Abstract
Primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC) and autoimmune hepatitis (AIH) are the major forms of autoimmune liver diseases each characterized by the destruction of a specific liver cell type and the presence of differing auto-antibodies. We took a proteomic approach utilizing in situ matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) to obtain profiles directly from liver samples of patients with PBC, PSC, AIH and controls. The ability to precisely localize the region for acquisition of MALDI MS allowed us to obtain profiles from bile ducts, inflammatory infiltrates and hepatocytes from each biopsy sample. Analysis tools developed to identify peaks and compare peaks across diseases and cell types were used to develop models to classify the samples. Using an initial set of testing samples from PBC patients and controls, we identified unique peaks present in bile ducts, inflammatory infiltrates and hepatocytes that could classify samples in a validation cohort with 88–91% accuracy. Interestingly, profiles of PSC and AIH did not differ significantly from PBC. Identification of proteins in these peaks may represent novel autoantigens or effector molecules. These findings illustrate the potential of a proteomic approach to autoimmune diseases with in situ MALDI MS.
Keywords: autoimmune hepatitis, mass spectrometry, primary biliary cirrhosis, primary sclerosing cholangitis
Introduction
Primary biliary cirrhosis (PBC), primary sclerosing cholangitis (PSC) and autoimmune hepatitis (AIH) encompass the three major forms of autoimmune liver diseases. PBC is a chronic inflammatory disease of the liver characterized by a destruction of small bile ducts and loss of tolerance to the E2 component of the mitochondrial pyruvate dehydrogenase complex (PDC-E2). In addition to the well-characterized anti-mitochondrial antibodies (AMAs) to PDC-E2, autoreactive CD4 and CD8 T-cell responses are enriched in the livers of patients with PBC.1 The target of PSC is also the bile ducts but this disease affects primarily the medium and large ducts resulting in an obliterative fibrosis and structuring of the ducts.2 The association between inflammatory bowel disease and PSC has lead to the concept that the liver damage of PSC is due to the homing of intestinal lymphocytes to the liver. Although autoantibodies including antineutrophil antibodies, have been reported in PSC, they have yet to be fully characterized and their role in disease pathogenesis has not been established. In contrast to PBC and PSC, AIH is a disease affecting the parenchymal cells of the liver and is often associated with antinuclear, antineutrophil and anti-actin antibodies.3 Together, these diseases demonstrate how a single organ may be selectively targeted by different autoimmune diseases.4 A proteomic approach to cataloging the proteins including antigens and effector molecules that are differentially expressed in these diseases and in health would be a tremendous advance towards understanding autoimmune liver diseases.
In situ matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI MS) is a novel method that allows the analysis of proteins directly in tissue.5, 6, 7 There are several advantages to in situ MALDI MS over traditional MALDI MS of whole tissue or isolated cells including the ability of in situ MALDI MS to analyze specific areas of tissue affected or unaffected by disease and specific cells types without the requirement for cell isolation which can lead to changes in the MALDI MS profiles. Herein we demonstrate the application of in situ MALDI MS to PBC, PSC and AIH to compare the protein expression profiles of bile ducts, inflammatory infiltrates and hepatocytes.
