenclature.
The nomenclature of the zinc metalloproteases used in the present study is that of Oggioni et al. (6) with reference to genes cloned and characterized in previous studies: IgA1 protease (4, 10, 11, 38), ZmpB (9), and ZmpC (6). The fourth zinc metalloprotease gene was identified next to the iga locus of the published genome of S. pneumoniae G54 (1) and was recently named zinc metalloprotease D (ZmpD) by Camilli et al. (33). Initially, all zmp genes were annotated as IgA1 protease genes in the first genome sequence of S. pneumoniae TIGR4 (3) and in several subsequently released pneumococcal genomes. Accordingly, our analysis of zinc metalloprotease genes retrieved from genome sequences at the NCBI website revealed that many are mislabeled.
Distribution and phylogenetic analysis of the zinc metalloprotease genes.
Putative zinc metalloprotease sequences corresponding to a product of >500 amino acids, including the C terminus, which comprises the proteolytic domain, were retrieved by BLAST searches using representatives of each gene as the query sequence and used for phylogenetic analyses. In total, 297 sequences representing 154 strains and 20 different bacterial species were used for further analyses. Among the draft genome sequences, a few of the zmp sequences were truncated in the N terminus. This was caused by either a split of the sequence into two contigs or to differences in the number of nucleotides in A or T polymeric tracts, which presumably represent sequencing errors. The analyses showed that the overall structure of the four Zmp proteins (Zmps) is very similar (Fig. 2). The IgA1 protease, ZmpB, and ZmpC all have a signal peptide predicted by SignalP in their N termini. For all the IgA1 proteases and some of the ZmpBs and ZmpCs, this is of the YSIRK type characteristic for streptococcal and staphylococcal proteins, containing a Gram-positive anchor domain with the LPXTG sortase processing site. The ZmpDs have a transmembrane helix in the N terminus predicted by TMpred. However, this was not recognized by SignalP and may represent an atypical signal peptide. Following this, all the Zmps have a copy of the LPXTG motif. In all the IgA1 proteases, except for one, the X is an Asn, whereas it varies among the other proteases. The N-terminal location is atypical among anchor motifs, which usually are located in the C termini of cell wall proteins. After the anchor motif, two transmembrane segments were predicted by TMpred. The latter of these may function as a second “signal peptide” cleaved by the signal peptidase in accordance with the proposed secretion model (7, 8). Immediately downstream of the signal peptide region, repeat regions followed in all Zmps. Many of the repeats were imperfect and varied in sequence, length, and number in different Zmps and strains, comprising from 131 (ZmpD in S. pneumoniae strain BS458) to 1,561 amino acids (ZmpC in S. sanguinis SK160) located between the more-conserved regions. In the C-terminal part, which comprises the proteolytic domain, all the Zmps harbored a motif characteristic of zinc metalloproteases (Fig. 2). Because of the extreme diversity in the repeat region, the N-terminal region was excluded from the multiple amino acid sequence alignment, which resulted in sequences of approximately 1,200 amino acids.
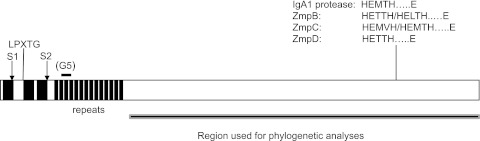
FIG 2 .
Schematic structure of the zinc metalloproteases. The three black boxes at the left of the structure indicate hydrophobic regions proposed to serve as transmembrane domains in the signal peptide and in combination with the cell wall anchor motif LPXTG. S1 and S2 indicate sites presumably cleaved by the signal peptidase. The G5 domain conserved in the IgA1 proteases and the region used for phylogenetic analyses are indicated. The sequence of the active site is shown for the individual Zmp proteins.
Cluster analyses were based on nucleotide sequence alignments of the respective zmp genes. All putative zinc metalloprotease sequences were annotated according to cluster analysis. The phylogenetic reconstruction of the zinc metalloprotease genes revealed three distinct clusters, supported by significant bootstrap values (97 to 100%) (Fig. 3). Two clusters included the reference of iga and zmpC, respectively, whereas the third cluster included both zmpB and zmpD references. The iga and zmpC genes were closer to each other than they were to the zmpB and zmpD genes (see Table S2 in the supplemental material). These relationships were also reflected in the active site sequences (Fig. 2). Sequences of zmpD, which clustered with zmpB, were primarily recognized as a separate cluster because of their unique genome location (see below). Thus, sequences tentatively annotated as zmpB clustered into multiple subgroups, which show no more homology to each other than to the cluster of sequences annotated as zmpD (Table S3).
FIG 3 .
Phylogenetic tree based on zmp gene sequences as described in the text and using the minimum evolution algorithm. Bootstrap values, based on 500 replicates, are shown for major evolutionary lineages. The species are indicated by colored dots.
The iga gene was exclusively found in species of the Mitis group of Streptococcus and in the available genome sequences of G. haemolysans. In accordance with previous studies, the iga gene was found in all available genome sequences of S. pneumoniae and S. sanguinis and was variably present in genomes of strains of S. mitis, whereas it was absent in S. gordonii genomes (Fig. 1) (31). We previously reported that IgA1 protease activity is a variable characteristic in S. oralis (39). However, MLSA analysis of the strains included in this study (Fig. 1) showed that the cluster of IgA1 protease-negative strains represent the recently described species S. oligofermentans (40). Thus, all strains of S. oralis examined had IgA1 protease activity, and all genome sequences of this species contained the iga gene. The genome sequence of S. pseudopneumoniae contained the iga gene, and all four S. pseudopneumoniae strains tested, including the type strain, had IgA1 protease activity. IgA1 protease activity was variable among S. infantis strains but absent from all strains of S. parasanguinis, S. australis, S. peroris, S. cristatus, and the two Salivarius group species S. vestibularis and S. salivarius. In addition to species of the genus Streptococcus, all available genome sequences of G. haemolysans (ATCC 10379, CCUG 4815, and M341) contained an iga gene in accordance with our previous observations (8).
Substantial diversity was observed within the iga cluster (mean distance, 0.342 ± 0.007) (see Fig. S1 in the supplemental material), which consisted of numerous subgroups (Fig. 3). Most of these subgroups were related to species, indicating limited interspecies recombination. However, significant diversity (mean distance, 0.351 ± 0.007) was observed among the iga sequences of pneumococcal strains, which were separated into two major subclusters, one of which contained strains that possessed ZmpD. This suggests the existence of distinct subpopulations of pneumococci. The iga genes of S. mitis were distributed all over the iga cluster (mean distance, 0.399 ± 0.008), reflecting the significant genetic diversity and population structure of this species (39). In contrast, iga genes of S. oralis and especially of S. sanguinis were nearly identical (mean distances of 0.070 ± 0.003 and 0.062 ± 0.002, respectively). Interestingly, the newly reported iga gene of S. suis did not cluster with iga genes of the other streptococci but formed a distinct cluster which, based on our phylogenetic analysis, is closer to the zmpC cluster (Fig. 3).
