Abstract
Pseudoxanthoma elasticum (PXE) is a heritable ectopic mineralization disorder caused by loss-of-function mutations in the ABCC6 gene which is primarily expressed in the liver. There is currently no effective treatment for PXE. In this study, we characterized bone marrow derived mesenchymal stem cells (MSCs) and evaluated their ability to contribute to liver regeneration, with the aim to rescue PXE phenotype. The MSCs, isolated from GFP-transgenic mice by magnetic cell sorting, were shown to have high potential for hepatic differentiation, with expression of Abcc6, in culture. These cells were transplanted into the livers of 4-week-old immunodeficient Abcc6−/− mice by intrasplenic injection one day after partial hepatectomy, when peak expression of the stromal cell derived factor-1 (SDF-1) in the liver was observed. Fluorescent bioimaging analyses indicated that transplanted MSCs homed into liver between day 1 and 7, and significant numbers of GFP-positive cells were confirmed in the liver by immunofluorescence. Moreover, enhanced engraftment efficiency was observed with MSCs with high expression levels of the chemokine receptor Cxcr4, a receptor for SDF-1. These data suggest that purified MSCs have the capability of differentiating into hepatic lineages relevant to PXE pathogenesis and may contribute to partial correction of the PXE phenotype.
1. Introduction
Pseudoxanthoma elasticum (PXE), a life-altering and frequently devastating disease, affects the skin, the eyes, and the cardiovascular system with ectopic mineralization [1, 2]. PXE is caused by the mutations in the ABCC6 gene, which encodes a member of the C-family of ATP-binding cassette transporters [3, 4]. Surprisingly, this gene appears to be expressed primarily in the liver and the kidneys, tissues not clinically affected in PXE [5]. We have developed an Abcc6 −/− mouse model by targeted ablation of the Abcc6 gene [6]. These mice recapitulate histopathologic features of human PXE, and serve as an excellent model system to study pathomechanisms leading to tissue mineralization as a result of Abcc6 inactivation, and they serve as a platform to evaluate the curative effects of different treatment modalities. Recently, we have demonstrated that PXE is a metabolic disorder with the primary pathology in the liver and with secondary involvement of elastic fibers in soft tissues [7, 8]. However, the precise function of ABCC6 protein, consequences of the ABCC6 mutations at the mRNA and protein levels, and the pathomechanisms leading to mineralization of the elastic fiber structures are largely unknown. Currently, there are no treatment modalities available for this disorder.
Various therapeutic strategies have been explored in clinical trials based on cutting-edge basic research on liver metabolic diseases. Gene therapy and cellular therapy are overlapping fields of biomedical research with similar therapeutic goals of tissue regeneration. However, relatively little progress has been made in gene therapy since the first clinical trial in 1990 [9]. Short-lived nature of gene therapy and problems of safety with viral vectors have kept gene therapy from becoming an effective treatment for many genetic diseases [10, 11]. Liver transplantation might be an effective therapy for severe liver diseases, but few patients can benefit from this procedure due to the shortage of donor organs. Moreover, the whole organ transplantation involves major surgery, is highly invasive and requires lifelong immunosupression. Currently, cellular therapy with stem cells and their progeny is a promising new approach capable of addressing mostly unmet medical needs. The considerable excitement surrounding the stem cell field is based on the unique biological properties of these cells and their capacity to self-renew and regenerate tissue and organ systems. Specifically, bone marrow stromal cells are an attractive source for cell-based gene therapy to genetic liver disorders, and their capability of differentiating into hepatocyte lineage has been demonstrated previously [12, 13]. The cell transplantation has been performed in several patients with modest liver metabolic correction, such as in the patients with Crigler-Najjar syndrome and with advanced liver failure [14–16]. It appears, therefore, that PXE would be an appropriate candidate disease to test cell-based therapeutics.
Hereby, we preliminarily evaluated a stem-cell-based therapeutic approach for PXE by assessment of the potential of MSCs in liver reconstitution with the aim to rescue the PXE phenotype in Abcc6 −/− mice and eventually on patients.
2. Materials and Methods
2.1. Mice and Cell Transplantation
An immunodeficient PXE mouse model [8], generated by crossbreeding the traditional Abcc6 −/− mouse [6] with a well-established immunodeficient Rag1 −/− mouse in C57BL/6 background (strain: 002216F; The Jackson Laboratory, Bar Harbor, ME), was used in this study. four-week-old mice were anesthetized and 50% partial hepatectomy (PHx) was performed following the standard protocol of Higgins and Anderson [17]. For the administration of cells, approximately 5 × 105 Cxcr4-MSCs or unmodified MSCs (see below) at passage 6, isolated from GFP-transgenic mice (C57BL/6-Tg UBC-GFP; The Jackson Laboratory) as the source of donor cells, were delivered into the recipient mouse liver by intrasplenic injection at 24 hours after PHx. The mice were maintained under pathogen-free conditions and were handled in accordance with the guidelines for animal experiments by the Institutional Animal Care and Use Committee of Thomas Jefferson University.
2.2. Bone Marrow Derived Stem Cell Isolation and Characterization
The method of magnetic cell sorting (MACS) was utilized to obtain desired populations of bone marrow derived mesenchymal stem cells (MSCs) as described by manufacturer (Miltenyi Biotec, Cambridge, MA). Briefly, wild-type mouse MSCs were harvested from 4-week-old GFP-transgenic mice and enriched by immunomagnetic separation strategies using cocktails of antibodies that deplete the differentiated cells of hematolymphoid lineages (Lin), such as the cells expressing the following lineage antigens: CD5, CD45R, CD11b, Gr-1, 7-4, and Ter-119. To obtain a pure population of stem cells, positive selection with Sca-1 antibody was utilized to further sort cells by MACS. These cells were maintained in the MSC medium containing MesenCult MSC Basal Medium (Stemcell Technologies, Vancouver, Canada), 20% Mesenchymal Stem Cell Stimulatory Supplements (Stemcell Technologies), 100 unit/mL penicillin (Invitrogen, Carlsbad, CA), 100 μg/mL streptomycin (Invitrogen), and 2.5 μg/mL amphotericin B (Invitrogen). The culture medium was changed every 3 days. Mouse MSCs at passage 6, identified as Lin−CD45−CD31−Sca-1+, were used as MSCs for the experiments.