Martial and methods
Liver sample preparation
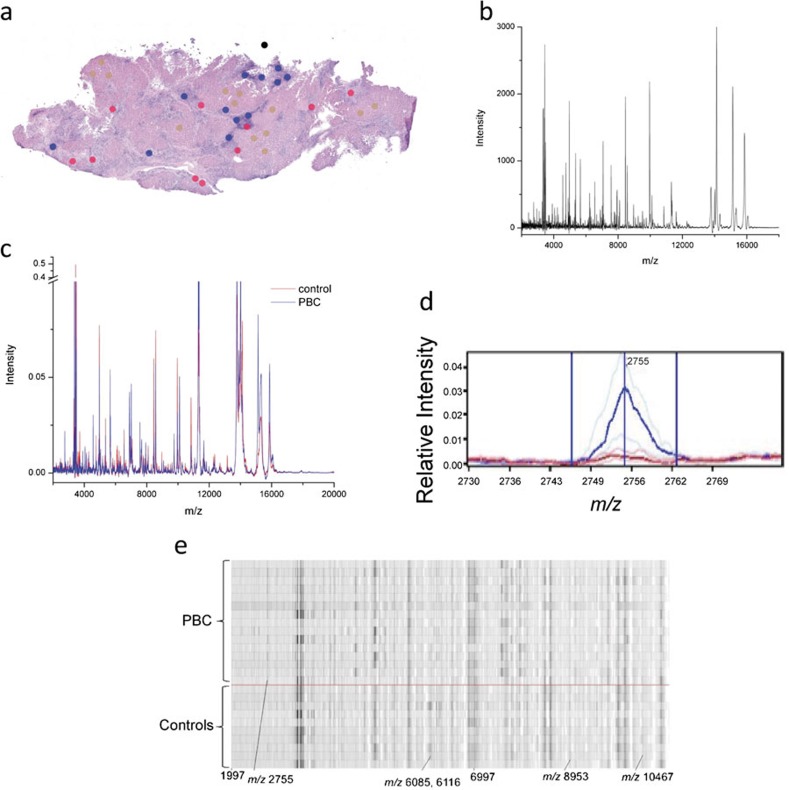
Liver tissue was obtained from liver explanted at the time of liver transplantation from 29 PBC patients, 11 PSC patients and 7 AIH patients. Nineteen healthy control livers were obtained from organ donors. Frozen sections (12 µm thick) were cut immediately adjacent to the MALDI–MALDI MS sections (see the section on ‘MALDI MS'), stained with hematoxylin and eosin (H&E), and analyzed. The resulting image files were then marked with color-coded 200-µm circles, the size of the applied matrix spots MALDI MS to designate bile ducts, inflammatory infiltrates and hepatocytes (Figure 1a).
Figure 1.
Overview of data acquisition and analysis of MALDI MS data obtained from liver tissue. (a) Hematoxylin and eosin stained tissue sections were imaged and areas for MALDI MS are marked for bile duct (red), hepatocyte (brown) and inflammatory infiltrate (blue) profiling. Each dot represents the size of the matrix droplet deposited on the serial section utilized to acquire the MADLI MS data. (b) Raw data are acquired and processed with subtraction of background and normalization before averaging of replicas for each individual sample. (c) Processed spectra aligned and analyzed for differentially expressed peaks. Average (per group) spectra for controls (red line) and PBC (blue line). (d) Representative median spectra of a differentially expressed peak. Heavy and faint blue lines represent the median PBC peak and twenty-fifth and seventy-fifth percentiles, respectively. Similar red lines represent the control peaks. (e) Representative gel plot of PBC (above red line) and controls (below red line). MALDI MS, matrix-assisted laser desorption/ionization mass spectrometry; PBC, primary biliary cirrhosis.
MALDI MS
Frozen tissues were sectioned (12 µm thick) at −15 °C on a Leica cryostat (Leica Microsystems Inc., Bannockburn, IL, USA). The sections were transferred and thawed onto gold-coated stainless steel MALDI target plates. Consecutive sections (12 µm) were cut, picked up on glass slides and stained with H&E for pathological classification.