Almost all of the species included in this study harbored zmpB with the exception of strains of S. cristatus and S. suis. In addition, only two out of five complete genomes of S. oralis contained a gene that was part of the zmpB cluster (Fig. 1 and 3). The putative zmpB genes exhibited the most significant diversity of any of the zmp genes (mean distance, 0.555 ± 0.011) (see Fig. S1 in the supplemental material) and consisted of multiple subgroups mostly related to species. Like iga genes, zmpB genes of S. pneumoniae formed multiple subgroups (mean distance, 0.545 ± 0.009) each consisting of nearly identical zmpB alleles (see below). zmpB genes of S. sanguinis were nearly identical (mean distance, 0.051 ± 0.002) and formed a separate cluster together with zmpB genes from S. gordonii in accordance with the close genetic relationship between these species. In contrast, a significant diversity was observed among zmpB genes of S. mitis (mean distance, 0.359 ± 0.006). Although most of them are closely related, some S. mitis strains (SK321, M334, SK1080, SK597, and SK564) contained zmpB genes more related to zmpB from S. oralis and S. pneumoniae. The S. mitis strains SK597 and SK564 contained two genes that fall within the zmpB cluster. One of these genes is closely related to zmpB genes of other S. mitis strains, whereas the other showed closer relationship to the zmpB genes of pneumococci, although they represent independent lineages. Interestingly, we previously concluded that strain SK597 is a chimera of S. mitis and S. oralis and observed that strain SK564 possesses several genes usually considered characteristic of S. pneumoniae such as lytA, ply, and a complete cap locus (39). The observed relationships of zmpB of “S. mitis biovar 2” strains support the notion that this taxon is distinct and closer to S. oralis than to S. mitis (37, 39) (Fig. 1). In general, sequence relationships of zmp were in accordance with relationships revealed by MLSA (Fig. 1). Interestingly, three zmpB fragments with significant homology to the corresponding gene in S. salivarius and S. vestibularis were present in genomes of the nonhuman member of the Salivarius group, S. thermophilus. Notably, a close relationship was observed neither between zmpB genes of the different Gemella species nor between the two strains of Gemella haemolysans, suggesting that these genes were acquired by independent horizontal transfer events. The putative zmpB gene observed in Granulicatella elegans showed homology to pneumococcal zmpB genes, whereas the zmpB genes from Granulicatella adiacens represent a distinct lineage in the zmpB cluster.
ZmpC was identified in several species of the Mitis group of Streptococcus, although variable within most species except for S. sanguinis, S. gordonii, G. haemolysans, and S. suis, which were all positive (Fig. 1 and see Fig. S1 in the supplemental material). Extensive diversity was observed within the zmpC cluster (mean diversity, 0.408 ± 0.008) (Fig. S1). However, zmpC genes of S. sanguinis were very conserved (mean distance, 0.058 ± 0.003) and formed a distinct cluster together with zmpC genes from strains of the closely related species S. gordonii. Surprisingly, three out of the five complete genomes of S. oralis contained two genes that fell within the zmpC gene cluster. Some of these genes formed a distinct cluster, consisting only of putative zmpC genes from strains of S. oralis. The zmpC genes found in a limited number of S. pneumoniae strains were conserved and formed a separate cluster, whereas zmpC genes of the closely related S. mitis strains were distributed all over the zmpC cluster (mean distance, 0.426 ± 0.011). The single putative zmpC gene from G. haemolysans was not closely related to any of the other zmpC genes. A putative zmpC gene in one of the other G. haemolysans genomes was divided into several contigs and was excluded from the analysis. The zmpC gene of S. cristatus constituted a separate lineage, most closely related to zmpC genes of S. gordonii and S. sanguinis.
The more recently discovered zmpD gene was recognized only in genomes of S. pneumoniae and S. mitis and was found to be variably present in both species (Fig. 1). Limited diversity was observed in the zmpD cluster compared to the other zmp clusters (mean diversity, 0.295 ± 0.006) (see Fig. S1 in the supplemental material). zmpD genes of S. pneumoniae formed three subgroups each consisting of nearly identical alleles (see below). The zmpD genes of S. mitis all represented divergent lineages within the zmpD cluster.
To test whether exclusion of the N-terminal sequence biased the conclusions, we performed separate cluster analysis of the region corresponding to amino acids 1 to 153 in the IgA1 protease of S. pneumoniae TIGR4 (Fig. 2). The pattern of clustering (not shown) was identical to that shown in Fig. 3 except for the N-terminal sequence of the iga gene of G. haemolysans, which formed a distinct subgroup, suggesting a distinct or more ancient origin. Notably, the N-terminal sequence of ZmpC in G. haemolysans ATCC 10379 was most closely related to the N-terminal sequences of the zmpB genes in G. haemolysans, suggesting homologous recombination between different zmp genes in closely related strains. In the zmpD genes, the N-terminal sequence formed a distinct cluster supported by a high bootstrap value (100%), thus supporting the separate annotation of ZmpD.
Comparison of the sequence diversity of Zmps.
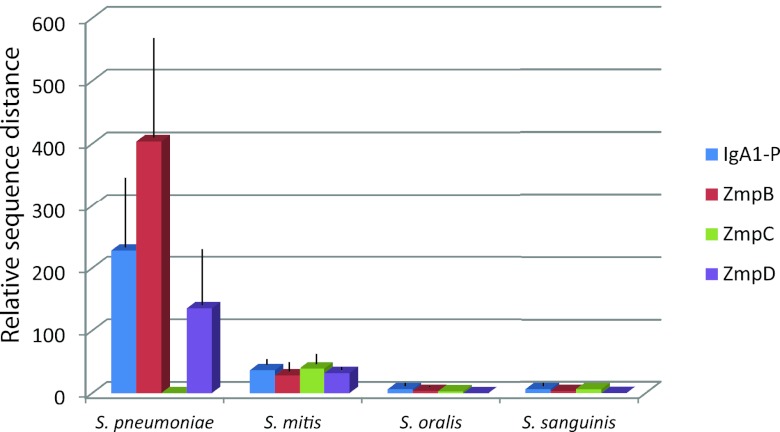
To estimate the selection for diversity of the individual Zmp proteins in S. pneumoniae, S. mitis, S. oralis, and S. sanguinis, we calculated the within-species mean distances of amino acid sequences of each of the Zmp proteins relative to the corresponding distances of amino acid sequences of translated concatamers of the seven genes used for the MLSA analysis. Identical sequences in otherwise genetically distant strains resulting from recent homologous transfers among S. pneumoniae strains (see below) were excluded from these analyses. The results, shown in Fig. 4, revealed significant differences between the species. The genetic diversity of Zmps of S. sanguinis and S. oralis were close to that seen for the seven housekeeping genes used for MLSA. Among S. mitis strains, the amino acid sequence distances were 28 to 36 times larger than those of the corresponding housekeeping proteins, but these values were not statistically significantly different from the values for S. oralis and S. sanguinis. However, Zmps of S. pneumoniae showed remarkably higher relative sequence diversity than that observed for the three commensal species (P < 0.0001). The only exception was the virtually identical ZmpC alleles present in the few pneumococcal strains that carried the corresponding gene.
FIG 4 .
