Abstract
Comprehensive knowledge of proteome complexity is crucial to understanding cell function. Amino termini of yeast proteins were identified through peptide mass spectrometry on glutaraldehyde-treated cell lysates as well as a parallel assessment of publicly-deposited spectra. An unexpectedly large fraction of detected amino-terminal peptides (35%) mapped to translation initiation at AUG codons downstream of the annotated start codon. Many of the implicated genes have suboptimal sequence contexts for translation initiation near their annotated AUG, and their ribosome profiles show elevated tag densities consistent with translation initiation at downstream AUGs as well as their annotated AUGs. These data suggest that a significant fraction of the yeast proteome derives from initiation at downstream AUGs, increasing significantly the repertoire of encoded proteins and their potential functions and cellular localizations.
Keywords: Protein translation initiation sites
Introduction
The size and complexity of eukaryotic proteomes has been of considerable interest, especially in light of the unexpected reductions in the estimates of gene number predicted after sequencing of human and other genomes (1, 2). However, recent ribosome profiling data (3, 4) and observations of non-standard translation initiation mechanisms, such as leaky scanning or translation reinitiation (5–11) (Fig. 1D), have led to suggestions that eukaryotic proteomes may be significantly more complex than currently annotated. For example, translation initiation at AUG codons downstream (dnAUGs) of the annotated start codon (annAUG) would lead to truncated proteins, or if out of frame, proteins with different amino acid sequence, with potentially new functions or cellular localizations and targeting.
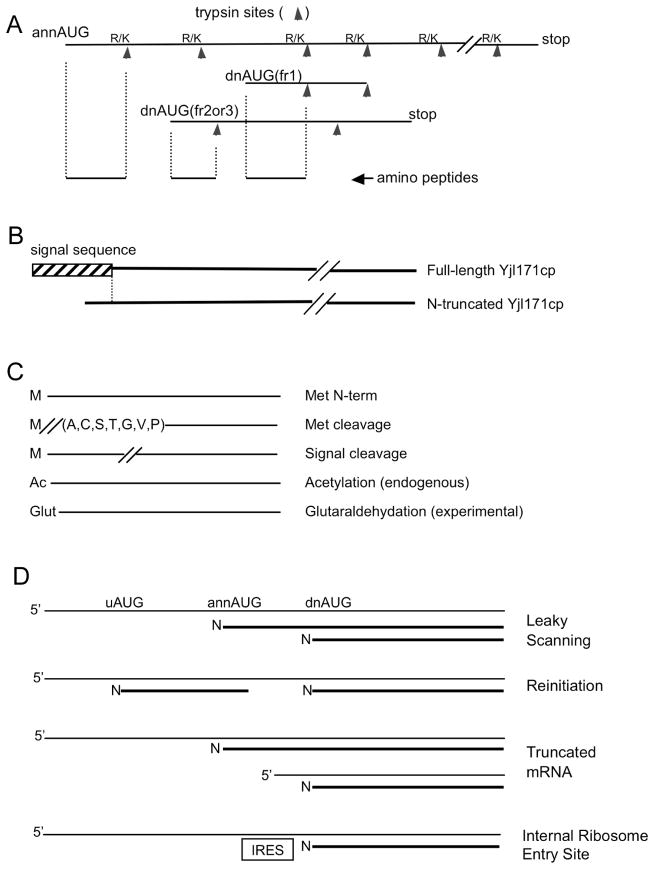
Figure 1. Protein products from initiation at dnAUGs.
(A) Amino-terminal derived peptides from translation initiation at dnAUGs can be distinguished from annAUG initiation by the absence of an amino-terminal trypsin site. (B) An example of dnAUG initiation. Yjl171cp is annotated as a GPI-anchored cell wall protein, and the detected full-length Yjl171cp protein has an ER signal sequence; however, the signal sequence is not predicted in the detected N-terminal truncated protein from initiation at a dnAUG. N-terminal peptides for both the full-length and truncated proteins were also detected following partial purification of C-terminal TAP-tagged Yjl171cp protein. (C) The sequence databases for peptide mass spectrometry included products for terminal methionine and signal peptide cleavage. Optional mass increases for acetylation (+42 Da) or glutaraldehydation (+68 Da) were also assessed. (D) Potential mechanisms for translation initiation at dnAUGs.
