Abstract
Proteomic analyses of readily obtained human fluids (e.g., serum, urine, and saliva) indicate that the diagnosis of complex diseases will be enhanced by the simultaneous measurement of multiple biomarkers from such samples. This paper describes the development of a nanoparticle-based multiplexed platform that has the potential for simultaneous readout of large numbers of biomolecules. For this purpose, we have chosen pancreatic adenocarcinoma (PA) as a test bed for diagnosis and prognosis. PA is a devastating form of cancer in which an estimated 86% of diagnoses resulted in death in the United States in 2010. The high mortality rate is due, in part, to the asymptomatic development of the disease and the dearth of sensitive diagnostics available for early detection. One promising route lies in the development of a serum biomarker panel that can generate a signature unique to early stage PA. We describe the design and development of a proof-of-concept PA biomarker immunoassay array coupled with surface enhanced Raman scattering (SERS) as a sensitive readout method.
1. Introduction
Diagnostic tests play a vital role in disease detection. The results from such tests provide data essential to the timely identification of a disease, informed decisions on patient treatment, and tracking the response to therapy.1 Some diseases, however, are asymptomatic early in their progression, manifest symptoms not easily linked to a specific disease, or have symptoms and biomarkers that differ from patient to patient (e.g., ovarian2 and pancreatic3 cancers). These diseases are all too often detected at late stages of progression, resulting in high mortality rates.
A key component to addressing this situation requires a paradigmatic shift in the capabilities of today’s diagnostic test platforms. Most of these platforms measure the presence of one disease marker in a single specimen, which unfortunately can limit the ability to accurately identify the disease and monitor its progression and response to therapy.
Strategies for early detection require high clinical sensitivity (i.e., the ability of a test to correctly identify those patients with a disease) and extremely high clinical specificity (i.e., the ability of a test to correctly identify those patients without the disease) to attain a clinically actionable positive predictive value.4, 5 When functioning alone, conventional markers fall short of this required clinical sensitivity or specificity.6 Furthermore, a recent theoretical treatment has shown that if the detection strategy is based on a blood-based biomarker shed by a developing tumor burden, it may take upwards of ten years before in vivo concentrations are detectable by current clinical blood assays.7 As a result, a movement toward developing multiple markers for one disease has been gaining momentum.8–14 The potential to provide superior diagnostic and prognostic information using multimarker strategies has stimulated the development of assays capable of parallel measurements of multiple biomarkers on the same specimen – multiplexed assays. The ability to measure multiple protein markers simultaneously is attractive for several reasons. First, combining protein markers in panels can theoretically achieve acceptable levels of clinical sensitivity and specificity. Second, specimen conservation is critical due to the difficulty and expense in obtaining the well-characterized disease cohorts necessary to develop screening tools for disease markers. Third, analytical performance may be improved as a result of reduced sample handling. Fourth, combining several measurements into one test effectively lowers costs and turnaround-time. However, hurdles remain in developing a platform able to realize the benefits multiplexed analysis including: (1) the inability of existing platforms to rapidly and simultaneously detect the appearance of multiple cancer markers at exceedingly low concentrations and to accurately track subtle, but potentially important longitudinal progression in marker concentrations; (2) the high capitalization/labor costs and turn-around times associated with such platforms for translational use; and (3) the paucity of available clinical samples from early disease stages to identify markers of diagnostic value. It is therefore apparent that improvements in early diagnosis require the development of assay platforms that can assess a panel of biomarkers for the discovery, validation, and application of a biomarker signature unique to disease stage and that can also detect biomarkers at levels below those achievable today.
In this paper, we demonstrate that a multiplexed platform using surface-enhanced Raman scattering (SERS) as a readout tool has the potential to meet these requirements. To these ends, the following describes the construction, calibration, and preliminary testing of an array designed to detect two pancreatic adenocarcinoma (PA) biomarkers. As a proof-of-concept demonstration, multiplexed SERS results are compared to individually obtained ELISA (enzyme linked immunosorbent assay) results.