For MALDI MS profiling, two serial 12-µm sections were cut from each sample. One section was mounted onto a charged glass slide and stained with H&E following standard protocols. An adjacent section was thaw mounted directly onto a gold-coated MALDI target followed by fixation in graded ethanol washes (70, 90 and 95% for 30 s each). Photomicrographs of H&E-stained tissue sections were obtained with an Olympus BX-50 microscope at a magnification of ×10. Using Photoshop all histology images were annotated using a small circular shape (200 µm) to mark areas of interest for matrix spotting. Where available, a minimum of 10 spots was placed on each of bile ducts, hepatocytes and lymphocytes. Annotated histology images were then overlaid with the target plate images through Photoshop. Annotated marks are also placed into at least four distinctive features that are visible in the MALDI target image, which provides landmarks for registering the MALDI plate to the robotic spotter. The coordinates from the annotated histology and MALDI plate images are extracted and registered to the robotic spotter. An acoustic robotic spotter (LabCyte, Sunnyvale, CA, USA) was used to place crystalline matrix (20 mg/ml sinapinic acid in 1∶1 acetonitrile: 0.2% trifluoroacetic acid) spots (∼120 pl) that were typically 180–220 µm in diameter. A total of 78 drops were placed at each location to achieve optimal signal. Tissue profile spectra were acquired using an Autoflex II (Bruker Daltonics, Billerica, MA, USA) MALDI mass spectrometer and run using an automated linear-mode acquisition method optimized for 2–40 kDa with an extraction voltage of 20 kV, an acceleration voltage of 18.65 kV and a lens voltage of 6 kV. Delayed extraction was optimized for resolution at 12 kDa. A total of 400 laser shots were collected for each profile spectrum (Figure 1b).
Data processing and analysis
Spectral preprocessing
Preprocessing of spectra is necessary to render spectra comparable and to ensure the reproducibility of the statistical analysis procedure (Figure 1c–e). Preprocessing was performed using proprietary analysis tools developed by Biodesix Inc. (Steamboat Springs, CO, USA), and described elsewhere.8, 9, 10 In brief, the background and noise were estimated based on local noise estimators using a local (in m/z) robust asymmetric estimator and then subtracted from each spectrum and normalized to total ion current. Peaks were detected using a signal-to-noise ratio cut-off of 4.0. Replicate spectra from the same tissue sample were used to create an average spectrum for each individual sample. Average spectra were aligned using a set of common peaks. The preprocessing procedure was optimized using the training set and held fixed for the classification of all test sets.
Training and classifier optimization
Samples from each clinical group of interest were randomly split into training and test sets.
Each spectrum can be characterized by a set of features. Features are defined as integrated, background-subtracted and normalized intensities over a chosen m/z range containing a peak. A classifier is a combination of a set of features and the algorithm, which allows for discrimination between groups.
Selection of features for the classifier is performed using a variant of a floating search,11 based on calculation of univariate P values from Mann–Whitney U (Wilcoxon), minimization of leave one-out cross-validation errors in the training set and visual inspection of features. The classification algorithm we use is a straightforward implementation of a k-nearest-neighbor (KNN) algorithm,12 which requires as parameters a set of representative and labeled instances (i.e., list of selected feature values from a training sample set).
To classify a new spectrum, the KNN algorithm first calculates the Euclidean distance of the feature values of the new spectrum to those of the representative spectra. This calculation yields a list of distances from the test spectrum to each representative spectrum. For the KNNs (those with the k smallest distances) the labels are compared. The assigned label is a simple majority vote over the KNN labels.
Cross-validation was used for the assessment of performance during the training of the classifier. A fixed number (one for leave one-out cross-validation or a prescribed number N for LNOCV) of instances were removed from the training set, a classifier was generated using the remaining instances, and the performance of this classifier was evaluated by applying it to the omitted instances and comparing classifier and true labels. Classifier parameters and the set of selected features were optimized using these cross-validation procedures.
Validation of a classifier on the independent test set
After training and optimization, all parameters were frozen. No changes in the classification algorithm were allowed during the analysis of the test set. For each test spectrum, the required feature values for the features comprising the classifier were determined and are presented to the fixed KNN classifier, which then returned a label.