To characterize the cell surface markers by flow cytometric analysis, harvested MSCs were incubated at 4°C for 30 minutes with rat anti-mouse CD11b, CD45, CD105, CD106, Sca-1, CD29, or MHC-1 antibody (R&D system, Minneapolis, MN), followed by 30 minutes incubation with APC-labeled rabbit anti-rat IgG antibody (BD Biosciences, San Jose, CA). Cells were examined by using FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ) and data were analyzed with Flowjo software (Tree Star Inc., Ashland, OR).
2.3. Transfection and Selection of MSCs Expressing Exogenous Cxcr4
MSCs at approximately 80% confluency were transfected with pCMV6-Kan/Neo carrying a full length cDNA of mouse Cxcr4 (Origene, Rockville, MD) using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. The transfected cells were maintained in the MSC medium and subjected to the selection by G418 at the dose of 1,500 μg/mL, and switched to the maintenance dose of 500 μg/mL in 2 weeks. The positive transfected cells were described as Cxcr4-MSCs in subsequent experiments.
2.4. In Vitro Hepatic Differentiation
Prior to starting the hepatic differentiation, MSCs at passage 5 were maintained in the regular MSC culture medium until at 80–90% confluence. The hepatic differentiation was elicited in the differentiation-inducing medium, which consisted of DMEM (Invitrogen) supplemented with 10% FBS, 10 ng/mL hepatocyte growth factor (HGF) (PeproTech Inc, Rocky Hill, NJ), 10 ng/mL basic fibroblast growth factor (bFGF) (PeproTech INC) and 10 ng/mL oncostatin M (R&D system). The medium was changed every 3 days, and the cells were cultured for 8 days.
2.5. Immunofluoresence
To analyze the protein expression in the differentiated MSCs, the cells either in the regular MSC culture medium or in the differentiation-inducing medium were fixed in 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100 at day 8. The cells were incubated with mouse anti-human mouse albumin (Alb) antibody (R&D system) or rabbit anti-mouse cytokeratin (CK)-18 antibody (Novus, Littleton, CO), followed by the incubation with the second antibody, Texas Red conjugated anti-rabbit IgG (Molecular Probe, Eugene, Oregon). DAPI was used for nuclear counterstaining.
To examine the presence of GFP positive transplanted MSCs in the liver, the engrafted livers were removed, fixed with 4% paraformaldehyde, processed in a gradient sucrose, and then subjected to immunofluorescent analysis. The processed livers were embedded in Tissue-tec OCT Compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan), and stored at −80°C until use. Six-μm-thick frozen sections of the liver were incubated with rabbit anti-GFP antibody (Invitrogen). Subsequently, sections were stained with FITC goat anti rabbit IgG secondary antibody (Invitrogen).
2.6. RT-PCR and qPCR
To examine the liver specific gene expression in differentiated MSCs, total RNA was extracted from the cultured regular MSCs or the differentiated MSCs at day 8 of culture using RNeasy Mini Kit (Qiagen, Hilden, Germany). RNA samples were subjected to random-primed reverse transcription by using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). RT reaction products were used for PCR for amplifying mouse CK-18, hepatocyte nuclear factor (HNF)-3b and Abcc6, with actin as an internal control. To analyze the expression of Cxcr4 in the transfected cells with pCMV6-Cxcr4 plasmid, RNA isolation and reverse transcription procedures were performed as above with on-column DNA digestion. The PCR was performed to amplify mouse Cxcr4 gene in transfected or untransfected cells with Gapdh as an internal control.
To quantitate the percentage of migrated GFP positive MSCs into the liver by qPCR, total DNA was extracted from homogenized engrafted liver using the DNeasy Kit (Qiagen). SYBR Green PCR amplification of GFP was performed in ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) using SYBER Green PCR Master Mix (Applied Biosystems). The amount of GFP+ DNA in each 50 ng DNA sample was quantified and normalized to interleukin (IL)-2 DNA. The relative expression level of the target gene was calculated using the ΔΔCt method. To prepare the standard curve, genomic DNA from the wild type mouse was spiked with serial dilutions of genomic DNA from the GFP-transgenic mouse. The diluted GFP standard samples were subjected to the PCR amplification of GFP and IL-2, and the percentage of GFP transgene per 50 ng tissue-derived DNA was calculated.
2.7. ELISA for Murine SDF-1
Serum samples were collected from the mice before PHx, and at 2 hours, 1 day, 3 days, and 7 days after the surgery. SDF-1 levels of mouse serum were determined using the mouse SDF-1 ELISA Kit (R&D system) according to the manufacturer's instructions. Liver samples were harvested from mice at the designated time points after the surgery and homogenized using a homogenizer and a 27 gauge needle in the RIPA buffer (Sigma-Aldrich, St. Louis, MO) supplemented with the proteinase inhibitor cocktail (Complete, Mini; Roche Applied Science, Indianapolis, IN) at 4°C. The mixture was centrifuged for 20 minutes at 12,000 rpm at 4°C and the supernatant was collected. The SDF-1 concentration in the liver extracts was examined by using the mouse SDF-1 ELISA Kit (R&D system) and normalized by the total protein concentration measured by BCA Protein Assay Kit (Pierce, Rockford, IL).
2.8. Migration Determination In Vitro and In Vivo
In vitro migration assays were carried out in a 48-well transwell using polycarbonate membranes with 8-μm pores (Osmonics; Livermore, CA). After different concentrations of mouse recombinant SDF-1 (R&D system) were added to the lower chamber, 2.5 × 103 MSCs or Cxcr4-MSCs in 30 μL of DMEM with 0.5% FBS were placed in the upper chamber of the transwell assembly. After incubation at 37°C and 5% CO2 for 4 hours, the upper surface of the membrane was scraped gently to remove nonmigrating cells and washed with PBS. The cells on the membrane were fixed in 4% paraformaldehyde for 15 minutes and stained with Giemsa. The number of migrating cells was determined by counting 3 random fields per well under the microscope at 100x magnification. Experiments were performed in triplicate.