Given the inherent analytical sensitivity (i.e., the ability of a test to measure small differences in concentration or the slope of calibration curve for the substance under investigation) and potential for multiplexed detection, other research groups have begun investigating the utility of SERS for diagnostic uses.12, 15–18 In a recent report using the SERS detection method extended by the work detailed herein, we have shown that the analysis of a potential biomarker for PA (i.e., the mucin protein MUC4) in serum is feasible, but that this and possibly other potential early stage markers may be present at levels below the detection capabilities of ELISA.19 Several reports argue that panels composed of marker combinations will have increased power to accurately diagnose PA over any single marker alone.20–22 A panel approach may require multiple markers to realize an effective diagnostic algorithm because malignancies develop from different combinations of multiple genetic lesions. That is, one marker may correctly identify a subset of PA subjects, whereas other markers may identify another, overlapping subset. With sufficient overlap, a marker panel should have increased accuracy over the individual markers. Indeed, a recent meta-analysis of genomic and proteomic studies23 concluded there are at least 162 secreted proteins over-expressed in PA tumors that should be evaluated as potential markers and inclusion in a panel. We believe that the successful development of the proposed platform will prove invaluable in enabling these and many other related studies to become commonplace.
As a readout method for immunoassays, detection levels with SERS rivals those of fluorescence,24 and has been used in our laboratory for low-level detection of pathogens, biowarfare agents, biomarkers, and other molecules of interest to the medical and veterinary sciences.25–33
Two relevant and well-established markers were utilized to further the development of our platform for PA: serum carbohydrate antigen 19-9 (CA 19-9) and matrix metalloproteinase-7 (MMP-7). CA 19-9 is currently the only validated serum marker for pancreatic cancer. While currently finding use in postoperative surveillance, CA 19-9 is not clinically sensitive and/or clinically specific enough to serve as an early marker for PA.34–36 Moreover, ~10% of the population are unable to synthesize CA 19-9.10, 37 MMP-7 is a member of the protein family involved in the breakdown of extracellular matrix in normal physiological processes. Unlike other MMPs, which are primarily expressed by stromal cells, MMP-7 expression is restricted to glandular epithelium, and has been implicated in early tumor development.21, 38, 39 Serum levels of CA 19-9 and MMP-7 are presently assessed using individual commercially available ELISA kits. Although effective, this approach often requires at least 25 μL of serum for each test and may not detect low levels of either marker. For example, in the case of CA 19-9 serum levels, the minimum detectable level reported by the ELISA vendor is 5 U/mL (~3 ng/mL).40
Our preliminary results indicate that the SERS platform: (1) is amenable to analyte multiplexing, (2) only requires a tenth of the sample volume required by ELISA, and (3) has superior limits of detection (LOD) compared to ELISA. The following sections describe the results of this immunoassay array in comparison to those obtained using standard ELISA analysis.
2. Materials and methods
2.1. Reagents
Gold nanoparticles (AuNPs) (60 nm, ~1×1010 particles/mL) were purchased from BBInternational. Dithiobis(succinimidyl propionate) (DSP), bovine serum albumin (BSA), Tween 20, and NaCl were from Sigma Aldrich. Phosphate buffered saline (PBS: 10 mM; pH 7.4) and borate buffer packs (BB: 50 mM; pH 8.5) were acquired from Pierce. 5,5′-dithiobis(succinimidyl-2-nitrobenzoate) (DSNB) was synthesized per an earlier procedure.5 Instant nonfat powdered milk was obtained locally. The SERS assay used two different sets of monoclonal antibodies (mAbs). For MMP-7, the same lyophilized MMP-7 mAb was employed to modify the capture substrate and ERLS, along with recombinant human MMP-7 (rh-MMP-7; ~29 kDa) as the antigen; these reagents were purchased from R&D Systems. The work on CA 19-9 used 10-C04E and 10-C04C clones as respective capture and tracer mAbs and CA 19-9 antigen (~210 kDa); these reagents were obtained from Fitzgerald Industries International. Pooled human serum was from Innovative Research. Deidentified patient serum samples, designated as DIDM17, DIDM30, and DIDM199, were collected from healthy males over the age of 50 who were sent to the University of Utah clinic for prostate specific antigen tests.
Two sets of ELISA kits were used. The kit for MMP-7 was purchased from R&D Systems (Quantikine Human MMP-7 Immunoassay, Product number DMP700; Minneapolis, MN), and that for CA 19-9 was obtained from Diagnostic Automation (Gastrointestinal Cancer Antigen CA 19-9 Enzyme immunoassay, Product number 6909; Calabasas, CA).