Results
Comparison of MALDI MS from bile ducts, hepatocytes and infiltrates of PBC and healthy controls
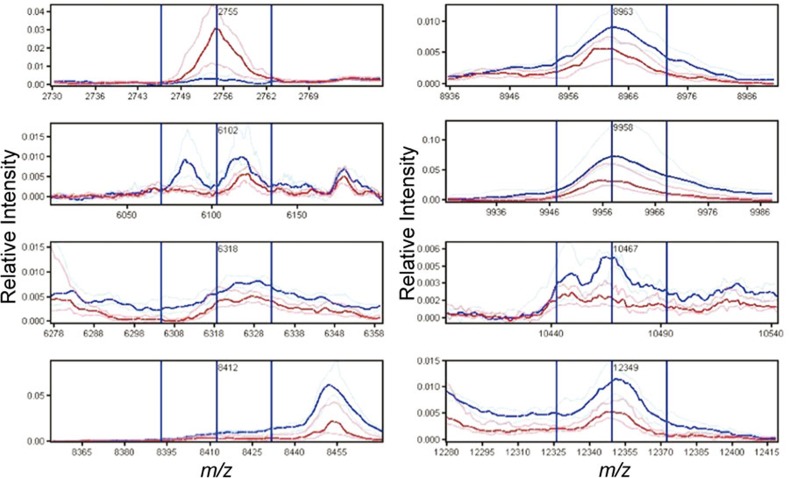
In our initial analysis, we compared the spectra obtained from liver of PBC patients (n=14) and controls (n=9). In the analysis of the bile duct training set, 101 peaks were identified of which eight were found to be differentially expressed and essential for the optimal classification of the training set, including a feature centered at m/z 6102 which was a result of the merging of two peaks at m/z 6085 and 6116 (Table 1 and Figure 2). Using these peaks to classify the test cases correctly identified 12 of 14 PBC cases and 8 of 9 controls resulting in an accuracy of 88%, sensitivity of 86% and specificity of 89%.
Table 1. Differentially expressed peaks between PBC and control bile ducts.
| m/z | PBC (n=14) | Controls (n=9) | P value | ||
|---|---|---|---|---|---|
| Mean | CV | Mean | CV | ||
| 2755 | 0.367 | 0.608 | 0.08 | 1.233 | 3.5×10−4 |
| 6102a | 0.121 | 0.59 | 0.627 | 1.3 | 1.2×10−3 |
| 6318 | 0.129 | 0.368 | 0.267 | 0.538 | 2.8×10−4 |
| 8412 | 0.118 | 0.552 | 0.273 | 0.25 | 2.3×10−4 |
| 8963 | 0.064 | 0.412 | 0.122 | 0.52 | 2.5×10−3 |
| 9958 | 0.434 | 0.459 | 0.988 | 0.519 | 1.7×10−3 |
| 10467 | 0.063 | 0.72 | 0.13 | 0.489 | 2.1×10−3 |
| 12349b | 0.126 | 0.608 | 0.254 | 0.413 | 3.0×10−3 |
Abbreviations: CV, covariance; PBC, primary biliary cirrhosis.
Feature m/z 6102 is a result of merging of two peaks: m/z 6085 and m/z 6116.
May represent macrophage inhibitory factor.
Figure 2.
Spectral views of median spectra (per group) of differentially expressed peaks between PBC (blue) and control (red) bile ducts identified in the training set. PBC, primary biliary cirrhosis.