To assess the in vivo migration ability of MSCs, fluorescence bio-imaging system was utilized. The mice transplanted with MSCs labeled with a fluorescent dye, Vybrant DiD (Molecular Probes, Eugene, OR) were monitored at multiple time points (day 1, week 1, week 2, and week 3) by using an IVIS Luminar XR live imaging system (Caliper, Hopkinton, MA) at excitation filter 644 nm and emission filter 665 nm.
2.9. Data and Statistical Analysis
Statistical analyses were performed with Student's t-test. P values <0.05 were considered statistically significant.
3. Results and Discussion
3.1. MSC Isolation and Cell-Surface Antigen Profile
Total bone marrow from GFP-transgenic mice was obtained by purging the medullar canal of the femurs and tibias with a 26 gauge needle, and MSCs were purified by magnetic cell sorting using antibodies against a panel of hematolymphoid lineage antigens for negative selection and then antibodies against Sca-1 for positive selection (see Section 2). Flow cytometric analysis of freshly isolated cells demonstrated that approximately 99% cell populations were both GFP and Sca-1 positive (data not shown). The cells were cultured in the regular MSC medium and passaged when they reached 80% confluence. Under the microscopic observation, the cells at early passage (passage 3) exhibited a thin spindle like shape and became large fibroblast-like in a swirl pattern at passage 5 (Figure 1(a)). Flow cytometric analyses showed a positive histogram peak for Sca-1 and CD29, but both positive and negative histogram peaks for CD11b, CD45, and CD106 were observed with the cells at passage 3. The histogram positive peak for CD11b and CD45, and negative peak for CD106 disappeared at passage 5 (Figure 1(b)). These observations indicated the presence of two populations: hematopoietic stem cells (HSCs) and MSCs, and that a significant enrichment in MSCs was gained during subsequent culturing of the cells. MSCs have been shown to be a resource of hepatic differentiation, but there are no specific markers of MSCs. Thus, to obtain purified population of MSCs, a further purification may be needed by in vitro culture following selection using by a group of surface markers.
Figure 1.
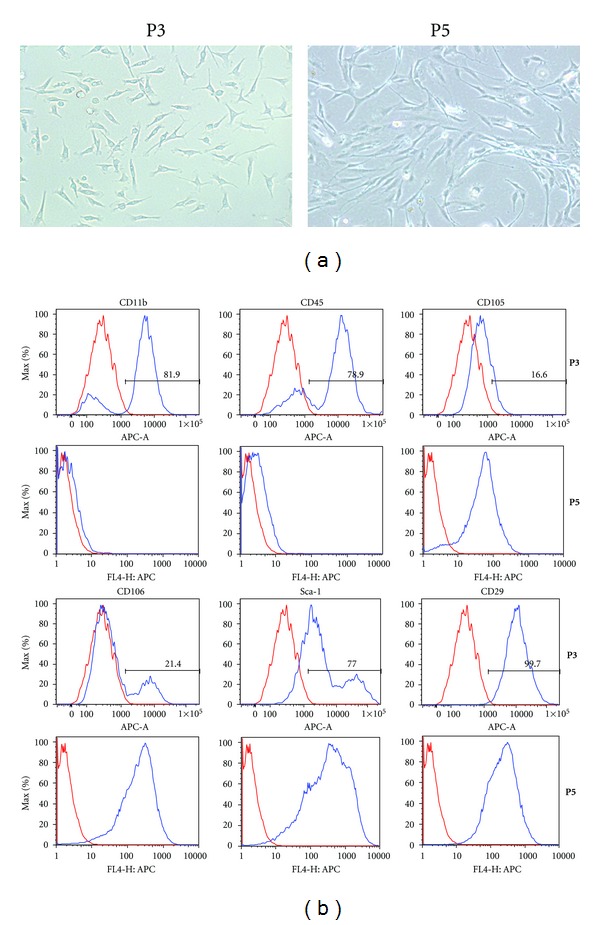
Characterization of the mouse bone marrow derived stem cells. The bone marrow derived stem cells were isolated by magnetic cell sorting using a Lin− cocktail of antibodies as well as stem cell antigen-1 (Sca-1) antibody and then cultured in the regular stem cell culture medium. (a) The cells were observed by phase contrast microscopy at passage 3 (left) or passage 5 (right). The cells at passage 5 became more fibroblastic appearing compared to passage 3. (b) Flow cytometry was utilized to examine the surface markers of cells at passage 3 (top panels) or passage 5 (bottom panels) for the purity of cell population.
The cells at passage 3, 5, or 7 were also analyzed by FACS for their immunologic properties using monoclonal antibody to mouse MHC class I. MSCs (passage 3, 5, and 7) derived from the same mouse expressed high levels of MHC class I when compared to melanoma derived cells (BL-TAC, negative control) and mouse fibrosarcoma derived cells (MC57G, positive control) (Figure 2). In the literature, it has been suggested that MSCs may be immunoprivileged and can be transplantable between MHC-incompatible individuals [18]. On the other hand, it has been suggested that MSCs are not intrinsically immunoprivileged and could not serve as a “universal donor” in immunocompetent MHC-mismatched recipients [19]. In our study, high expression of MHC class I supported the latter possibility and suggested that modulation of immune response may be of concern when these cells are employed in vivo.
Figure 2.
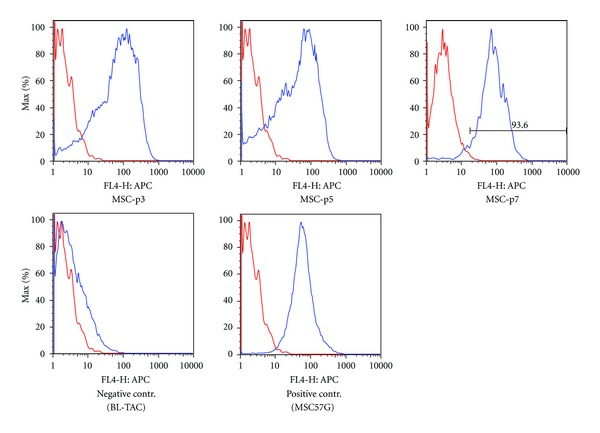
Flow cytometric characterization of MHC class I expression in bone marrow derived stem cells. The cells at passage 3 (MSC-p3), 5 (MSC-p5) or 7 (MSC-p7) were analyzed by FACS using monoclonal antibody to mouse MHC class I. Mouse melanoma derived cells (BL-TAC) and mouse fibrosarcoma derived cells (MSC57G) were used as negative and positive controls, respectively.