2.2. Preparation of ERLs
ERLs are designed to yield a strong Raman signal and demonstrate immunospecificity and their preparation has been previously described.31 Briefly, the pH of a 1.0 mL suspension of AuNPs was adjusted to 8.5 by the addition of 40 μL of 50 mM BB and then modified by the addition of 10 μL of 1.0 mM DSNB in acetonitrile, and mixed for 3 h. ERL preparation was completed by adding 20 μL of 1.0 μg/mL of either anti-MMP-7 or anti-CA 19-9 (10-C04C) to the suspension and allowing the immobilization reaction to proceed for 12 h. To stabilize the ERLs and to block unreacted succinimidyl esters, 10% BSA in 2 mM borate buffer (100 μL) was added to the suspension. After 3 h incubation, the suspension was centrifuged at 2000×g for 10 min. After discarding the colorless supernatant, the ERL pellet was resuspended in 1.0 mL of 2 mM BB containing 1% BSA. This procedure was repeated two more times, and suspended in 500 μL of BB. Finally, the suspension was modified with 50 μL of 10% NaCl and then passed through a 0.22 μm syringe filter (Millipore) to remove any large aggregates. In this assay DSNB is used as a Raman reporter, which yields a signal at 1336 cm−1 due to its symmetric nitro stretch [νs(NO2]. The concentration of each marker in a patient sample is then calculated from the corresponding standard curve using the average relative intensity of the Raman reporter molecule, DSNB
2.3 Capture substrate preparation
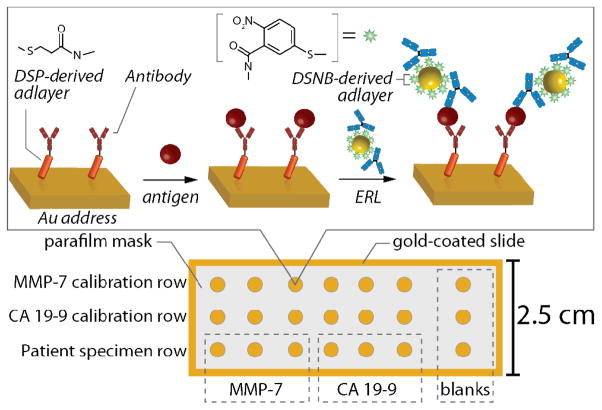
To carry out multiple assays in an array format (Fig. 1), 1×3 inch glass slides were used as the underlying substrate. These slides 5 were ultrasonically bathed in 10% Contrad 70 solution (Micro, Cole-Parmer), deionized water, and ethanol, sequentially for 30 min. The slides were then restively coated with chromium (~15 nm) and ~300 nm of gold (99.999% purity) at a rate of 0.1 nm/s. Prior to use, these substrates were cleaned in a 270 mTorr argon plasma (Harrick) at medium power for 15 min.
Fig. 1.
Schematic of SERS array for MMP-7 and CA 19-9.
The following steps were used to form the addressed array. First, a Parafilm mask with a 3×7 array of 3 mm holes was thermally sealed to the gold-coated glass slide for 1 min at 70 °C. This created an array of exposed gold “wells,” each serving as an individual capture address. This assembly was then immersed in a solution of 0.1 M ethanolic DSP for ~12 h, rinsed with ethanol and dried under a directed stream of nitrogen. Anti-MMP-7 (10 μL, 100 μg/mL), diluted in 50 mM borate buffer (pH 8.5), was applied to the seven sample areas in row 1 and first three sample areas 1–3 of row 3 of the capture substrate. Anti-CA 19-9 (10 μL, 100 μg/mL), diluted in 50 mM borate buffer (pH 8.5), was applied to row 2 and sample areas 4–6 of row 3. The capture antibodies were allowed to incubate for 3 h in a humidity chamber. In this step a capture antibody layer is formed by coupling of the terminal succinimidyl ester of the DSP derived monolayer with exposed antibody-amine groups. The antibody suspension was then aspirated from each well followed by twice rinsing and aspirating 10 μL of PBS. Each of these two rinse aliquots were reverse pipetted five times with the same pipette tip to avoid cross-contamination of the MMP-7 and CA 19-9 sample areas. Next, 20 μL of a 4% instant nonfat milk/0.1% Tween 20 blocking buffer in 10 mM PBS was pipetted onto each sample area and allowed to incubate overnight. Afterwards, the sample areas were individually rinsed with PBS via pipette displacement 3×10 μL as previously described.