Translation initiation is thought to depend on a ribosome scanning mechanism (12) in which ribosomes typically initiate translation at the first AUG they encounter when scanning from the 5′ cap of the mRNA; however, AUGs can be skipped if they are too close to the 5′ cap (13) or in a poor sequence context (11). Current annotations of proteomes in genome projects generally rely on the assumption that, in most cases, individual mRNAs each give rise to a single protein, typically encoded by the longest open reading frame (ORF) uninterrupted by stop codons. For example, the initial annotation of the yeast proteome identified protein coding regions as ORFs encoding more than 99 amino acids (14). The annotated translation start positions were in some cases subsequently adjusted to more C-terminal start sites when cross-species sequence comparisons revealed poor protein conservation at the site originally annotated (15). But is it correct to assume that scanning ribosomes generally initiate at only one site on an mRNA? By analyzing the amino-termini of proteins on a proteomic scale through peptide mass spectrometry, we show here that for many genes, ribosomes appear to use multiple start sites, including sites downstream of the annotated start codon, giving rise to protein products with different amino termini, and in some cases, different reading frames.
Methods
Glutaraldehyde Treatment of Proteins
To enhance detection, amino termini of proteins were modified by reductive glutaraldehydation based on the protocol of Russo et al. (16). A pellet of approximately 1000ug of protein lysate from YSH474 yeast cells was resuspended in 0.1% acetic acid (pH 4) to give a protein concentration of 5ug/ul. The resuspension was then brought to a concentration of 100mM sodium cyanoborohydride and 0.5% glutaraldehyde, and incubated at room temperature for 20 minutes. An equal volume of 1 M Tris-HCl was added and the sample was pelleted with 30% TCA, and digested with 10ug of trypsin (Promega); some samples were concentrated with Zip Tips (Millipore). The trypsin digest was loaded onto a nanospray column packed with C18 resin (Michrom Bioresources) and analyzed using a LCQ Deca XP mass spectrometer (Thermo Scientific).
TurboSequest Assessment
Peptides were identified with TurboSequest (Bioworks 3.2 package) using a Dell XPS server. For analysis of our glutaraldehyde-treated cell lysates, we included optional mass increases to peptides: (i) N-terminal amino acid +68 Da for glutaraldehyde modification; (ii) N-terminal amino acid +42 Da for acetylation modification (requiring a second TurboSequest run); (iii) any lysine +68; (iv) N-terminal proline +86 Da; and (v) N-terminal methionine oxidation +16 Da. Each TurboSequest parameter file included a precursor mass tolerance of 3.0 Da and a fragment mass tolerance of 1.0 Da. Precursor peptides for MS/MS analysis were prepared by trypsin digestion, which cuts after R or K, except when flanked by P. For the TurboSequest analysis, in addition to requiring trypsin-cleavage sites at both ends of the precursor peptides (or one end if a terminal peptide), an internal trypsin site was allowed in the Glutaraldehyde-treatment analysis (two internal sites were allowed in the PeptideAtlas analysis discussed below). TurboSequest matches to trypsin fragments with internal trypsin sites were subsequently discarded based on our decoy analysis (see below).
A sequence “database” file of downstream Open Reading Frames (dnORFs) in FASTA format was constructed as follows. Using an MS-SQL database of yeast genomic sequences (6,718 genes downloaded from SGD March 2008 (17)), translated dnORFs were computed for translation initiation at all AUG triplets within 100 nt downstream of the annotated translation start codon. DnORFs in the same reading frame as the annAUG (frame 1) included sequence from the initiation methionine to the following second trypsin site; frame 2 and 3 dnORFs included the full dnORF. We also included (i) N-terminal sequences that would result from amino peptidase cleavage of the initiation methionine if it was followed by amino acids A, C, S, T, G, V, or P (18); and (ii) N-terminal sequences following cleavage of a signal peptide predicted by the SignalP algorithm version 3.0 (19) (these sequences extended to the second trypsin site after their N-terminus).
The TurboSequest algorithm computes a probability score for each peptide match, and for each set of equivalent MS/MS experiments (e.g. in a single PeptideAtlas accession; Supplementary Table S1), spectrum matches were considered that were below a probability score threshold chosen to ensure that the target false identification rate was below 5%. False identification rates for detection of N-terminal peptides were calculated as follows. For each MS/MS run, we calculated the number of forward and reverse decoy N-terminal peptides matched. Individual peptides with more than one match were only counted once per MS/MS run, and these counts were then summed over all MS/MS runs. The target false identification rate was calculated as (# decoy peptides/# forward peptides) as described (20). The false identification rate was 5% ± 0.5% (± s.d. based on bootstrap analysis). In our glutaraldehyde treatment experiments, false identification rates < 5% were obtained for probability score thresholds of 0.28 for glutaraldehyde-modified and 0.06 for acetylated peptides (Supplementary Table S1). Matches to 35 unmodified N-terminal peptides were disregarded because the false identification rate for these matches was very high (30%), suggesting that many of these were false matches and that the glutaraldehyde treatment is efficient.