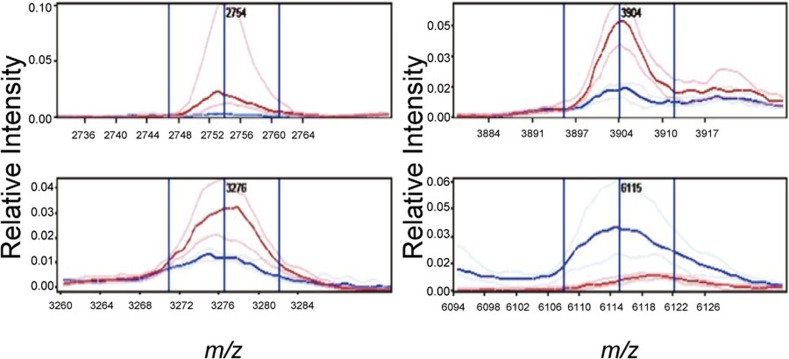
The analysis of hepatocytes identified four differentially expressed peaks which were incorporated in the classifier (Table 2 and Figure 3). Similar to the classification of bile ducts, the classifier for hepatocytes based on four peaks selected performed well and correctly identified 13 of 14 PBC cases and 8 of 9 controls. Six differentially expressed peaks were selected for the classifier of the inflammatory infiltrates of PBC cases and controls in the training set. The inflammatory infiltrate classifier correctly identified 14 of 15 PBC cases but only 5 of 8 controls in the test set.
Table 2. Differentially expressed peaks between PBC and control hepatocytes.
| m/z | PBC (n=14) | Controls (n=9) | P value | ||
|---|---|---|---|---|---|
| Mean | CV | Mean | CV | ||
| 2754 | 0.602 | 0.986 | 0.061 | 0.815 | 1.4×10−4 |
| 3276 | 0.412 | 0.568 | 0.175 | 0.343 | 2.3×10−4 |
| 3904 | 0.672 | 0.734 | 0.206 | 0.356 | 5.8×10−5 |
| 6115 | 0.211 | 2.393 | 0.531 | 0.714 | 5.3×10−4 |
Abbreviations: CV, covariance; PBC, primary biliary cirrhosis.
Figure 3.
Spectral views of median spectra (per group) of differentially expressed peaks between PSC and AIH bile ducts and inflammatory infiltrates. AIH, autoimmune hepatitis; PSC, primary sclerosing cholangitis.
Analysis of MALDI MS from PSC and AIH relative to PBC
To determine if the patterns observed in the bile ducts of PBC were unique or a shared feature of other autoimmune liver diseases, we compared the spectra from all 29 PBC cases, 11 PSC cases and 7 AIH cases. No statistically significant differences were found between the samples. The features having the lowest P values between the groups were generally between 3 and 5 kDa, with the lowest P values being of the order of 10−3 to 10−2. Several features in the range 3–4 kDa appeared with relatively low P values in more than one of the comparisons. For these m/z values, the average feature values increased from AIH through PBC to PSC. Hence, the differences were most marked between PSC and AIH.
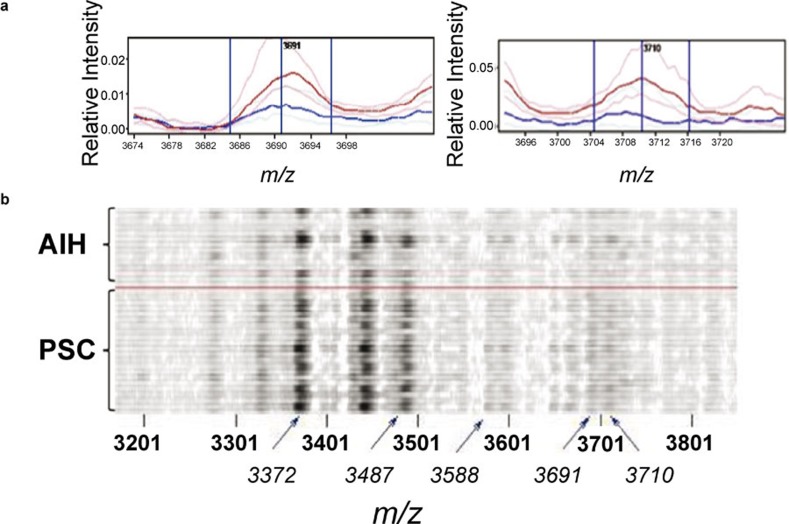
In the inflammatory infiltrate PSC and AIH typically centers on the portal tract. At the same time, the insufficient resolution of the spectra acquisition in situ may lead to overlap between the signals from these regions. To address this problem, we performed an analysis in which spectra from bile ducts and lymphocytes were combined (Table 3 and Figure 4). There were still no statistically significant differences, but the P values obtained were lower than in the other comparisons. In particular, features at m/z 3691 and 3710 stood out from the remaining features as having particularly low P values (1×10−3 and 3×10−3 respectively). However, as can be seen from the gelplot of the m/z 3200–3800 region (Figure 4b), there was a great deal of variation in the differentiating features within the groups, but overall there are more spectra with large amplitudes for these features in the PSC group than in the AIH group.