Once the capture substrate was prepared, rows one and two were exposed to 10-μL aliquots of rh-MMP-7-spiked (row 1) and CA 19-9-spiked (row 2) dilute pooled human serum (1:4 v/v pooled human serum:PBS) for 3 h at room temperature in a humidity chamber. Sample area 7 of rows 1 and 2 received 10-μL blanks (un-spiked 1:4 diluted pooled serum). 2.5-μL aliquots of DIDM17, DIDM30, and DIDM199 were diluted to 10 μL in PBS and applied to areas 1–3 of row three for MMP-7 determination and areas 4–6 for CA 19-9 determination. After the 3-h incubation, each sample area was rinsed via pipette displacement with 10 mM PBS containing 1% Tween 20. The captured antigens were then exposed to a 10 μL ERL suspension for ~12 h at room temperature in a humidity chamber. The substrates were rinsed with 10 mM PBS containing 1% Tween 20 and dried with a stream of high-purity nitrogen before measuring the SERS signal.
2.4. Preparation of Calibration Standards
MMP-7 antigen was diluted with 0.10 M PBS to produce a stock solution of 80.0 ng/mL. This solution was then used to prepare calibration standards by further dilutions in PBS and the addition of blank pooled human serum such that the ratio of serum to buffer was 1:4. The same procedure was used for calibrant solutions of CA 19-9, starting from a stock concentration of 73.8 ng/mL in PBS. The standards were then divided into 10 μL aliquots for SERS immunoassay and 50 μL portions for ELISA.
2.5. Preparation of Serum Samples
The three serum samples were thawed to room temperature and then diluted with PBS to a 1:4 ratio of serum:PBS. Each test by SERS used 10 μL of diluted sample, whereas that for ELISA employed 50 μL of diluted sample.
2.6. Instrumentation
2.6.1. SERS
The Raman spectra were collected with a NanoRaman I fiber-optic-based Raman system (Concurrent Analytical, Inc.), a portable, field-deployable instrument. The light source was a 30-mW, 632.8-nm He-Ne laser. The spectrograph consisted of an f/2.0 Czerny-Turner imaging spectrometer (6 – 8 cm−1 resolution) and a Kodak 0401E CCD thermoelectrically cooled to 0 °C. The incident laser light was focused to a 25-μm spot size on the substrate at normal incidence using an objective with a numerical aperture of 0.68; the power at the sample was ~3.7 mW. The same objective was used to collect the scattered radiation. All spectra were acquired with an integration time of 1 s.
2.6.2 ELISA
ELISA analyses were performed by following manufacturer instructions on 50 μL aliquots of the 1:4 serum:PBS samples and calibrants with the aforementioned kits for MMP-7 and CA19-9. Both kits used horseradish peroxidase-labeled monoclonal antibodies as the tracer antibody and tetramethylbenzidine (TMB) as the colorimetric reagent. After incubation with the tracer antibody and subsequent washing steps, a 100 μL aliquot of TMB substrate solution was added to each well, shaken for 20 s, and allowed to react with the peroxidase label for 20 min in the dark to generate tetramethylbenzidine diimine (TMBD). 100 μL of the kit-included stop-solution was then added to stop the colorimetric development. The well plates were gently shaken for 30 s prior to interrogation at 450 nm with a VMax Kinetic ELISA Microplate Reader (Molecular Devices, Sunnyvale, CA) and analyzed using SoftMax software (version 4.6). On average, these analyses required 3 h to complete.