Mean amino acid sequence diversities (with standard deviations [error bars]) of zinc metalloproteases relative to those of translated concatenated sequences of housekeeping genes used for MLSA (multilocus sequence analysis) in S. pneumoniae, S. mitis, S. oralis, and S. sanguinis. With the exception of ZmpC, the relative diversities of proteases of S. pneumoniae were significantly higher than those of the commensal Streptococcus species (P < 0.0001). IgA1-P, IgA1 protease.
Genomic localization of the zinc metalloproteases.
Comparative analyses of the zinc metalloprotease gene regions in selected genomes revealed a highly conserved gene order within species and among closely related species. Synteny was observed between the iga loci of S. pneumoniae, S. pseudopneumoniae, and S. mitis with the exceptions that several S. mitis strains lacked iga and that some iga-positive S. mitis strains and some pneumococci had the additional zmpD gene adjacent to the iga gene (Fig. 5). However, in the genomes of S. mitis SK137 and SK321, which both lack the iga gene, an inserted region of approximately 15 kb was identified. This region has significant homology to a genome region in the species Granulicatella elegans and conceivably represents a recombination event between these species. The GC content of 29.7% of this inserted region differs significantly from the 40% G+C of the S. mitis genome. The closer similarity to the GC content of G. elegans (33%) suggests that this region was transferred from G. elegans to S. mitis. Interestingly, S. mitis strain B6 lacks the iga gene but contains a gene encoding a pathogenicity protein at the position equivalent to the IgA1 protease gene (41). These observations suggest that the iga region is a target for recombination events. Interestingly, the iga locus of the closely related S. oralis (see below), S. sanguinis, and G. haemolysans showed no synteny with that of the above-described species. In agreement with previous observations (34), the genome of S. gordonii did not encode an IgA1 protease gene, although the region was otherwise syntenic to that of S. sanguinis in agreement with the close phylogenetic relationship between these two species.
FIG 5 .
Map of genome regions harboring the different zinc metalloproteases. (A) iga; (B) zmpB; (C) zmpC and the multiple zmp region revealed in strains of S. oralis. The map shows partially synteny between closely related species. Zmp genes are highlighted with red/brown colors. Genes not showing homology to other regions at the amino acid level are indicated by a dark gray color.
In general, synteny was observed between the zmpB loci of the closely related species S. pneumoniae, S. pseudopneumoniae, S. mitis, and S. oralis, whereas limited synteny was revealed between these species and S. infantis (Fig. 5). Synteny was also observed between the zmpB gene regions of S. sanguinis and S. gordonii, although not complete, but in analogy with the iga regions, no synteny was observed between the zmpB regions of S. sanguinis/S. gordonii and the zmpB regions of S. pneumoniae/S. pseudopneumoniae/S. mitis/S. oralis. The close phylogenetic relationship of S. parasanguinis and S. australis (Fig. 1) was reflected in complete synteny between their zmpB regions, which were distinct from all the other investigated species. Interestingly, S. parasanguinis contains a region with complete homology to the zmpB region of S. sanguinis and S. gordonii, but without zmpB, suggesting gene loss or genome rearrangements in S. parasanguinis. Although S. vestibularis and S. salivarius are closely related, no synteny was seen between the zmpB loci of the two species. In S. thermophilus, the mentioned zmpB fragments were located equivalent to zmpB of S. vestibularis. No synteny was observed between the zmpB loci of G. elegans and G. adiacens or any of the Streptococcus species. Only limited synteny was observed between the zmpB loci of the two examined genomes of G. haemolysans, which may suggest genome reorganization or acquisition of zmpB by separate events within strains of the same species. In fact, the G. haemolysans strains showed a higher degree of homology to either G. sanguinis or G. moribillorum than to each other. To confirm the identity of the Gemella strains, their relationships to designated type strains of the recognized species was investigated by MLSA. These analyses confirmed the species assignments of the genomes. However, as some of the investigated genomes are draft genomes, the possibility that the observed lack of synteny is due to incorrect assembly cannot be excluded.
Partial synteny was observed between the zmpC region of S. pneumoniae and some strains of S. mitis (Fig. 5). However, S. mitis SK569 contains a zmpC locus with no synteny to any other zmpC locus observed in the present study. Interestingly, S. oralis harbors a region with multiple zmp genes containing the IgA1 protease gene, the zmpC gene, and an additional zinc metalloprotease gene forming a distinct subgroup within the zmpC cluster (tentatively termed zmpC2) (see Fig. S1 in the supplemental material). Insertion sequences were observed near the additional zmpC gene, which may explain the variable presence of this gene. Strain 73H25AP was assigned to the species S. oligofermentans, a close relative of S. oralis, and did not contain the region with multiple zmp genes but contained genes with homology to flanking genes of this region. Partial synteny was observed between the zmpC loci of S. sanguinis and S. gordonii. No synteny was observed between the zmpC loci of G. haemolysans and S. suis or any of the other zmpC loci.
Evidence of homologous recombination within species.
The separate phylogenetic analyses of each of the zmp genes revealed species-specific subgroups in each of the zmp clusters (see Fig. S1 in the supplemental material). Comparison with the overall genetic relatedness of the strains, as revealed by MLSA of housekeeping genes (Fig. 1), demonstrated clear evidence of recent and frequent recombinational events within S. pneumoniae (Fig. 6). Thus, virtually identical alleles of the IgA1 protease gene often combined with zmpD, and zmpB genes were identified in genetically distinct S. pneumoniae strains. One striking example is strain MLV-016 which, as expected, shares a zmpB allele with the genetically closely related strains SP11-BS70 and AP200. However, MLV-016 harbors distinct iga and zmpD alleles, which are identical to the genes in the strains in the same group of the tree, SP195, GA17570, and SP9-BS68, and to the genetically distinct strains CDC1087-00, GA47368/CDC1873-00, and G54. Likewise, the genetically distant strains JJA, CDC3059-06, and CDC0288-04 share the entire iga-zmpD locus. This pattern shows that the iga gene and the flanking zmpD gene, spanning approximately 11 kb, are transferred together. The observed pattern of zmpB alleles also includes several examples of recent homologous recombination, e.g., the identical allele in the genetically distinct strains SP195/GA17570/SP9-BS68, GA17545, 670-6B, and CDC1087-00, and in the strains INV104/SPN061370/SPN033038/SPN032672, 70585, SP11-BS70/MLV-016/AP200, and GA47901.
FIG 6 .
Phylogenetic tree representing the genetic relatedness of S. pneumoniae strains demonstrating evidence of homologous recombination. The tree was constructed with the minimum evolution algorithm with partial sequences of the housekeeping genes map, pfl, ppaC, pyk, rpoB, sodA, and tuf. Different colors indicate different Zmp alleles.