In addition to our glutaraldehyde-treatment experiments, we also analyzed publicly deposited spectra from yeast cell lysates downloaded from http://www.peptideatlas.org/repository/ and analyzed as follows. TurboSequest was run using the Sequest parameter files posted for each experimental series (Supplementary. Table S1). The parameter files were modified to include optional acetylation (+42 Da) at the N-terminus of peptides. TurboSequest was run using a FASTA file sequence “database” composed of all annORFs, as well as all dnORFs mapping within 100 nt of the annAUG, and with the methionine aminopeptidase and signal peptide cleavage modifications as described above. We analyzed 20 experimental series uploaded by multiple research groups (Supplementary Table S1). For each experimental series, we computed a TurboSequest probability threshold that gave a false identification rate < 5% and only considered TurboSequest matches below this probability threshold (Supplementary Table S1). Matches to N-terminal, internal, and C-terminal peptides were used to calculate the probability threshold.
In this analysis, we excluded all matches with a TurboSequest initial ranking (RSp rank) > 1 because in our glutaraldehyde-treatment experiments, matches with RSp rank > 1 had a false identification rate of 18% instead of 5% suggesting that they were less reliable (also see (21)). We also excluded all matches to peptides with internal trypsin sites since these matches were unreliable; for example, N-terminal peptides with a single internal trypsin site had a false identification rate of 28% consistent with reports (22) that trypsin digestion can be highly efficient. The TurboSequest algorithm also reported N-terminal modifications (+68 Da or +42 Da) to internal peptides, but these matches were expected to be false since N-terminal glutaraldehydation prior to trypsin treatment or in vivo acetylation should be restricted to the N-termini of proteins, and correspondingly, the decoy detection rate was very high (749 decoy internal peptides; 760 forward internal peptides). We also performed control experiments (not shown) in which cell lysates were partitioned into gel slices of known molecular weight size ranges (25–37 kDa, 37–50 kDa). Although the TurboSequest algorithm had no knowledge of the parent protein sizes before trypsin digestion, peptide matches conformed to the correct parent size ranges, the frequency of incorrect matches being a little higher than the decoy frequencies in these experiments. (This analysis took into account the portion of the proteome in each size range that would be detected randomly.) These controls provide confidence in the observed matches to N-terminal peptides and their association with translation initiation at annAUGs and dnAUGs.
All outputs from TurboSequest runs were uploaded into a relational database and analyzed using stored procedures written in MS-SQL to compute false identification rates and integrate the MS/MS data with other data sets including the yeast genome, ribosome profile, mRNA and protein expression, and Gene Ontology data.
Results and Discussion
To assess the repertoire of amino termini of proteins in the budding yeast proteome, tandem peptide mass spectrometry (MS/MS) was performed on yeast cell lysates. Proteins in cell lysates were digested in vitro with trypsin endopeptidase, and the resulting peptides were sampled and further fragmented by collision induced dissociation (CID). The Sequest algorithm (TurboSequest (23)) was used to determine the sequences of tryptic peptides through computational comparison of experimental spectra with theoretical spectra computed from an exhaustive sequence database of annotated proteins in the yeast proteome. By also including in the database sequences expected from initiation at dnAUGs within 100 nucleotides (nt) of the annAUG, we identified peptides resulting from initiation at both dnAUGs and annAUGs. The peptides originating from the N-termini of proteins, including those initiated at dnAUGs, can be distinguished from internal and C-terminal tryptic fragments because they do not have a trypsin site at their N-terminus (Fig. 1A). The sequence database included amino-terminal sequences from signal peptide cleavage predicted by the SignalP algorithm (19) and methionine cleavage by methionine aminopeptidase (18) (Fig. 1C).
Output from the TurboSequest algorithm was filtered by decoy analysis (20, 21) to ensure a target false identification rate below 5%. Confidence in the TurboSequest matches was confirmed in control experiments that tracked parent protein masses before trypsin digestion (see Methods), and separate control experiments using partial purification of C-terminal-epitope-tagged proteins (see below).
The detection efficiency of N-terminal peptides was increased through treatment of proteins in cell lysates with glutaraldehyde which modifies their amino termini by reductive alkylation (16). The reductive alkylation results in a molecular weight increase of +68 Da on the amino-terminal amino acid of proteins as well as internal lysines, and increases MS/MS detection of a1 fragment ions. Proteins that had been acetylated in vivo would not be modified at their N-terminus by glutaraldehyde. Therefore, tryptic peptides were assessed for potential molecular weight increases of +68 Da (glutaraldehydation) or +42 Da (acetylation). TurboSequest matches were observed to 69 glutaraldehyde-modified and 54 acetylated N-terminal peptides, including one observed in both forms (Table 1).