3. Results and discussion
3.1. Construction of calibration lines
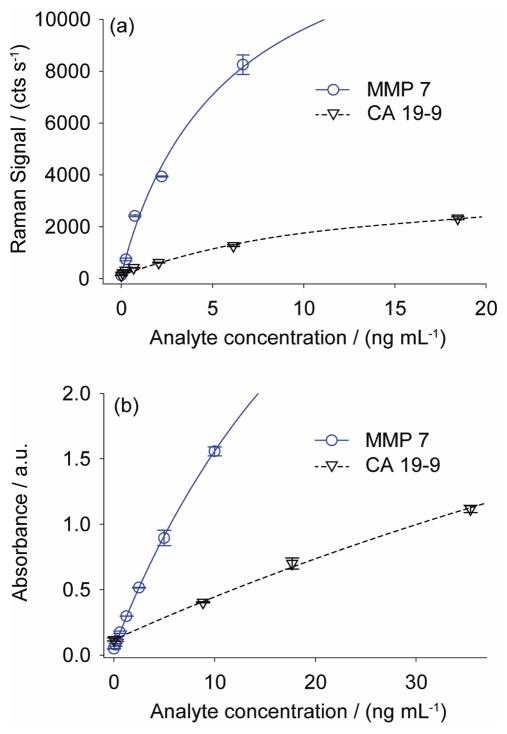
Spectra from each of the calibration wells were used to establish a set of standard curves for the SERS measurements, and a similar set of data was collected for this purpose for the ELISA tests. The concentrations for the standards contained antigen levels up to ~20 ng/mL. The resulting calibration lines for the four sets of measurements are shown in Fig. 2. The SERS calibration lines (Fig. 2a) were constructed by plotting the relative intensity of the DSNB symmetric nitro stretch at each standard concentration. Each point represents the average of five readings across the surface of each address. A similar set of data was collected in duplicate for the ELISA measurements (Fig. 2b), in which the absorbance of TMBD was monitored at 450 nm. The analysis for best-line fits employed a single-site binding isotherm (Eq. 1), which is derived from an overall immnoassay equilibrium between a ligand (protein biomarker) and receptor (Ab) (see Appendix) and incorporates a constant background signal (Ns). In Eq. 1, I(Ci) is the intensity of the signal at antigen concentration Ci, Bmax is the apparent binding capacity, and Kd is the apparent dissociation constant for the overall assay.
Fig. 2.
(a) SERS Calibration plots for MMP-7 and CA 19-9 antigen spiked into 1:4 pooled human serum:PBS buffer. Each point represents the average of 5 measurements and error bars are ± 1 standard deviation. In some cases, the error bars are smaller than the data point. (b) ELISA Calibration plots for the same MMP-7 and CA 19-9 standards used in (a). Each point represents the average of two measurements. Error bars for these data are ± absolute deviation.
| Eq. 1 |
The theoretical LOD for the assays were calculated using the parameters in Table 1 (the blank signal plus 3×standard deviation of the blank signal) to be 2.28 pg/mL for MMP-7 and 34.5 pg/mL for CA 19-9, which are lower than those for ELISA: 31.8 pg/mL for MMP-7 and 987 pg/mL for CA 19-9. These results point to the possibility of using SERS as a readout method for detecting concentrations lower than those previously explored by ELISA, which is an area of intense interest for early diagnosis of PA.
Table 1.
Figures of Merit Comparison for SERS and ELISA.
| Parameter | SERS | ELISA | ||
|---|---|---|---|---|
|
| ||||
| MMP-7 | CA 19-9 | MMP-7 | CA 19-9 | |
| Bmaxa | 14900 | 3810 | 6.14 | 5.77 |
| Kd (M) | 1.96×10−10 | 6.52×10−11 | 1.06×10−9 | 7.95×10−10 |
| Nsa | 131 | 130 | 0.0480 | 0.116 |
| R2 | 0.989 | 0.991 | 0.999 | 0.999 |
| Blank σa | 2.00 | 3.19 | 0.00212 | 0.0113 |
| LOD (pg/mL) | 2.28 | 34.5 | 31.8 | 987 |
| LOD (M) | 78.6×10−15 | 164×10−15 | 1.10×10−12 | 4.70×10−12 |
Units for these parameters are cts s−1 for SERS and absorbance units for ELISA.
We also find that for each detection method, the LOD for MMP-7 was about three times better than that for CA 19-9. The largest contributors to this intra-method difference in LOD are the noise of the blank signal, Blank σ, (different levels of nonspecific adsorption from sample to sample) and the apparent maximum label surface concentration, Bmax, which can be affected by the surface concentration of “active” capture mAbs, the affinity constants of both label and capture mAbs to the marker, and the surface concentration of label mAb on the gold nanoparticle label.
3.2. Comparison of SERS and ELISA for patient samples
After completing the construction and analysis of the calibration plots for best fit equations for the four sets of measurements, the responses from the patient samples were evaluated for levels of MMP-7 and CA 19-9. Note that these specimens were diluted to a 1:4 ratio of serum:PBS, matching the preparation and matrix of the calibrant suspensions.