In sharp contrast to the observations for S. pneumoniae genes, the observed species-specific subgroups leave no evidence of recent interspecies recombination between zmp genes presumably because the requirements for regions with high sequence homology are no longer satisfied. However, the homology of one of the two clusters of zmpB genes in pneumococci to zmpB genes from S. sanguinis conceivably reflects ancient recombination between these two species. Likewise, the iga sequences from G. haemolysans constituted distinct lineages within the cluster of iga genes of pneumococcal strains positive for the ZmpD protease, in support of our previous suggestion that the iga gene of G. haemolysans originated by horizontal transfer from the genus Streptococcus (8). In agreement with previous observations (9), the amino acid sequences of the N-terminal region of ZmpB in S. pneumoniae TIGR4 and CGSP14, which represent the two different pneumococcal clusters, were nearly identical. However, a significant drop in similarity was thereafter observed and the C-terminal region of CGSP14 showed more homology to zmpB from S. sanguinis SK36 (58%) than to TIGR4 (41%). In addition, the active site motif of S. pneumoniae CGSP14 is identical to the motif in S. sanguinis SK36 (HELTH) but distinct from that present in S. pneumoniae TIGR4 (HETTH).
ZmpC protease activity.
Distribution of ZmpC has so far only been studied within strains of S. pneumoniae (33). However, in the present study, we identified the presence of potential zmpC genes in other bacterial species, including several Streptococcus spp. and in Gemella haemolysans. To extend our knowledge of the distribution of zmpC, we screened a selection of well-characterized strains for ZmpC protease activity (see Table S4 in the supplemental material) by use of a previously described method (6). In brief, we assayed the specific cleavage of the human gelatinase MMP-9, the only recognized substrate for ZmpC, by using gelatinase zymogram gels. As references, S. pneumoniae TIGR4 mutants for iga (Δiga), zmpC (ΔzmpC), and zmpB (ΔzmpB) were included in the zymogram assay. To assay for MMP-9 cleavage, whole-cell cultures and supernatants were incubated with MMP-9 prior to zymogram analysis. Cleavage of MMP-9 was evidenced by a supplementary band of gelatinolytic activity (see representative examples in Fig. 7). Three out of four clinical pneumococcal strains were positive for cleavage of MMP-9, and two out of 12 tested S. mitis strains cleaved MMP-9, indicating the presence of ZmpC. However, only for S. pneumoniae and S. mitis strains did the activity detected by the zymogram assay correlate with the presence of a gene in the zmpC cluster. None of the 7 S. sanguinis strains tested or the 10 S. oralis strains tested showed the ability to cleave MMP-9 in the assay, although the respective genomes included a putative zmpC gene. Interestingly, the zmpC genes of S. sanguinis and S. oralis and the zmpC gene of S. mitis SK569, also negative for MMP-9 cleavage, share the active site motif HEMTH and fall within a cluster distinct from the zmpC genes of S. mitis and S. pneumoniae with the active site motif HEMVH and the ability to cleave MMP-9 (see Fig. S1 in the supplemental material). In addition, the zmpC gene of G. haemolysans ATCC 10379, which was unable to cleave MMP-9 in the assay, falls within the cluster of S. pneumoniae and S. mitis zmpC genes but share the active site motif HEMTH with zmpC from S. sanguninis, S. oralis, and S. mitis SK569. This finding may indicate substrate differences in the putative ZmpC proteases or may indicate that MMP-9 is not the principal substrate of ZmpC and thus, is cleaved only by a subfraction of the ZmpC proteases. Furthermore, it indicates that differences in the active site motif may result in different substrate specificities.
FIG 7 .
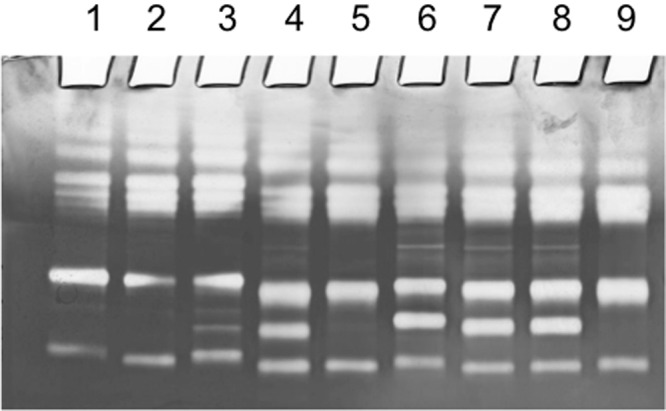
ZmpC protease activity assay. Zymogram gel showing gelatin cleavage by MMP-9 (matrix metalloprotease 9) and the proteolytic effect of ZmpC. MMP-9 was preincubated with different bacteria prior to electrophoresis. Lane 1, control (no bacteria); lane 2, S. mitis SK597 (negative for ZmpC); lane 3, S. mitis SK142 (positive for ZmpC); lane 4, S. pneumoniae 276/95 (positive for ZmpC); lane 5, S. pneumoniae TIGR4 ΔzmpC mutant; lane 6, S. pneumoniae TIGR4 ΔzmpB mutant; lane 7, S. pneumoniae TIGR4 Δiga mutant; lane 8, S. pneumoniae TIGR4 (positive for ZmpC); lane 9, S. pneumoniae R6 (negative for ZmpC).
IgA1 protease activity of Streptococcus suis.
IgA1 protease activity in S. suis was recently reported (36). This was unexpected, because swine are the primary habitat of S. suis and the fact that streptococcal IgA1 proteases do not cleave porcine IgA. Our phylogenetic analysis showed that the gene encoding the S. suis protease forms a distinct cluster, which is closely related to zmpC genes from other streptococci. We compared the human IgA1 cleaving activity of S. suis proteases with the IgA1 protease from S. pneumoniae TIGR4. Three S. suis strains, strains CCUG7986, CCUG7984, and CCUG42755 obtained from the Culture Collection, University of Göteborg, were examined. Although all were positive for the putative iga gene by PCR as previously described (36), we observed no evidence of IgA1 cleavage after overnight incubation with harvested bacteria or culture supernatants even when extreme bacterium-to-substrate ratios were used (not shown). S. pneumoniae TIGR4 was used as a positive control, and a TIGR4 mutant for iga (Δiga) was used as a negative control.
DISCUSSION
The existence of four paralogs of the zinc metalloprotease (zmp) family of genes that may be present in S. pneumoniae and other members of the Mitis group of streptococcal bacteria suggest that these genes have been evolutionarily successful. One of the genes (iga or zmpA) encodes the IgA1 protease. Interestingly, the genetically distinct but functionally identical serine IgA1 protease of pathogenic Neisseria species and Haemophilus influenzae are also encoded by genes that are part of a family of paralogous genes that have evolved into different functions (42). To obtain information on origin and significance, we studied the phylogenetic relationship and distribution of the streptococcal zinc metalloproteases, the IgA1 protease, ZmpB, ZmpC, and ZmpD. Our cluster analysis, which included sequences of all homologous zinc metalloproteases available in public databases as of October 2011, clearly demonstrated three distinct clusters corresponding to the three proteases observed in S. pneumoniae TIGR4 (IgA1 protease, ZmpB, and ZmpC) (Fig. 3). In contrast, the zmpD gene of S. pneumoniae G54 and its closest homologs were closely related to the zmpB sequences. The zmpD sequences, primarily identified by their position in the genome next to the iga gene (21), formed a separate subgroup within the zmpB cluster, which is no more genetically divergent from any zmpB subgroup than zmpB subgroups are from each other. However, our analyses show that the separate annotation of ZmpD suggested by Camilli et al. (33) is further supported by a unique structure of the N-terminal region of the protease. Originally, ZmpD was suggested to represent a fourth class of enzymes due to its low level of homology to the other proteases (6). Unfortunately, substrate specificities of ZmpD and ZmpB have not yet been defined. Identification of their functions will also elucidate whether the presence of two homologs of ZmpC in some strains of S. oralis functionally compensates for the lack of ZmpB homologs (Fig. 1).