Table 3. Differentially expressed peaks between AIH and PSC bile ducts and inflammatory infiltrates.
| m/z | AIH (n=14) | PSC (n=22) | Wil P | ||
|---|---|---|---|---|---|
| Mean | CV | Mean | CV | ||
| 2754 | 0.136 | 1.245 | 0.269 | 0.828 | 1.1×10−2 |
| 3372a | 1.954 | 1.576 | 3.968 | 0.925 | 1.2×10−2 |
| 3487b | 0.856 | 1.499 | 1.987 | 0.795 | 9.0×10−3 |
| 3588 | 0.176 | 0.716 | 0.279 | 0.724 | 1.9×10−2 |
| 3691 | 0.091 | 0.638 | 0.2 | 0.504 | 1.1×10−3 |
| 3710 | 0.116 | 1.449 | 0.228 | 0.769 | 3.3×10−3 |
| 4124 | 0.233 | 0.665 | 0.36 | 0.497 | 1.9×10−2 |
| 4749 | 0.264 | 0.825 | 0.429 | 0.526 | 1.4×10−2 |
| 15127c | 3.046 | 0.721 | 1.772 | 1.138 | 9.9×10−3 |
| 15284 | 1.837 | 0.273 | 1.392 | 0.374 | 1.9×10−2 |
Abbreviations: AIH, autoimmune hepatitis; CV, covariance; PSC, primary sclerosing cholangitis, Wil, Wilcoxon.
May represent human neutrophil defensin-2.
May represent human neutrophil defensin-3.
Hemoglobin α-chain.
Figure 4.
Spectral views of median spectra (per group) (a) and gel plot (b) of peaks m/z 3200–3800 region from combined analysis of bile ducts and inflammatory infiltrates from AIH (blue) and PSC (red) livers. AIH, autoimmune hepatitis; PSC, primary sclerosing cholangitis.
Discussion
In this study, we directly profiled protein expression from liver tissue of PBC, PSC and AIH patients as well as healthy controls with MALDI MS, and defined profiles that enabled classification of PBC and healthy controls. In addition, we found that there were no significant differences in profiles between PBC, PSC and AIH. Previous studies have investigated the gene expression patterns in autoimmune liver diseases.13, 14, 15, 16, 17 However, mRNA expression in liver cannot always indicate which proteins are expressed or how their activity might be modulated after translation.18 Accordingly, analysis of the proteome in serum, tissues and other clinical materials might better reflect the underlying pathological state in autoimmune diseases rather than gene expression patterns. Further, mRNA expression of liver generally has been done on whole tissue samples and although individual liver cell types can be isolated by various procedures, they generally require manipulations that could alter gene expression patterns. Our method utilized freshly frozen tissue limiting the possibility of changes in protein composition during the acquisition of MALDI MS profiles.
The complex protein mixtures of tissues lead to MALDI MS profiles containing thousands of data points derived from real protein signatures but contaminated with electronic and chemical noise, variability in instrumentation and variable crystallization of matrix, necessitating the use of analytical techniques developed for this application. Accordingly, we used a sophisticated tool to preprocess, align and detect peaks from hundreds of protein profiles and used advanced statistical methods to confidently identify the most significant proteins associated with the diseases and cell type of interest. The use of validation cohorts reaffirms the significance of the peaks identified in the initial analysis of the training cohorts.