The results from these analyses are summarized in Table 2. For MMP-7, the levels measured by both SERS and ELISA are a few nanograms per milliliter, with precisions of ~6% or less. These levels are at the upper end of the expected range of healthy individuals.16, 41 Moreover, the levels found for samples DIDM199 and DIDM30 by SERS and ELISA differ by less than 10%. There is a much greater difference in the two levels for sample DIDM17.
Table 2.
MMP-7 and CA 19-9 Levels in Patient Samples.
| Specimen | Antigen | Mean Concentration (ng/mL)
|
|
|---|---|---|---|
| SERS | ELISA | ||
| DIDM199 | MMP-7 | 3.53±0.12 (3.4%)a | 3.91±0.16 (4.1%) |
| DIDM30 | 4.77±0.12 (2.5%) | 4.36±0.26 (5.9%) | |
| DIDM17 | 1.92±0.08 (4.2%) | 2.79±0.16 (5.7%) | |
|
| |||
| DIDM199 | CA 19-9 | 50.4±2.9 (5.8%) | 30.7±2.0 (6.5%) |
| DIDM30 | NDb | ND | |
| DIDM17 | ND | ND | |
Percent variation is reported as standard deviation/mean value.
ND: not detected.
The results for the measurements of CA 9-19 in the three samples are also in reasonable agreement. Interestingly, the levels found by both techniques for sample DIDM199 are at the upper end of that typical for a healthy individual.15, 23 In contrast, the amounts of this marker in samples DIDM17 and DIDM30 appear to fall below the LODs reported in Table 1. While only speculative, this situation may reflect the inability of these patients to express the CA 19-9 antigen, a condition affecting about 10% of the population.8, 35 Taken together, these results begin to place the SERS platform on firm footing for potential clinical use.
4. Conclusions
Overall, the first generation of a SERS immunoassay array for two PA biomarkers, CA 19-9 and MMP-7, was successful. The LODs using this platform were calculated to be 2.28 pg/mL for MMP-7 and 34.5 pg/mL for CA 19-9 from spiked serum. In comparison to ELISA, the current diagnostic of choice for these markers, the SERS based assay has lower LOD for CA 19-9 and MMP-7 (×29 and ×14 lower respectively), and points toward the possibility of using SERS for low-level detection of PA biomarkers. The SERS and ELISA determined levels of CA 19-9 and MMP-7 in serum samples were comparable, pointing to the use of SERS in real-world samples. An additional advantage to using a SERS based array format is that less sample is necessary for the assay. Preparation of serum samples for the SERS immunoassay requires ~2.5 μL/well, whereas ELISA and similar strategies can use ~20 μL sample sizes. Future development of a 96 well format for this type of analysis will provide a footprint compatible with fluid handlers and robotic spotting systems allowing the platform to be used in today’s automated laboratories.
Acknowledgments
This work was supported by the National Institutes of Health (U01CA151650 and R33CA155586 to M.D.P. and S.J.M.), P30CA042014 from the Huntsman Cancer Institute for support of core facilities, the Huntsman Cancer Institute Pancreas Cancer Research Program, the Utah Science Technology and Research Initiative, and the Huntsman Cancer Foundation. M.D.P. has a financial interest in Concurrent Analytical, Inc. We thank Patti Larrabee for performing the ELISA assays.
Appendix
The fitting parameters for the single-site binding isotherm used herein are apparent constants for the entire sandwich assay, which is a multi-step, multi-equilibria assay. When used in this fashion, two assumptions are made: (1) the initial equilibrium between the antigen (Ag) and Ab (capture antibody) is not affected by subsequent rinse or labelling steps; and (2) there is a 1:1 binding stoichiometry between Ag and label (the label is an ERL in the SERS assay and a peroxidase-labeled monoclonal antibody for the ELISA assay).