In agreement with previous results, the iga gene was found in members of the Mitis group of streptococci and in genomes of G. haemolysans (8, 31, 39). The improved identification of Streptococcus species by MLSA analysis (37) applied in this study provides new details about the distribution of IgA1 proteases. Thus, the activity is present in all strains of S. pneumoniae, S. pseudopneumoniae, S. sanguinis, and S. oralis, is variably present in S. mitis and S. infantis, and absent in S. gordonii, S. oligofermentans, S. parasanguinis, S. australis, and S. cristatus (Fig. 1). All these species are part of the Mitis group of streptococci. Recently, the existence of an IgA1 protease in S. suis capable of cleaving human IgA1 was reported (36). In accordance with the clustering of the described IgA1 protease gene of S. suis with zmpC genes, we were unable to confirm IgA1 protease activity even at extreme enzyme-substrate ratios. S. suis is a major swine pathogen, and although it causes zoonotic infections in humans, it is more likely that the zinc metalloprotease of S. suis, conserved in all complete genome sequences of this species, has other functions adapted to its natural porcine habitat.
Previously, the distribution of ZmpB, ZmpC, and ZmpD was described only among pneumococci (33). Our study supports the former findings in pneumococci and reveals the presence of these zinc metalloproteases in several other Streptococcus species, in three Gemella species, G. haemolysans, G. moribillorum, and G. sanguinis, and in two Granulicatella species, G. elegans and G. adiacens, previously known as nutritionally deficient streptococci. With the exception of S. thermophilus (see below) and the exception of some strains of S. oralis, zmpB is present in all examined Streptococcus species of the Mitis and Salivarius groups and in Gemella and Granulicatella species. These results support the importance of these zinc metalloproteases, and the ubiquitous presence of ZmpB suggests that this enzyme plays a crucial, but yet undefined, role in the physiology of these streptococci. The finding that zmpB was present as three fragments in all available genomes of S. thermophilus, in contrast to the very closely related S. salivarius and S. vestibularis, may reflect the possibility that the associated function is irrelevant in the nonhuman habitat of S. thermophilus.
The variable presence of IgA1 protease activity in some commensal species was previously explained by loss of the function as a result of adaptation to a commensal lifestyle and the ensuing subtle relationship with the adaptive immune system of the host in contrast to that of the pathogenic pneumococci (39, 42). The observation that none of the iga genes found in strains of S. mitis shows significant homology with pneumococcal iga genes but reflects the overall sequence divergence of S. mitis supports the hypothesis that the variable presence in S. mitis is a result of loss rather than acquisition by some strains. Conversely, the occurrence of the zmpC gene in a limited number of pneumococcal strains but in virtually identical alleles suggests a recent origin of zmpC in pneumococci, most likely due to independent gene acquisition of zmpC by individual strains from a common donor, which may be a strain of S. pneumoniae.
The demonstrated ability of ZmpC to cleave MMP-9 (6) may constitute an advantage in invasion of various tissues, but the results of our ZmpC protease assay suggest that there are other natural substrates for this enzyme. Thus, MMP-9 was cleaved by ZmpC-positive S. mitis and S. pneumoniae strains, whereas strains of other species carrying a zmpC homolog all failed to cleave MMP-9 in the assay.
The present phylogenetic analyses show significant diversity within each of the protease clusters of species-specific subgroups. However, in S. pneumoniae, virtually identical alleles of iga, zmpB, zmpC, and zmpD genes were observed in several otherwise distinct phylogenetic lineages, as revealed by comparison with the genetic relationships based on housekeeping gene sequences (Fig. 6). This is evidence of a remarkably high frequency of horizontal transfer of zmp genes and combinations of genes (iga and zmpD) in today’s population of pneumococci.
Previously, mosaic structures were identified in IgA1 protease genes of pneumococci and related streptococci indicating that homologous recombination has played a significant role in the genetic diversification of these proteases (10, 20, 43). Interestingly, the pneumococcal iga genes were divided into two distinct subclusters, one representing strains that harbored the additional zmpD gene located just downstream of iga and the other representing strains that lacked zmpD. This might be explained by the existence of distinct evolutionary clades of S. pneumoniae that are unable to exchange genetic material due to a yet unknown restriction mechanism against homologous recombination. However, this explanation is contradicted by the MLSA-based observation that strains harboring zmpD were no more closely related to each other than to strains lacking zmpD (Fig. 6). A more likely explanation is horizontal transfer of the entire iga locus, including zmpD spanning approximately 11 kb among part of the pneumococcus population. The virtually identical alleles in genetically distinct strains indicate that this is still a frequently occurring event similar to capsular type switches, which often involve transfer of the complete capsule locus (44, 45) spanning between 10 and 30 kb (46). The demonstrated lack of recent recombination between species, as indicated by the species-specific subgroups of the individual zinc metalloprotease genes (see Fig. S1 in the supplemental material), is likely to reflect the extent of sequence divergence, which conceivably has become a barrier to efficient homologous recombination (47). Two notable exceptions demonstrating more ancient recombination events are the zmpB alleles of pneumococci, which form two major clusters, one of which shows more homology to zmpB genes from S. sanguinis, and the homology between the iga gene of S. pneumoniae and G. haemolysans.
The three zinc metalloproteases IgA1 protease, ZmpB, and ZmpC showed a remarkably different degree of amino acid sequence diversity relative to that of the housekeeping proteins encoded by genes used in the MLSA. While genes in S. sanguinis and S. oralis have remained relatively conserved, the Zmps of S. pneumoniae clearly have been exposed to an intense diversifying selection pressure, not working on Zmps of S. sanguinis and S. oralis (Fig. 4). It is conceivable that this reflects differences in the relationship of the species to the human host. While the pathogen S. pneumoniae induces a strong antibody response both in the mucosal and systemic compartments of the immune system to surface-exposed components of the bacterium, the immune system is largely nonresponsive to commensal bacteria (48). The notable exception from this pattern is S. mitis, which showed a somewhat higher relative diversity (8 to 11 times) than S. sanguinis and S. oralis though significantly lower than that of S. pneumoniae (P < 0.0001). We hypothesize that this increased diversity of zmp genes in the commensal S. mitis is the result of a selection pressure exerted by antibodies induced by S. pneumoniae proteases which, at least for the IgA protease, are known to cross-react with those of S. mitis (49).