3.2. Differentiation Potential of Bone Marrow Stem Cells into Hepatocytes In Vitro
To investigate the potential of hepatic differentiation of MSCs, the purified MSCs at passage 6 were cultured for 8 days in differentiation-inducing medium, that is, DMEM containing 10% fetal bovine serum, 10 ng/mL HGF, 10 ng/mL bFGF, and 10 ng/mL oncostatin M. The cell morphology changed as they became extended and larger (Figure 3(a), top panel). Immunofluorescence analysis was performed for the expression of the hepatocyte-specific proteins and demonstrated that CK-18 and Alb were positively expressed in the differentiated cells but not in the undifferentiated ones (Figure 3(a), middle and bottom panels). HNFs and CK-18, key players for the hepatogenesis, were expressed in the differentiated cells by RT-PCR, while negative in control cells cultured with the MSC culture medium for the same period (Figure 3(b)). MSCs, maintained either in differentiation-inducing medium or under basal condition, expressed Abcc6 mRNA at comparable levels.
Figure 3.
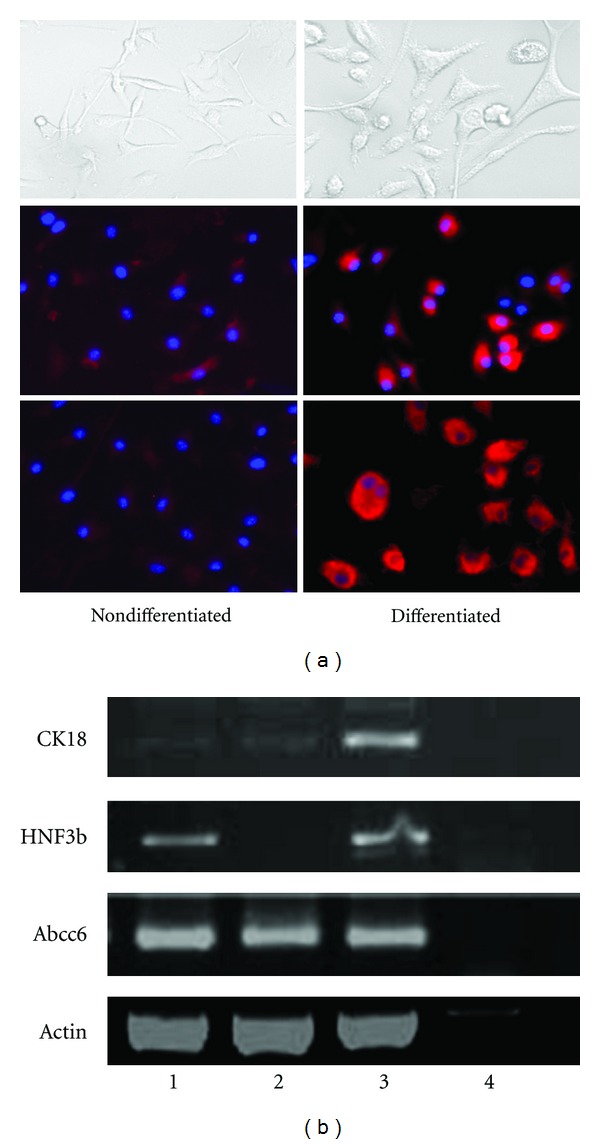
Hepatocytic differentiation capacity of mouse bone marrow stem cells in vitro. The cells at passage 5 were cultured either in the regular stem cell culture medium (a, left panels) or in the differentiation-inducing medium (a, right panels). As observed by phase contrast microscopy at day 8 (top panels), the cells became extended and larger in the differentiation-inducing medium (a, right top). Immunofluorescent analysis was performed at day 8 of differentiation. Cells were stained with antibodies to liver-specific marker proteins, albumin (a, middle panels), or CK-18 (a, bottom panels). DAPI staining was used to identify the nuclei (blue). Strong signals were observed in the cells cultured in the differentiation-inducing medium (a, right middle and bottom panels), while weak signal (a, left middle and bottom panels), if any, was detected in the cells cultured in the regular stem cell medium. (b) RT-PCR was conducted and the mRNA levels of different liver-specific genes were examined. They are cytokeratin-18 (CK-18), hepatocyte nuclear factor 3b (HNF3b) and Abcc6, respectively. Lane 1: cultured cells in differentiation-inducing medium; lane 2: cultured cells in stem cell culture medium; lane 3: MLE-10 (mouse liver epithelial cell line); lane 4: H2O blank.
The differentiation process of stem cells requires a specific microenvironment. The results clearly demonstrated that the isolated mouse MSCs are able to differentiate into hepatocytes under in vitro conditions in medium containing growth factors HGF, bFGF and oncostatin M, which have been reported to be important components for liver development and differentiation [20, 21]. In addition, our differentiated cells could show the expression of the Abcc6 gene, which is lost in PXE and may be needed to restore its phenotype. These data provide critical information about stem cell biology that should contribute to the development of regenerative medicine for liver diseases in general and for PXE in particular.
3.3. Migration Capability of Bone Marrow Stem Cells
3.3.1. MSCs Cxcr4 Expression and In Vitro Migration
MSCs at passage 4 were transfected with a murine Cxcr4 expression vector driven by CMV promoter, pCMV6-mCxcr4, and then selected with G418 to obtain cells with stable expression (Cxcr4-MSCs). The Cxcr4-MSCs were analyzed for expression of Cxcr4 first by RT-PCR. High level of Cxcr4 mRNA was observed in these cells while lower, barely detectable levels were present in untransfected MSCs (Figure 4).
Figure 4.