The initial equilibrium is written as
| Eq. 2 |
where the dissociation constant, Kd, can be represented as
| Eq. 3 |
In Eq. 3, the square brackets represent molar concentrations of the species enclosed. The fraction of bound Ab, θ, can be represented by the ratio of Ab:Ag to the total amount of Ab
| Eq. 4 |
This can be recast in terms of Kd and [Ag] by multiplying the numerator and denominator by [Ag], dividing both by [Ab:Ag], and substituting the definition for the dissociation constant.
| Eq. 5 |
The amount of occupied Ab, and as a consequence bound Ag, is the fractional occupancy multiplied by Bmax, the total number of Ab available to bind (apparent binding capacity).
| Eq. 6 |
We typically observe a constant background due to nonspecific adsorption of label, Ns. Therefore, the total amount of label (both associated with Ag and that due to Ns) gives rise to a signal, I(Ci), indicative of a specific concentration of Ag (Ci).
| Eq. 7 |
In this treatment, the apparent Kd is equal to the concentration of Ag that gives rise to a signal half the intensity of Bmax.
References
- 1.Fu E, Yager P, Floriano PN, Christodoulides N, McDevitt JT. Pulse, IEEE. 2011;2:40–50. doi: 10.1109/MPUL.2011.942766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi YP, Shim HS, Gao MQ, Kang S, Cho NH. Cancer Lett. 2011;307:62–71. doi: 10.1016/j.canlet.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 3.Hidalgo M. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 4.Poruk KE, Firpo MA, Mulvihill SJ. Ann Surg. 2012 in press. [Google Scholar]
- 5.Firpo MA, Boucher KM, Mulvihill SJ. Sci Transl Med. 2012 in preparation. [Google Scholar]
- 6.Ren J, Cai H, Li Y, Zhang X, Liu Z, Wang JS, Hwa YL, Zhang Y, Yang Y. Expert Rev Mol Diagn. 2010;10:787–798. doi: 10.1586/erm.10.39. [DOI] [PubMed] [Google Scholar]
- 7.Hori SS, Gambhir SS. Sci Transl Med. 2011;3:109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Clin Chem. 2010;56:186–193. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastarache JA, Koyama T, Wickersham NL, Mitchell DB, Mernaugh RL, Ware LB. J Immunol Methods. 2011;367:33–39. doi: 10.1016/j.jim.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang S, Zahn J, Horecka J, Kunz P, Ford J, Fisher G, Le Q, Chang D, Ji H, Koong A. J Transl Med. 2009;7:105. doi: 10.1186/1479-5876-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pla-Roca M, Leulmi RF, Tourekhanova S, Bergeron S, Laforte V, Moreau E, Gosline SJC, Bertos N, Hallett M, Park M. Mol Cell Proteomics. 2012;11:M111.011460. doi: 10.1074/mcp.M111.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti KK, Dinish US, Samanta A, Vendrell M, Soh K-S, Park S-J, Olivo M, Chang Y-T. Nano Today. 2012;7:85–93. [Google Scholar]
- 13.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotech. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 14.Kingsmore SF. Nat Rev Drug Discovery. 2006;5:310–321. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen K, Han H, Luo Z. Analyst. 2012;137:1259–1264. doi: 10.1039/c2an15997j. [DOI] [PubMed] [Google Scholar]
- 16.Lee M, Lee S, Lee J, Lim H, Seong GH, Lee EK, Chang SI, Oh CH, Choo J. Biosens Bioelectron. 2011;26:2135–2141. doi: 10.1016/j.bios.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Tripp RA, Dluhy RA, Zhao Y. Nano Today. 2008;3:31–37. [Google Scholar]
- 18.Dougan JA, Faulds K. Analyst. 2012;137:545–554. doi: 10.1039/c2an15979a. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Lipert RJ, Jain M, Kaur S, Chakraboty S, Torres MP, Batra SK, Brand RE, Porter MD. Anal Chem. 2011;83:2554–2561. doi: 10.1021/ac102829b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koopmann J, Fedarko NS, Jain A, Maitra A, Iacobuzio-Donahue C, Rahman A, Hruban RH, Yeo CJ, Goggins M. Cancer Epidemiol Biomarkers Prev. 2004;13:487–491. [PubMed] [Google Scholar]
- 21.Kuhlmann KF, van Till JW, Boermeester MA, de Reuver PR, Tzvetanova ID, Offerhaus GJ, Ten Kate FJ, Busch OR, van Gulik TM, Gouma DJ, Crawford HC. Cancer Epidemiol Biomarkers Prev. 2007;16:886–891. doi: 10.1158/1055-9965.EPI-06-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeone DM, Ji B, Banerjee M, Arumugam T, Li D, Anderson MA, Bamberger AM, Greenson J, Brand RE, Ramachandran V, Logsdon CD. Pancreas. 2007;34:436–443. doi: 10.1097/MPA.0b013e3180333ae3. [DOI] [PubMed] [Google Scholar]
- 23.Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LDN, Goel R, Mathivanan S, Marimuthu A, Kashyap M, Vizza RF, Mayer RJ, DeCaprio JA, Srivastava S, Hanash SM, Hruban RH, Pandey A. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.For a recent review see the SERS thematic issue in Chem Soc Rev. 2008;37:873–1076.