Apart from the diversification resulting from a substantial selection pressure, the significant accumulation of nucleotide substitutions observed within each of the proteases indicate an ancient origin of all zinc metalloprotease genes. This is supported by the primary function of the IgA1 protease. Conceivably, the IgA1 protease evolved by gene duplication in response to emergence of the immunoglobulin A1 (IgA1) subclass, the principal mediator of adaptive immunity in the upper respiratory tract, in the common ancestor of humans, chimpanzees, and gorillas (39, 50) which, according to recent calculations, existed 6 to 7 million years ago (51). The more extensive diversity of ZmpB and its ubiquitous presence in multiple species suggest that zmpB is the most ancestral gene of the paralogous proteases and that it was present in the common ancestor of the Mitis and Salivarius groups of streptococci. The finding that homologous genes are located at genome positions specific to each group of closely related species (Fig. 5) suggests that the paralogs of zmpB evolved independently by gene duplication in common ancestors of the groups of species known today. Subsequent loss of function is likely to explain the lack of the IgA1 protease in S. gordonii and S. oligofermentans and the fragmented zmpB in S. thermophilus. Conversely, S. suis is likely to have acquired the zmpC homolog from a member of the Mitis group of streptococci.
Due to the genome location of zmpD, just downstream of the IgA1 protease gene, it is tempting to imagine the emergence of zmpD as the result of a duplication event of iga. However, on the basis of the results of the present cluster analyses, it is clear that zmpD shows more identity to zmpB genes than to iga. This is also reflected in the active site sequences (Fig. 2). Analysis of the intergene region downstream of the IgA1 protease gene in S. pneumoniae TIGR4 revealed sequences with homology to bacteriophage attachment sites and transposons. Thus, we favor the scenario that zmpD originated by an ancient duplication of the zmpB gene in a common ancestor of S. pneumoniae and S. mitis. Subsequently, it was transferred and integrated into its current position just downstream of the IgA1 protease gene. This theory fits well with the observed insertion of a G. elegans gene region in some strains of S. mitis at the iga genome location and suggests that the iga locus and its neighbor regions may be a hot spot for recombination events.
The results presented from analyses of the paralogous zinc metalloproteases, including the IgA1 protease, show that they are ancestral properties in Mitis group streptococci and support the idea that they play a crucial, yet partly unknown role in the relationship of S. pneumoniae with its human host. This is supported by the ubiquitous presence of ZmpB and the IgA1 protease combined with evidence of an intense selection for antigenic diversification resulting from substantial sequence divergence and very frequent transfers of entire genes and combinations of iga and the flanking zmpD. While the IgA1 protease is conserved in some of the related species, it has been lost by others presumably as a result of their adaptation to a commensal lifestyle. Yet, the activity has been transferred to distant species of Gemella. ZmpB is ubiquitous to members of the Mitis and Salivarius groups of streptococci and is likely to serve an important but yet unknown housekeeping function associated with the human host. This is supported by the finding that ZmpB is undergoing elimination from the genome of S. thermophilus, the only species with no relationship to humans or other mammals.
MATERIALS AND METHODS
Data set of sequences.
Nucleotide and amino acid sequences encoding the zinc metalloproteases IgA1 protease, ZmpB, ZmpC, and ZmpD were retrieved from publicly available databases at NCBI using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein sequences encoding IgA1 protease, ZmpB, and ZmpC from the complete genome of S. pneumoniae strain TIGR4 and the recently discovered ZmpD from the genome of S. pneumoniae strain G54 were used as query sequences. To be able to validate the species origin of the sequences and to disclose the genetic relationships of the strains from which the sequences originated, we retrieved sequences of seven housekeeping genes (map, pfl, ppaC, pyk, rpoB, sodA, and tuf) and performed MLSA according to the principles described by Bishop et al. (37).
Predictions, phylogenetic analyses, and genome localization.
Signal peptides were identified using SignalP (http://www.cbs.dtu.dk/services/SignalP/) and transmembrane regions predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html). Due to extensive variation in nucleotide sequences, multiple amino acid sequence alignments were performed using MAFFT version 6.843 (52). L-INS-I was the applied method used with default parameters. The amino acid sequence alignment was converted into a nucleotide alignment using PAL2NAL (http://www.bork.embl.de/pal2nal/). All sequences contained a region of repeat sequences within the N-terminal sequence. These regions interfered with the alignment and therefore were excluded, resulting in sequences of approximately 1,200 amino acids. Phylogenetic tree construction was conducted using MEGA V5 (53) using the minimum evolution algorithms and the Kimura two-parameter model with pairwise deletion of gaps or missing data. Bootstrap analyses based on 500 replicates were conducted. Mean molecular distances were determined using the Kimura two-parameter method, and the standard error (SE) was determined for each parameter. Only single copies of identical alleles were included in mean distance calculations. In order to determine the genome localization of the individual zinc metalloproteases, open reading frames (ORF) flanking the iga or zmpD, zmpB, and zmpC genes were retrieved from complete and draft genome sequences of selected strains available at NCBI (http://www.ncbi.nlm.nih.gov/) and compared using BLASTP.
Relative amino acid sequence distances.
As a measure of the selection for diversity, we determined the within-species mean distances of amino acid sequences of each of the Zmp proteins for S. pneumoniae, S. mitis, S. oralis, and S. sanguinis relative to the corresponding distances of amino acid sequences of translated concatamers of the seven genes used for MLSA analysis. The mean distances were calculated in MEGA V5, and differences between species were statistically analyzed by unpaired analysis of variance (ANOVA) test using the GraphPad InStat software (GraphPad Software, Inc., La Jolla, CA).
Construction of deletion mutants.
To construct iga, zmpB, and zmpC deletion mutants of S. pneumoniae TIGR4, regions flanking the genes were amplified by PCR (Expand High Fidelity PCR system; Roche) using genomic DNA from S. pneumoniae strain TIGR4 as the template and cloned into the plasmid vector pCR2.1-TOPO (TOPO TA cloning kit; Invitrogen). Appropriate restriction sites (underlined) for further subclonings were incorporated at the ends of the PCR primers. Primer 5′ AAGCTTCCTCAATTCCTATTTTCGTGA 3′ combined with 5′ GGATCCAGACCTTTATTATATTAGTGTATT 3′ and 5′ CTCGAGGATGCAGAACACAATTACTAC 3′ combined with 5′ GGGCCCAAACGGTTCATCAGAGTTGAT 3′ were used to construct the iga deletion mutant. Primer 5′ AAGCTTCTAGTGAGGTGCTAGGTGGTC 3′ combined with 5′ GGTACCTTTTATCTCCTTTATTCATTCTCAAAAC 3′ and 5′ CTCGAGATTGAAATCTCTCATCTGCTTTGCGG 3′ combined with 5′ TCTAGATGCTTAAGTTGGACGGTG TAGT 3′ were used to construct the zmpB deletion mutant. Primer 5′ AAGCTTTGGCTGTGAGCACAACAAC 3′ combined with 5′ GGATCCTGAGTACTTTTCAACAATTACTTATC 3′ and 5′-CTCGAGATTGTAGAGTTTCATTGTTGAG-3′ combined with 5′ TCTAGACCAGAACGACCTCTGTATCCG 3′ were used to construct the zmpC deletion mutant. DNA constructs with these regions flanking the cat gene cassette from pR326 encoding chloramphenicol resistance (amplified by PCR using primers BM18 and BM19 and cloned into pCR2.1-TOPO) and in the same orientation as the gene to be replaced were used for insertion mutagenesis in S. pneumoniae (54). Transformation of S. pneumoniae TIGR4 with linearized plasmid DNA was performed as described using synthetic competence-stimulating peptides CSP-1 and CSP-2 (55, 56). Transformants were selected on agar plates containing 2 µg/ml of chloramphenicol. Gene disruptions were confirmed by PCR.