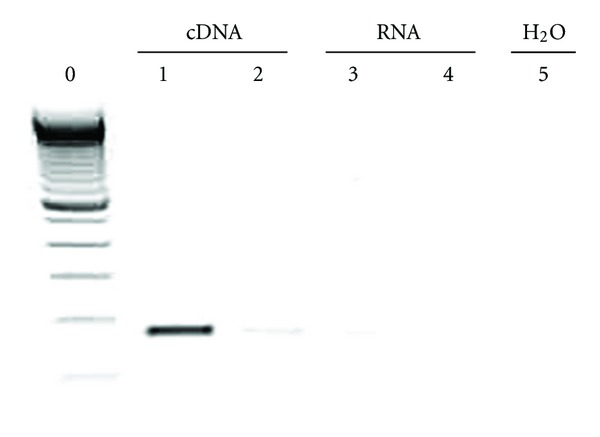
Overexpression of Cxcr4 in bone marrow derived stem cells. The cells at passage 5 were transfected with a mouse Cxcr4 expression vector (pCMV6-Cxcr4) and the stable cell clones expressing Cxcr4 were established. RT-PCR was conducted to examine the mRNA levels of mouse Crcx4 in the transfected or untransfected stem cells at the same passage. Lane 0: 100 bp DNA ladder; lane 1: reverse transcribed sample from the transfected cells; lane 2: reverse transcribed sample from the untransfected cells; lane 3: nonreverse transcribed sample from the transfected cells; lane 4: nonreverse transcribed sample from the untransfected cells; lane 5: H2O blank.
To evaluate cell migration in response to SDF-1, the ligand for Cxcr4, we examined whether exogenous overexpression of Cxcr4 enhances the chemotaxis of MSCs towards an SDF-1 gradient in a transwell migration assay. Cxcr4 modification of MSCs increased the number of cells migrating towards SDF-1 in a dose-dependent pattern (Figure 5(a)). Specially, the number of migrated cells significantly increased at the dose of 30 and 60 ng/mL when compared to that without adding SDF-1 (Figure 5(b)). Few migrated cells were found in untransfected MSCs with all different dose settings of SDF-1 (data not shown). These data indicate that overexpression of Cxcr4 enhances the ability of MSCs to respond to SDF-1 induced chemotaxis.
Figure 5.
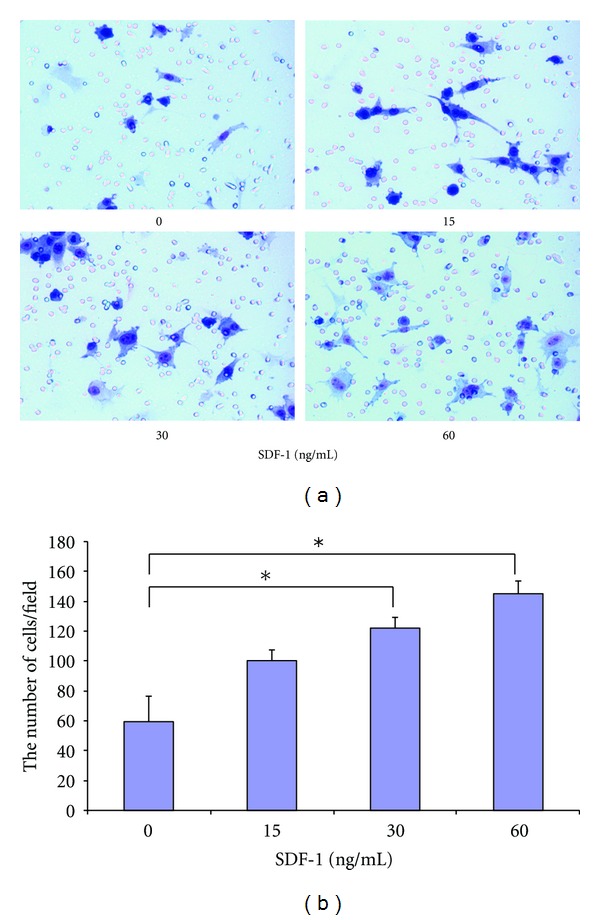
In vitro migration of Cxcr4-MSCs. (a) Representative images of transmigrated Mesenchymal stem cells (MSCs) stably overexpressing Cxcr4 in response to stromal-derived factor-1 (SDF-1) at concentrations of 0, 15, 30, or 60 ng/mL in transwell assay. (b) Average number of cells migrated in transwell migration assay counted in 100x magnification field. Results are mean ± SEM of 3 different fields from 3 independent experiments. The asterisks indicate statistically significant differences, P < 0.05 (Student's t-test).
3.3.2. The Levels of SDF-1 in the Liver and the Serum after Partial Hepatectomy
To investigate how partial hepatectomy affects the levels of SDF-1 in the liver tissue or in the blood, we first determined the temporal expression of SDF-1 mRNA in the hepatectomized livers by quantitative RT-PCR. SDF-1 increased at day 1 by more than 2-fold and then decreased gradually to reach the baseline level as before partial hepatecomy at day 7 (Figure 6(c)). Using the total liver protein extracts, ELISA showed a significant increase in the hepatectomized livers at day 1 and then the levels of SDF-1 quickly returned down to the baseline at day 3 and 7 after partial hepatectomy (Figure 6(d)), consistent with the changes at mRNA levels. Conversely, the serum levels of SDF-1 as measured by ELISA were decreased at 2 and 24 hours, and then returned to the baseline by 180 hours (day 7) (Figure 6(b)).
Figure 6.