These technologies enabled us to detect proteomic patterns that could be used to accurately classify PBC and healthy bile ducts, inflammatory infiltrates and hepatocytes from small biopsy samples. Our prediction models were found to classify cell types from PBC versus normal liver with 83–91% accuracy in the validation cohort. These profiles have the potential for both clinical and basic science applications.
Clinically, the ability to accurately diagnosis PBC on liver biopsy would be particularly useful in cases where the AMA is negative. Although the sensitivity of the AMA assay has improved and can now detect all but approximately 5% of PBC patients, the diagnosis of AMA-negative patients remains difficult.19 Perhaps more important would be the use of these profiles to establish prognosis in PBC, which currently is variable and difficult to predict. Our current study used only patients with end-stage disease but as has been done for non-small cell lung cancer, future studies could be performed on cases at various stages and ideally with prospectively collected outcomes to develop prognostic models based upon in situ MALDI MS profiles.5, 20
More relevant to the present study are the peaks identified and their potential to add to the understanding of autoimmune liver disease pathophysiology. Identities of the proteins that make up these profiles are unknown at present. However, a previous study using this technique in non-small cell lung cancer was able to distinguish tumors based upon prognosis and identified several proteins responsible for differentially expressed peaks.5 Identifying the cell and disease specific proteins in autoimmune liver diseases would have obvious implications. We recently demonstrated that, in contrast to other epithelial cells, small biliary epithelial cells translocate the PBC auto-antigen PDC-E2 immunologically intact to apoptotic bodies during apoptosis and based upon these in vitro experiments and the recurrence of PBC after liver transplantation, we propose that the biliary ‘apotope' is a generic feature of biliary epithelial cells and not unique to PBC patients. The proteins identified in the classifying peaks from the bile duct and hepatocyte profiles would be candidates for further study into their roles as novel auto-antigens in PBC and AIH, respectively. Alternatively, these proteins may represent effector molecules involved in processes such as apoptosis, lymphocyte homing or immune responses. We note that even in PBC in which the hepatocytes are relatively spared from injury, we were able to identify peaks that accurately differentiate PBC from controls. While much focus in PBC is aimed at the bile duct epithelial cells, these findings suggest that hepatocyte protein expression may be important in understanding the pathogenic mechanisms of PBC.
There were some statistical limitations to the current study. The sample size was small, particularly with the PSC and AIH groups, and a larger scale study to confirm our findings is necessary. In addition, the number of peaks reported in this paper was based on the smallest number of statistically significant peaks that could discriminate the classes. The possibility of achieving similar misclassification rates based on different subsets of peaks does exist. These limitations can easily be addressed by larger studies in the future.
In conclusion, we have used a novel proteomic approach to autoimmune liver diseases providing a new method that can be applied to these and other tissue-specific disorders of immunity. The peaks identified suggest the differential expression of proteins in bile ducts, inflammatory infiltrates and hepatocytes and may represent novel antigens in PBC, PSC and AIH. This study illustrates the promise and feasibility of in situ MALDI MS in the study of autoimmunity.
Acknowledgments
This work was supported by NIH/NIGMS 5R01 GM58008, Vanderbilt Ingram Cancer Center Core Support Grant P30 CA068485, NIH DK39588 and National Foundation for Cancer Research: Vanderbilt Center for Proteomics and Drug Action.