IgA1 protease activity assay.
Streptococcal strains identified by MLSA were examined for IgA1 protease activity by incubating a loop full of bacteria from a blood agar culture in 40 µl of a solution of myeloma IgA1 (1.8 mg/ml) overnight. Cleavage of IgA1 into characteristic fragments of the alpha chain was demonstrated by Western blotting as described below. For selected strains, bacteria were grown in 2× YT medium (57) overnight at 37°C in an atmosphere supplemented with 5% CO2. To distinguish between cell-associated and secreted IgA1 protease activity, 1-ml portions of cultures were centrifuged. Two samples of pellets were resuspended in 100 µl and 1 ml of phosphate-buffered saline (PBS), pH 7.4, respectively, and compared to the supernatant. Each sample of 100 µl was mixed with 10 µl of human myeloma IgA1 (1.8 mg/ml) and incubated overnight at 37°C. Characteristic cleavage fragments were detected by Western blotting. In brief, samples were mixed with sample buffer and boiled for 5 min, and 10-μl reaction mixture was subjected to reducing SDS-PAGE on 7% Tris-acetate Novex gels (Invitrogen) and electrotransferred to a polyvinylidene difluoride (PVDF) membrane (Immobiline Pseq; Millipore) using Tris-HCl-glycine buffer (pH 8.4) (192 mM glycine, 25 mM Tris, 20% [vol/vol] methanol). Nonspecific reactivity was blocked with blocking buffer (460 mM NaCl, 0.1% [wt/vol] bovine serum albumin [BSA], 50 mM Tris-HCl [pH 7.4], 1.5% Tween 20). The membranes were incubated for 2 h with the primary antibody, rabbit anti-human IgA (α chain) (catalog no. A262; Dako) diluted 1:1,000 in blocking buffer. After the membranes were washed, an additional 2 h of incubation was performed with the secondary antibody alkaline phosphatase-conjugated swine anti-rabbit immunoglobulins (Dako) diluted 1:2,000. Finally, the blot was developed with BCIP/NBT (5-bromo-4-chloro-3′-indolyl phosphate/Nitro Blue tetrazolium) substrate.
ZmpC protease activity assay.
Bacterial strains were grown overnight in Todd-Hewitt broth supplemented with Na pyruvate (Difco) at 37°C in an atmosphere supplemented with 5% CO2. Detection of MMP-9 cleaving activity was performed essentially by the method of Oggioni and coworkers (6). Briefly, 30 µl of the cell culture was incubated with 20 µl of recombinant human proenzyme MMP-9 (1 ng/μl) (Calbiochem) for 2 h. After centrifugation, 5 µl of the supernatant was mixed with an equal volume of 2× loading buffer, electrophoresed on a 10% zymogram (gelatin) gel (Invitrogen), and developed as recommended by the manufacturer. Finally, the gel was stained with Coomassie brilliant blue for 18 h and destained with 10% acetic acid. In this assay, areas of protease activity appear as clear bands against a dark blue background.
SUPPLEMENTAL MATERIAL
Phylogenetic trees based on the minimum evolution algorithm, representing the evolutionary relationships within each of the four zinc metalloproteases. IgA1 protease (A), ZmpB (B), ZmpC (C), and ZmpD (D). Species are indicated by colored dots. The asterisks in panel A indicate strains that contain the additional zmpD gene just downstream of iga. Bootstrap values exceeding 80 are shown. Download Figure S1, EPS file, 5.2 MB.
Strains represented by the 297 sequences used in the present study.
Mean molecular distances between groups. Standard errors are shown in grey.
Mean molecular distances between ZmpB subgroups and the ZmpD cluster. Standard errors are shown in grey.
Strains screened for ZmpC protease activity.
ACKNOWLEDGMENTS
This study was supported by a grant from the Danish Medical Research Council (10-083748).
We thank Jean-Pierre Claverys for providing the pR326 plasmid for gene disruption mutagenesis in S. pneumoniae. We are grateful to Lise Hall Schultz for technical assistance and to Christian Scholz for help with retrieval of sequences.
Footnotes
Citation Bek-Thomsen M, Poulsen K, Kilian M. 2012. Occurrence and evolution of the paralogous zinc metalloproteases IgA1 protease, ZmpB, ZmpC, and ZmpD in Streptococcus pneumoniae and related commensal species. mBio 3(5):e00303-12. doi:10.1128/mBio.00303-12.
REFERENCES
- 1. Dopazo J, et al. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microb. Drug Resist. 7:99–125 [DOI] [PubMed] [Google Scholar]
- 2. Hoskins J, et al. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tettelin H, et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506 [DOI] [PubMed] [Google Scholar]
- 4. Gilbert JV, Plaut AG, Wright A. 1991. Analysis of the immunoglobulin A protease gene of Streptococcus sanguis. Infect. Immun. 59:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Romanello V, et al. 2006. Cloning, expression, purification, and characterization of Streptococcus pneumoniae IgA1 protease. Protein Expr. Purif. 45:142–149 [DOI] [PubMed] [Google Scholar]
- 6. Oggioni MR, et al. 2003. Pneumococcal zinc metalloproteinase ZmpC cleaves human matrix metalloproteinase 9 and is a virulence factor in experimental pneumonia. Mol. Microbiol. 49:795–805 [DOI] [PubMed] [Google Scholar]
- 7. Bender MH, Weiser JN. 2006. The atypical amino-terminal LPNTG-containing domain of the pneumococcal human IgA1-specific protease is required for proper enzyme localization and function. Mol. Microbiol. 61:526–543 [DOI] [PubMed] [Google Scholar]
- 8. Takenouchi-Ohkubo N, Mortensen LM, Drasbek KR, Kilian M, Poulsen K. 2006. Horizontal transfer of the immunoglobulin A1 protease gene (iga) from Streptococcus to Gemella haemolysans. Microbiology 152:2171–2180 [DOI] [PubMed] [Google Scholar]
- 9. Bergé M, et al. 2001. The puzzle of zmpB and extensive chain formation, autolysis defect and non-translocation of choline-binding proteins in Streptococcus pneumoniae. Mol. Microbiol. 39:1651–1660 [DOI] [PubMed] [Google Scholar]
- 10. Poulsen K, Reinholdt J, Kilian M. 1996. Characterization of the Streptococcus pneumoniae immunoglobulin A1 protease gene (iga) and its translation product. Infect. Immun. 64:3957–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wani JH, Gilbert JV, Plaut AG, Weiser JN. 1996. Identification, cloning, and sequencing of the immunoglobulin A1 protease gene of Streptococcus pneumoniae. Infect. Immun. 64:3967–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bateman A, Holden MT, Yeats C. 2005. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics 21:1301–1303 [DOI] [PubMed] [Google Scholar]
- 13. Fischetti VA, Pancholi V, Schneewind O. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603–1605 [DOI] [PubMed] [Google Scholar]
- 14. Kilian M, Mestecky J, Schrohenloher RE. 1979. Pathogenic species of the genus Haemophilus and Streptococcus pneumoniae produce immunoglobulin A1 protease. Infect. Immun. 26:143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Male CJ. 1979. Immunoglobulin A1 protease production by Haemophilus influenzae and Streptococcus pneumoniae. Infect. Immun. 26:254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Plaut AG. 1988. Production and isolation of neissereal IgA proteases. Methods Enzymol. 165:117–120 [DOI] [PubMed] [Google Scholar]