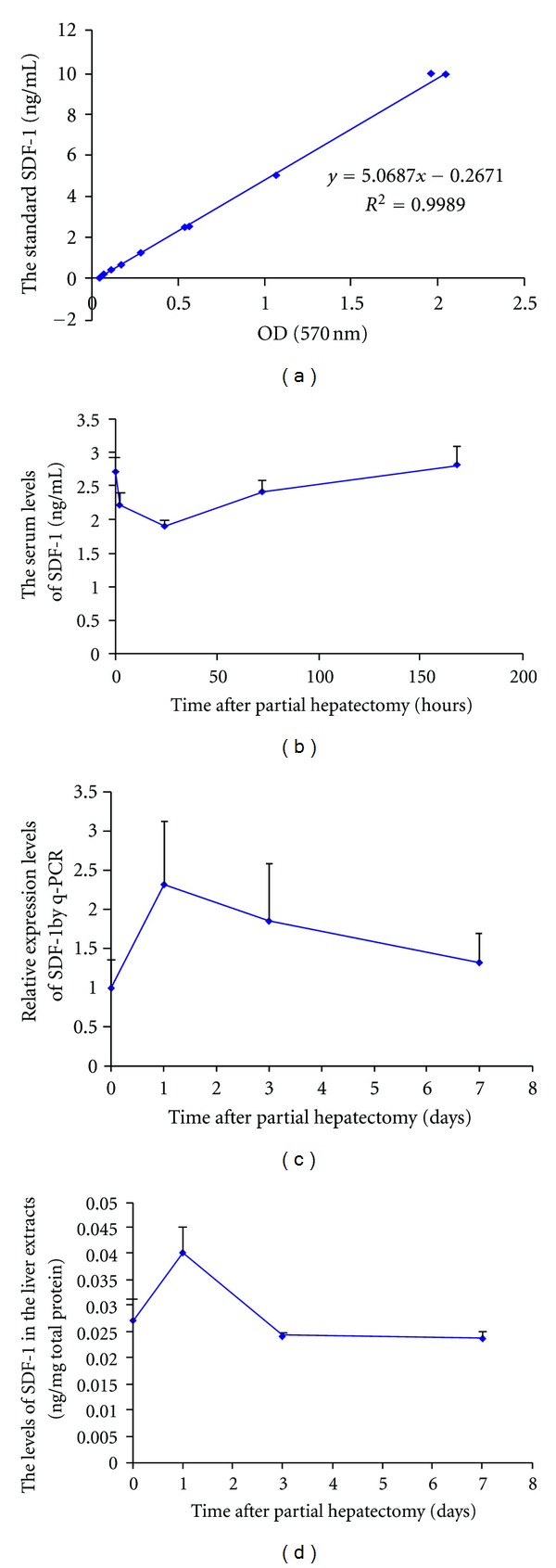
The levels of SDF-1 in the liver or the serum after partial hepatectomy. Serum samples (b) or liver extracts (d) were obtained from the mice at the different time points after partial hepatectomy, and analyzed to determine the levels of SDF-1 using ELISA (n = 3–5). The levels of SDF-1 were calculated based on the standard curve (a). RT-PCR was performed to examine the mRNA level of SDF-1 in the hepatectomized livers (c).
3.3.3. Homing of Transplanted MSCs towards the Liver
At 24 hours after partial hepatectomy in immunodeficient Abcc6 −/− mouse model, Abcc6 −/−; Rag1 −/−, 5 × 105 Cxcr4-MSCs or unmodified MSCs were administrated by intrasplenic injection. The transplanted mice were monitored for DiD-labeled MSCs for their migration towards the liver by a live IVIS Luminar XR imaging system. DiD is a fluorescent tracer which allows cells to be marked in distinctive color detectable by the imaging machine. DiD labeled MSCs were detected in the liver and spleen at day 1 and day 7 after transplantation (Figure 7(a)). The presence of GFP-positive MSCs at day 7 in the liver was confirmed by immunofluoresence using anti-GFP antibody (Figure 7(b)). Moreover, to quantitate the percentage of migrated cells, genomic DNA was isolated from the harvested liver at day 7 and qPCR was performed. Using a serially diluted GFP DNA samples as the standards, the average percentage of GFP-positive Cxcr4-MSCs in the liver was 0.22%, whereas it was 0.15% in the liver from mice transplanted with MSCs without Cxcr4 transfection (data not shown).
Figure 7.
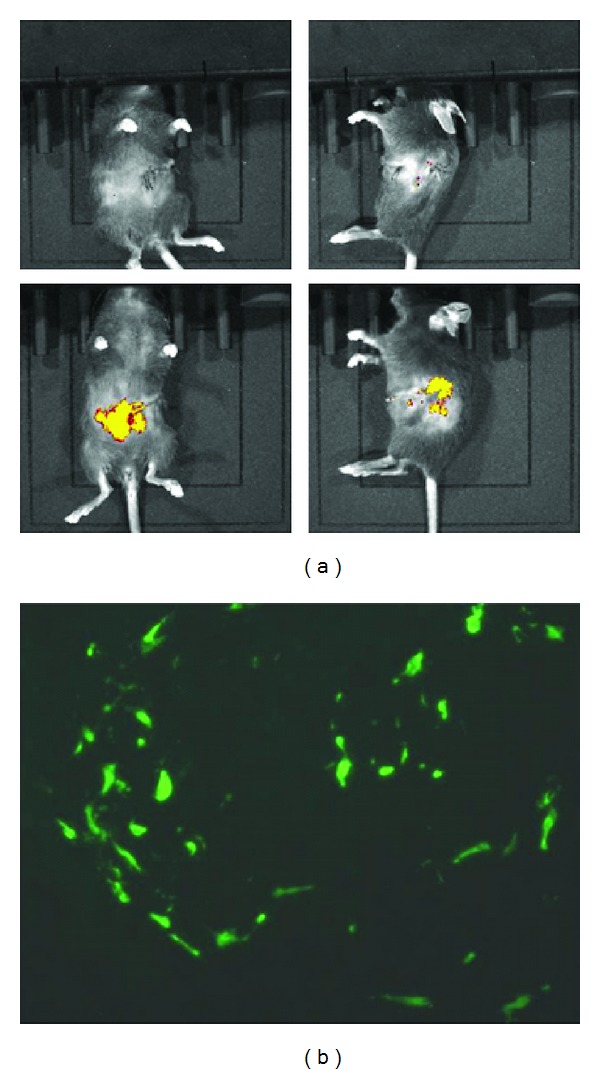
Migration of MSCs towards the hepatectomized liver through splenic vein. 5 × 105 Cxcr4-MSCs labeled with Vybrant DiD dye were administered to Abcc6 −/−; Rag1 −/− mice via spleen 24 hours after partial hepatectomy, and the livers were examined at day 7 of transplantation for the homing of the transplanted cells. (a) Representative fluorescence images of hepatectomized mice after MSCs transplantation at Day 7 (lower panels). The images were examined from the ventral (left panels) and lateral (right panels) sides in comparison with PBS injected hepatectomized mice as negative control (upper panels). (b) Six-μm frozen sections were examined by immunofluoresence with an anti-GFP antibody, demonstrating the presence of GFP positive cells in the liver of mice at 7 days after intrasplenic transplantation.