Table 1.
Detected Amino-terminal Peptides
| Experiments
|
||
|---|---|---|
| Amino peptide property | GLUT1 | PeptideAtlas2 |
| Total N-term peptides3 | 122 | 818 |
| annAUG peptides4 | 99 | 519 |
| downAUG peptides5 | 23 | 299 |
| Acetylated peptides | 53 | 376 |
| Not acetylated peptides | 68* | 406 |
| Acetylated and non-acetylated peptides | 1 | 36 |
| N-term met cleaved | 73 | 294 |
| Signal peptide cleaved | 8 | 49 |
Glutaraldehyde-treated cell lysates; detection efficiency of N-terminal peptides, compared to internal peptides, was elevated due to glutaraldehyde treatment
PeptideAtlas publicly deposited data at http://www.peptideatlas.org/repository/
A total of 903 N-terminal peptides were detected in 830 genes; 37 N-terminal peptides were detected in both experimental series
583 annPeptides were detected in 559 genes; 22 genes had both Met N-terminal-cleaved and uncleaved peptides, 2 genes had signal-peptide-cleaved and uncleaved peptides
320 downPeptides were detected in 299 genes; 14 genes had different downPeptides that mapped to different dnAUGs, and 7 to the same dnAUG (with and without cleavage modifications)
Glutaraldehyde modified peptides
Of the 122 different N-terminal peptides observed in the glutaraldehyde-treatment experiments, 99 corresponded to N-terminal peptides expected from translation initiation at annAUGs (Table 1). A surprisingly large fraction (19%, 23 peptides) of the detected N-terminal peptides corresponded to peptides expected from translation initiation at dnAUGs (Table 1; examples are illustrated in Fig. 2 and Supplementary Fig. S1). Given this unexpected result, the analysis was expanded to include an assessment of publicly-deposited spectra for yeast cell lysates (http://www.peptideatlas.org/repository). Based on the public data, a total of 818 different N-terminal peptides were detected including 299 peptides corresponding to initiation at dnAUGs (37%; Table 1). Combining the data for the glutaraldehyde treatment experiments with those of the publicly-deposited spectra, we observed 583 peptides for annAUG initiation (“annPeptides”), and 320 peptides for dnAUG initiation (35% “downPeptides”; Table 1, Supplementary Table S2). Although for most genes with detected N-termini, we observed either annPeptides (531 genes) or downPeptides (271 genes), but not both, it is likely that some of these genes express both forms even though only one was detected in this sampling of the proteome. Indeed, 26 genes had different N-terminal peptides that mapped to the annAUG and a dnAUG; and two additional genes had peptides for the annAUG and two different dnAUGs; moreover, 12 genes had peptides for two dnAUGs but not their annAUG. The frequencies of annPeptide and downPeptide detection were similar for many of the 28 genes for which both classes were detected (Supplementary Table S3). Detection of five of the frame-1 downPepides (HOM3, YJL171C, EFT2, PRS5, and URA1) was confirmed following partial purification of C-terminal-epitope-tagged versions of the proteins. For four of these (all except URA1), we confirmed detection of both an annPeptide and a downPeptide.
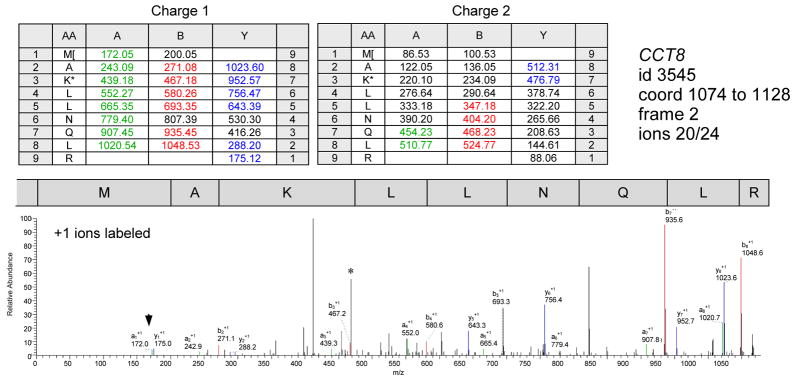
Figure 2. MS/MS spectrum of CCT8 downPeptide.