- 17. Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen EV. 1996. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS 104:321–338 [DOI] [PubMed] [Google Scholar]
- 18. Kilian M, Mestecky J, Russell MW. 1988. Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol. Rev. 52:296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reinholdt J, Kilian M. 1987. Interference of IgA protease with the effect of secretory IgA on adherence of oral streptococci to saliva-coated hydroxyapatite. J. Dent. Res. 66:492–497 [DOI] [PubMed] [Google Scholar]
- 20. Weiser JN, et al. 2003. Antibody-enhanced pneumococcal adherence requires IgA1 protease. Proc. Natl. Acad. Sci. U. S. A. 100:4215–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chiavolini D, et al. 2003. The three extracellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiol. 3:14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polissi A, et al. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620–5629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin L, et al. 1997. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol. Microbiol. 24:1083–1094 [DOI] [PubMed] [Google Scholar]
- 24. Beck SC, Meyer TF. 2000. IgA1 protease from Neisseria gonorrhoeae inhibits TNFalpha-mediated apoptosis of human monocytic cells. FEBS Lett. 472:287–292 [DOI] [PubMed] [Google Scholar]
- 25. Senior BW, Stewart WW, Galloway C, Kerr MA. 2001. Cleavage of the hormone human chorionic gonadotropin, by the type 1 IgA1 protease of Neisseria gonorrhoeae, and its implications. J. Infect. Dis. 184:922–925 [DOI] [PubMed] [Google Scholar]
- 26. Binscheck T, et al. 1995. IgA protease from Neisseria gonorrhoeae inhibits exocytosis in bovine chromaffin cells like tetanus toxin. J. Biol. Chem. 270:1770–1774 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y, Hayashida A, Bennett AE, Hollingshead SK, Park PW. 2007. Streptococcus pneumoniae sheds syndecan-1 ectodomains through ZmpC, a metalloproteinase virulence factor. J. Biol. Chem. 282:159–167 [DOI] [PubMed] [Google Scholar]
- 28. Govindarajan B, et al. 2012. A metalloproteinase secreted by Streptococcus pneumoniae removes membrane mucin MUC16 from the epithelial glycocalyx barrier. PLoS One 7:e32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blue CE, et al. 2003. ZmpB, a novel virulence factor of Streptococcus pneumoniae that induces tumor necrosis factor alpha production in the respiratory tract. Infect. Immun. 71:4925–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389–1406 [PMC free article] [PubMed] [Google Scholar]
- 31. Kilian M, Mikkelsen L, Henrichsen J. 1989. Taxonomic study of viridans streptococci: description of Streptococcus gordonii sp. nov., and emended description of Streptococcus sanguis (White and Niven 1946), Streptococcus oralis (Bridge and Sneath 1982), and Streptococcus mitis (Andrewes and Horder 1906). Int. J. Syst. Bacteriol. 39:471–484 [Google Scholar]
- 32. Lomholt H. 1995. Evidence of recombination and an antigenically diverse immunoglobulin A1 protease among strains of Streptococcus pneumoniae. Infect. Immun. 63:4238–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Camilli R, et al. 2006. Zinc metalloproteinase genes in clinical isolates of Streptococcus pneumoniae: association of the full array with a clonal cluster comprising serotypes 8 and 11A. Microbiology 152:313–321 [DOI] [PubMed] [Google Scholar]
- 34. Reinholdt J, Tomana M, Mortensen SB, Kilian M. 1990. Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect. Immun. 58:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang A, et al. 2011. IgA1 protease contributes to the virulence of Streptococcus suis. Vet. Microbiol. 148:436–439 [DOI] [PubMed] [Google Scholar]
- 36. Zhang A, et al. 2010. Identification and characterization of IgA1 protease from Streptococcus suis. Vet. Microbiol. 140:171–175 [DOI] [PubMed] [Google Scholar]
- 37. Bishop CJ, et al. 2009. Assigning strains to bacterial species via the internet. BMC Biol. 7:3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kilian M, Mestecky J, Kulhavy R, Tomana M, Butler WT. 1980. IgA1 proteases from Haemophilus influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Streptococcus sanguis: comparative immunochemical studies. J. Immunol. 124:2596–2600 [PubMed] [Google Scholar]
- 39. Kilian M, et al. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tong H, Gao X, Dong X. 2003. Streptococcus oligofermentans sp. nov., a novel oral isolate from caries-free humans. Int. J. Syst. Evol. Microbiol. 53:1101–1104 [DOI] [PubMed] [Google Scholar]
- 41. Denapaite D, et al. 2010. The genome of Streptococcus mitis B6—what is a commensal? PLoS One 5:e9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kilian M, Reinholdt J. 2005. Immunoglobulin A1 proteases of pathogenic and commensal bacteria of the respiratory tract, p 119–129 In Natarro JP, Cohen PS, Mobley HLT, Weiser JN, Colonization of mucosal surfaces. ASM Press, Washington, DC [Google Scholar]
- 43. Poulsen K, et al. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect. Immun. 66:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Coffey TJ, et al. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73–83 [DOI] [PubMed] [Google Scholar]
- 45. Croucher NJ, et al. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bentley SD, et al. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. 2012. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol. Rev. 245:132–146 [DOI] [PubMed] [Google Scholar]
- 49. Lomholt H, Kilian M. 1994. Antigenic relationships among immunoglobulin A1 proteases from Haemophilus, Neisseria, and Streptococcus species. Infect. Immun. 62:3178–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kawamura S, Saitou N, Ueda S. 1992. Concerted evolution of the primate immunoglobulin alpha-gene through gene conversion. J. Biol. Chem. 267:7359–7367 [PubMed] [Google Scholar]
- 51. Hobolth A, Christensen OF, Mailund T, Schierup MH. 2007. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 3:e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537:39–64 [DOI] [PubMed] [Google Scholar]
- 53. Tamura K, et al. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Claverys JP, Dintilhac A, Pestova EV, Martin B, Morrison DA. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123–128 [DOI] [PubMed] [Google Scholar]
- 55. Håvarstein LS, Hakenbeck R, Gaustad P. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pozzi G, et al. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic trees based on the minimum evolution algorithm, representing the evolutionary relationships within each of the four zinc metalloproteases. IgA1 protease (A), ZmpB (B), ZmpC (C), and ZmpD (D). Species are indicated by colored dots. The asterisks in panel A indicate strains that contain the additional zmpD gene just downstream of iga. Bootstrap values exceeding 80 are shown. Download Figure S1, EPS file, 5.2 MB.
Strains represented by the 297 sequences used in the present study.
Mean molecular distances between groups. Standard errors are shown in grey.
Mean molecular distances between ZmpB subgroups and the ZmpD cluster. Standard errors are shown in grey.
Strains screened for ZmpC protease activity.