The interaction of Cxcr4 and its ligand SDF-1 is reported to play a major role in homing of MSCs into tissues [22]. Cells in the injured organs highly express SDF-1, causing an elevation of localized SDF-1 levels, and the gradient of SDF-1 recruits MSCs into the injury site via chemotactic attraction through the interaction between SDF-1 and Cxcr4 [23, 24]. Thus, Cxcr4 has been recently used to enhance homing and engraftment of stem cells through increasing cell invasion in response to SDF-1 [23]. In this study, the local expression of SDF-1 was induced by PHx and attracted MSCs, especially Cxcr4-MSCs, toward the hepatectomized liver. Increased SDF-1 concentration was shown in the liver, but decreased SDF-1 in the blood, at 24 hours after PHx, at which time point we delivered the cells. In the mouse model of myocardial infarction, Abbott et al. reported that the SDF-1 expression increased in the peri-infarct zone, but its serum level decreased after the infarction, consistent with our result [22]. This gradient of SDF-1 may contribute to the recruitment of circulating progenitor cells expressing Cxcr4 into the hepatectomized liver (Figure 8).
Figure 8.
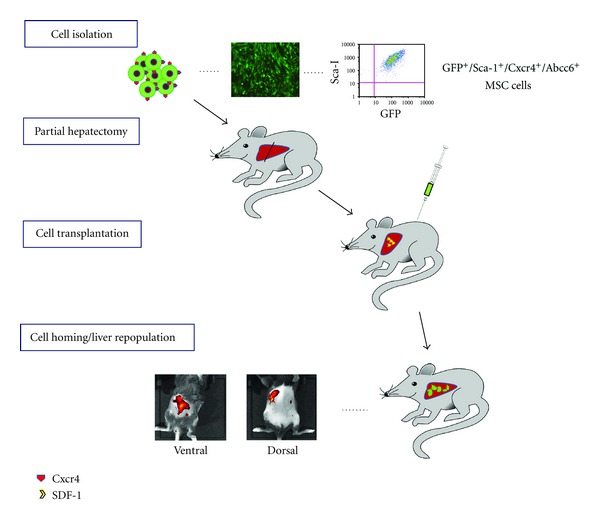
Schematic depiction of liver targeting of MSCs through intrasplenic injection. GFP-transgenic mouse bone marrow derived MSCs, which are positive for GFP, Sca-1, Cxcr4, and Abcc6, were injected through spleen 24 hours after partial hepatecotomy inducing release of SDF-1, a ligand of Cxcr4. The presence of transplanted cells was observed by a live imaging system at day 7 posttransplantation through ventral view (liver) and dorsal view (spleen).
4. Conclusions
The purpose of our study was to evaluate the potential of MSCs to differentiate into hepatic cells and for their efficient recruitment to the liver in the setting of partial hepatectomy. We intrasplenically delivered Lin−Sca1+ MSCs from Abcc6 +/+ GFP+ mice and identified donor cells by the presence of GFP in the liver. We did confirm the capability of MSCs to differentiate toward hepatocytic lineage in vitro by examining hepatic specific gene expression at mRNA and protein levels, including Abcc6 gene expression. We also observed homing response to SDF-1, and overexpression of Cxcr4 resulted in a significant additional increase in donor-cell migration both in vitro and in vivo studies. The transplanted MSCs reside in the recipient liver for at least up to 10 days.
Over the decades, many studies have utilized bone marrow-derived cells for cell transplantation therapy in animal models of various disorders. The reported treatment effects are variable, which may be related to differences in cell type and quantity of transplanted cells, timing and approach of cell transplantation and disorder model selection. Aurich et al. demonstrated the presence of functional transplanted cells in the recipient liver at 14 weeks after administration of human bone marrow MSCs through intrasplenic injection using an immunodeficient Pfp−/−/Rag2−/− mouse model, which has malfunction of NK cells and depletion of mature B and T cells [25]. The long term presence of transplanted cells in recipient liver may depend on the background of recipient mice and kind of transplanted MSCs, which requires further long term study.
Immune rejection is a potential problem of cell transplantation for treatment of patients. To overcome immune rejection, or to avoid it, the stem cells must either be derived from the recipient or identical twin, or the stem cells must be engineered in a way to circumvent the immune reaction. Currently, the approach that appears most promising entails using embryonic stem cells whose DNA has been replaced with the recipient's DNA, thus becoming “self”, the so-called therapeutic cloning [26].
In summary, these data suggest that purified MSCs have the capability of differentiating into hepatic lineages and homing to the liver. The interactions between SDF-1 and Cxcr4 could be a critical mechanism to recruit MSCs for liver targeting and regeneration, and further studies will define the optimal conditions to maintain and propagate MSCs in the target organs, such as in the liver in case of PXE.
Conflict of Interests
The authors state no conflict of interests.
Acknowledgments
The authors thank Dian Wang and Bethany Reutemann for technical assistance. This study was supported by the NIH/NIAMS Grants K08AR057099 (QJ) and R01AR28450 (JU).
Abbreviations
- PXE:
Pseudoxanthoma elasticum
- MSCs:
Mesenchymal stem cells
- SDF-1:
Stromal cell-derived factor 1
- Cxcr4:
CXC chemokine receptor 4
- HSCs:
Hematopoietic stem cells
- PHx:
Partial hepatectomy.