- 25.Driskell JD, Kwarta KM, Lipert RJ, Vorwald A, Neill JD, Ridpath JF, Porter MD. J Virol Meth. 2006;138:160–169. doi: 10.1016/j.jviromet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Driskell JD, Uhlenkamp JM, Lipert RJ, Porter MD. Anal Chem. 2007;79:4141–4148. doi: 10.1021/ac0701031. [DOI] [PubMed] [Google Scholar]
- 27.Grubisha DS, Lipert RJ, Park H-Y, Driskell J, Porter MD. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 28.Kwarta KM, Driskell JD, Lipert RJ, Porter MD. manuscript in preparation. [Google Scholar]
- 29.Ni J, Lipert RJ, Dawson GB, Porter MD. Anal Chem. 1999;71:4903–4908. doi: 10.1021/ac990616a. [DOI] [PubMed] [Google Scholar]
- 30.Park H-Y, Driskell JD, Kwarta KM, Lipert RJ, Porter MD, Schoen C, Neill JD, Ridpath JF. Top Appl Phys. 2006;103:427–446. [Google Scholar]
- 31.Porter MD, Lipert RJ, Siperko LM, Wang G, Narayanan R. Chem Soc Rev. 2008;37:1001–1011. doi: 10.1039/b708461g. [DOI] [PubMed] [Google Scholar]
- 32.Yakes B, Lipert JR, Bannantine JJ, Porter PMD. Clin Vaccine Immunol. 2008;15:235–242. doi: 10.1128/CVI.00335-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakes BJ, Lipert JR, Bannantine JP, Porter MD. Clin Vaccine Immunol. 2008;15:227–234. doi: 10.1128/CVI.00334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duffy M, Sturgeon C, Lamerz R, Haglund C, Holubec V, Klapdor R, Nicolini A, Topolcan O, Heinemann V. Ann Oncol. 2010;21:441–447. doi: 10.1093/annonc/mdp332. [DOI] [PubMed] [Google Scholar]
- 35.Goonetilleke KS, Siriwardena AK. Eur J Surg Oncol. 2007;33:266–270. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Scaife Courtney L, Shea Jill E, Dai Q, Firpo Matthew A, Prestwich Glenn D, Mulvihill Sean J. J Gastrointest Surg. 2008;12:1074–1080. doi: 10.1007/s11605-007-0425-3. [DOI] [PubMed] [Google Scholar]
- 37.Humphris JL, Chang DK, Johns AL, Scarlett CJ, Pajic M, Jones MD, Colvin EK, Nagrial A, Chin VT, Chantrill LA, Samra JS, Gill AJ, Kench JG, Merrett ND, Das A, Musgrove EA, Sutherland RL, Biankin AV ftNPC Network. Ann Oncol. 2012 [Google Scholar]
- 38.Wilson CL, Heppner KJ, Labosky PA, Hogan BLM, Matrisian LM. Proc Natl Acad Sci USA. 1997;94:1402–1407. doi: 10.1073/pnas.94.4.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto H, Itoh F, Iku S, Adachi Y, Fukushima H, Sasaki S, Mukaiya M, Hirata K, Imai K. J Clin Oncol. 2001;19:1118–1127. doi: 10.1200/JCO.2001.19.4.1118. [DOI] [PubMed] [Google Scholar]
- 40.Diagnostic Automation, Inc. 2011 http://diagnosticautomation.com/inserts06/List_B/PDF/CA-19-9_6909-16_4-6-06_.pdf.
- 41.Garrett NL, Vukusic P, Ogrin F, Sirotkin E, Winlove CP, Moger J. J Biophotonics. 2009;2:157–166. doi: 10.1002/jbio.200810057. [DOI] [PubMed] [Google Scholar]