References
- Selmi C, Meda F, Kasangian A, Invernizzi P, Tian Z, Lian Z, et al. Experimental evidence on the immunopathogenesis of primary biliary cirrhosis. Cell Mol Immunol. 2010;7:1–10. doi: 10.1038/cmi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron JH, Bowlus CL. The immunobiology of primary sclerosing cholangitis. Semin Immunopathol. 2009;31:383–397. doi: 10.1007/s00281-009-0154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiki Y, Aoki CA, Bowlus CL, Shimoda S, Ishibashi H, Gershwin ME. T cell immunity in autoimmune hepatitis. Autoimmun Rev. 2005;4:315–321. doi: 10.1016/j.autrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Moritoki Y, Lian ZX, Ohsugi Y, Ueno Y, Gershwin ME. B cells and autoimmune liver diseases. Autoimmun Rev. 2006;5:449–457. doi: 10.1016/j.autrev.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Yanagisawa K, Shyr Y, Xu BJ, Massion PP, Larsen PH, White BC, et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet. 2003;362:433–439. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–2900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- Cornett DS, Mobley JA, Dias EC, Andersson M, Arteaga CL, Sanders ME, et al. A novel histology-directed strategy for MALDI-MS tissue profiling that improves throughput and cellular specificity in human breast cancer. Mol Cell Proteomics. 2006;5:1975–1983. doi: 10.1074/mcp.M600119-MCP200. [DOI] [PubMed] [Google Scholar]
- Amann JM, Chaurand P, Gonzalez A, Mobley JA, Massion PP, Carbone DP, et al. Selective profiling of proteins in lung cancer cells from fine-needle aspirates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Cancer Res. 2006;12:5142–5150. doi: 10.1158/1078-0432.CCR-06-0264. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Cornett DS, Caprioli RM. Molecular imaging of thin mammalian tissue sections by mass spectrometry. Curr Opin Biotechnol. 2006;17:431–436. doi: 10.1016/j.copbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Sanders ME, Dias EC, Xu BJ, Mobley JA, Billheimer D, Roder H, et al. Differentiating proteomic biomarkers in breast cancer by laser capture microdissection and MALDI MSJ Proteome Res200871500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis S, Koutroumbas K.Pattern recognition. Boston; Elsevier/Academic Press; 2006. [Google Scholar]
- Webb AR.Statistical pattern recognition. New Jersey; Wiley; 2002. [Google Scholar]
- Honda M, Kawai H, Shirota Y, Yamashita T, Kaneko S. Differential gene expression profiles in stage I primary biliary cirrhosis. Am J Gastroenterol. 2005;100:2019–2030. doi: 10.1111/j.1572-0241.2005.41662.x. [DOI] [PubMed] [Google Scholar]
- Aoki CA, Dawson K, Kenny TP, Gershwin ME, Bowlus CL. Gene expression by PBMC in primary sclerosing cholangitis: evidence for dysregulation of immune mediated genes. Clin Dev Immunol. 2006;13:265–271. doi: 10.1080/17402520600800085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Borozan I, Milkiewicz P, Sun J, Meng X, Coltescu C, et al. Gene expression profiling of early primary biliary cirrhosis: possible insights into the mechanism of action of ursodeoxycholic acid. Liver Int. 2008;28:997–1010. doi: 10.1111/j.1478-3231.2008.01744.x. [DOI] [PubMed] [Google Scholar]
- Furuta K, Sato S, Yamauchi T, Ozawa T, Harada M, Kakumu S. Intrahepatic gene expression profiles in chronic hepatitis B and autoimmune liver disease. J Gastroenterol. 2008;43:866–874. doi: 10.1007/s00535-008-2237-y. [DOI] [PubMed] [Google Scholar]
- Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246–253. doi: 10.1016/j.jaut.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Oertelt S, Rieger R, Selmi C, Invernizzi P, Ansari AA, Coppel RL, et al. A sensitive bead assay for antimitochondrial antibodies: chipping away at AMA-negative primary biliary cirrhosis. Hepatology. 2007;45:659–665. doi: 10.1002/hep.21583. [DOI] [PubMed] [Google Scholar]
- Bauer JA, Chakravarthy AB, Rosenbluth JM, Mi D, Seeley EH, de Matos Granja-Ingram N, et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin Cancer Res. 2010;16:681–690. doi: 10.1158/1078-0432.CCR-09-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]