References
- 1.Li Q, Jiang Q, Pfendner E, Váradi A, Uitto J. Pseudoxanthoma elasticum: clinical phenotypes, molecular genetics and putative pathomechanisms. Experimental Dermatology. 2009;18(1):1–11. doi: 10.1111/j.1600-0625.2008.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neldner KH. Pseudoxanthoma elasticum. Clinics in dermatology. 1988;6(1):1–159. doi: 10.1016/0738-081x(88)90003-x. [DOI] [PubMed] [Google Scholar]
- 3.Ringpfeil F, Lebwohl MG, Christiano AM, Uitto J. Pseudoxanthoma elasticum: mutations in the MRP6 gene encoding a transmembrane ATP-binding cassette (ABC) transporter. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):6001–6006. doi: 10.1073/pnas.100041297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergen AAB, Plomp AS, Schuurman EJ, et al. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nature Genetics. 2000;25(2):228–231. doi: 10.1038/76109. [DOI] [PubMed] [Google Scholar]
- 5.Matsuzaki Y, Nakano A, Jiang QJ, Pulkkinen L, Uitto J. Tissue-specific expression of the ABCC6 gene. Journal of Investigative Dermatology. 2005;125(5):900–905. doi: 10.1111/j.0022-202X.2005.23897.x. [DOI] [PubMed] [Google Scholar]
- 6.Klement JF, Matsuzaki Y, Jiang QJ, et al. Targeted ablation of the Abcc6 gene results in ectopic mineralization of connective tissues. Molecular and Cellular Biology. 2005;25(18):8299–8310. doi: 10.1128/MCB.25.18.8299-8310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Q, Endo M, Dibra F, Wang K, Uitto J. Pseudoxanthoma elasticum is a metabolic disease. Journal of Investigative Dermatology. 2009;129(2):348–354. doi: 10.1038/jid.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Q, Oldenburg R, Otsuru S, Grand-Pierre AE, Horwitz EM, Uitto J. Parabiotic heterogenetic pairing of Abcc6−/−/Rag1−/− mice and their wild-type counterparts halts ectopic mineralization in a murine model of pseudoxanthoma elasticum. American Journal of Pathology. 2010;176(4):1855–1862. doi: 10.2353/ajpath.2010.090983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culver KW, Culver KW. Measuring success in clinical gene therapy research. Molecular Medicine Today. 1996;2(6):234–236. doi: 10.1016/1357-4310(96)88803-4. [DOI] [PubMed] [Google Scholar]
- 10.Gardlík R, Pálffy R, Hodosy J, Lukács J, Turňa J, Celec P. Vectors and delivery systems in gene therapy. Medical Science Monitor. 2005;11(4):RA110–RA121. [PubMed] [Google Scholar]
- 11.Woods NB, Bottero V, Schmidt M, Von Kalle C, Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440(7088):p. 1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]
- 12.Kuo TK, Hung SP, Chuang CH, et al. Stem cell therapy for liver disease: parameters governing the success of using bone marrow mesenchymal stem cells. Gastroenterology. 2008;134(7):2111–e3. doi: 10.1053/j.gastro.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lysy PA, Campard D, Smets F, Najimi M, Sokal EM. Stem cells for liver tissue repair: current knowledge and perspectives. World Journal of Gastroenterology. 2008;14(6):864–875. doi: 10.3748/wjg.14.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellor S, Shupe T, Petersen B. Stem cell therapy for inherited metabolic disorders of the liver. Experimental Hematology. 2008;36(6):716–725. doi: 10.1016/j.exphem.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lysy PA, Najimi M, Stéphenne X, Bourgois A, Smets F, Sokal EM. Liver cell transplantation for Crigler-Najjar syndrome type I: update and perspectives. World Journal of Gastroenterology. 2008;14(22):3464–3470. doi: 10.3748/wjg.14.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matas AJ, Sutherland DER, Steffes MW. Hepatocellular transplantation for metabolic deficiencies: decrease of plasma bilirubin in Gunn rats. Science. 1976;192(4242):892–894. doi: 10.1126/science.818706. [DOI] [PubMed] [Google Scholar]
- 17.Higgins GM, Anderson RM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgerical removal. Archives of Pathology. 1931;12(2):186–202. [Google Scholar]
- 18.Le Blanc K, Tammik C, Rosendahl K, Zetterberg E, Ringdén O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Experimental Hematology. 2003;31(10):890–896. doi: 10.1016/s0301-472x(03)00110-3. [DOI] [PubMed] [Google Scholar]
- 19.Eliopoulos N, Stagg J, Lejeune L, Pommey S, Galipeau J. Allogeneic marrow stromal cells are immune rejected by MHC class I- and class II-mismatched recipient mice. Blood. 2005;106(13):4057–4065. doi: 10.1182/blood-2005-03-1004. [DOI] [PubMed] [Google Scholar]
- 20.Ishii K, Yoshida Y, Akechi Y, et al. Hepatic differentiation of human bone marrow-derived mesenchymal stem cells by tetracycline-regulated hepatocyte nuclear factor 3β. Hepatology. 2008;48(2):597–606. doi: 10.1002/hep.22362. [DOI] [PubMed] [Google Scholar]
- 21.Shi XL, Mao L, Xu BY, et al. Optimization of an effective directed differentiation medium for differentiating mouse bone marrow mesenchymal stem cells into hepatocytes in vitro. Cell Biology International. 2008;32(8):959–965. doi: 10.1016/j.cellbi.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Abbott JD, Huang Y, Liu D, Hickey R, Krause DS, Giordano FJ. Stromal cell-derived factor-1α plays a critical role in stem cell recruitment to the heart after myocardial infarction but is not sufficient to induce homing in the absence of injury. Circulation. 2004;110(21):3300–3305. doi: 10.1161/01.CIR.0000147780.30124.CF. [DOI] [PubMed] [Google Scholar]
- 23.Karp JM, Leng Teo GS. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4(3):206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Cheng Z, Ou L, Zhou X, et al. Targeted migration of mesenchymal stem cells modified with CXCR4 gene to infarcted myocardium improves cardiac performance. Molecular Therapy. 2008;16(3):571–579. doi: 10.1038/sj.mt.6300374. [DOI] [PubMed] [Google Scholar]
- 25.Aurich I, Mueller LP, Aurich H, et al. Functional integration of hepatocytes derived from human mesenchymal stem cells into mouse livers. Gut. 2007;56(3):405–415. doi: 10.1136/gut.2005.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadereit S, Trounson A. In vitro immunogenicity of undifferentiated pluripotent stem cells (PSC) and derived lineages. Seminars in Immunopathology. 2011;33(6):551–562. doi: 10.1007/s00281-011-0265-9. [DOI] [PubMed] [Google Scholar]