MS/MS spectrum of glutaraldehyde modified downPeptide fragment MAKLLNQLR from gene CCT8 (id3545); the dnAUG maps 24 codons downstream of the annAUG. The methionine and lysine are both modified by glutaraldehydation (+68 Da). This spectrum has matches to many of the possible CID fragments (19 resolable 1+ ions out of 24; in addition, the b6 fragment was detected among the +2 charged ions *). Matched +1 ion peaks are labeled with designations (a, b and y ions labeled with green, red and blue respectively); the a1 ion (arrowhead) is sometimes enhanced with glutaraldehyde treatment (16).
We examined post-translational modifications of the 903 detected N-termini: 393 N-termini showed methionine aminopeptidase cleavage of the first amino acid (339 cases) or signal peptide cleavage (54) (Table 1), 409 were acetylated, 458 were not acetylated, and 36 were detected in both forms, acetylated and not acetylated. In addition, the amino acids found in the first four positions of the acetylated and non-acetylated (glutaraldehyde modified) N-terminal peptides had similar frequency profiles to those observed by Polevoda and Sherman (21) who collated published yeast protein amino terminus data from multiple researchers (Supplementary Fig. S2). For example, both datasets showed that an N-terminal serine is typically acetylated. 105 (22%) of the detected N-termini correspond to amino termini reported by Polevoda and Sherman (21), and the acetylation characteristics were consistent for 102 of these (97%).
DownPeptides were detected in all three reading frames (Fig. 3; Supplementary Table S4). A significantly elevated percentage (41%) were in frame 1 even though only 32% of the screened dnAUGs were in this frame (chi-square p < 0.001). This result, and the observation that the frequencies of frame-2 and -3 dnAUGs are depressed close to the annAUG (Fig. 3; also see (6)), suggest that there is selection for expression of N-terminal truncations of frame-1 proteins, rather than frame 2 or 3 proteins. Relative instability of shorter frame-2 or -3 proteins may also contribute to the higher detection of frame-1 downPeptides. The ORF lengths of the implicated frame-2 and -3 proteins are significantly longer than randomly selected downstream ORFs, suggesting there has been selection against stop codons in these ORFs (Fig. 3D).
Figure 3. Frame analysis of dnAUGs.
(A–C) The distances (in nucleotides) between dnAUGs and annAUGs in downPeptide genes are graphed for all frames pooled (A) or partitioned by frame (C). Compared to the frequencies of all dnAUGs within 100 nt of the annAUG in the full genome (B), frame 1 downPeptides are over-represented (chi-square p < 0.001, all bins > 10). (D) The lengths of frame-2 or -3 ORFs, defined by dnAUGs of detected downPeptides (188 ORFs, mean 74 nt), are compared with the full set of theoretical ORFs of frame-2 or -3 dnAUGs that start and finish within the annotated frame-1 ORFs (86,088 ORFs, mean 60 nt). ORFs of detected downPeptides are significantly longer based on chi-squre analysis of the full distributions (chi-square p < 2*10−7), as well as bootstrap analysis of means (p < 0.002).
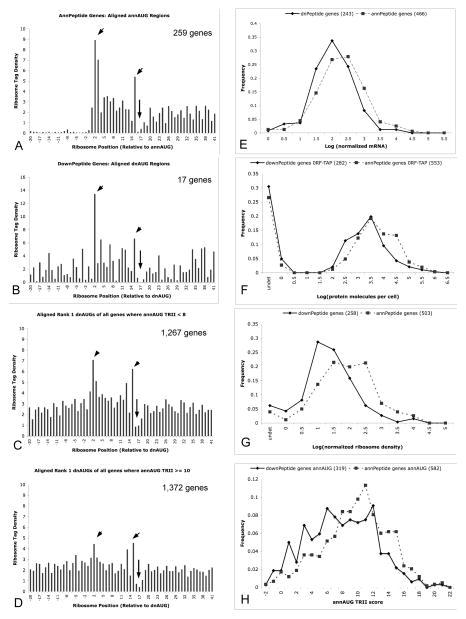
High frequencies of frame 1, 2 and 3 translation initiation upstream and downstream of the annAUG have also been suggested by transcriptome-scale ribosome profiling experiments in yeast (3) and mouse cells (4) that describe mRNA sequences protected by ribosomes. We analyzed downPeptide genes using ribosome-profile sequence reads of Ingolia et al. (3). DownPeptide genes aligned at the dnAUG showed ribosome profiles consistent with translation initiation at these sites (Fig. 4A,B). The characteristic initiation profile was particularly pronounced for downPeptide genes with poorer sequence context surrounding their annAUGs (“Kozak consensus” (24, 25) measured by Translation Relative Individual Information (TRII) score (17, 26)), suggesting a trend towards dnAUG initiation for these genes. Indeed, alignment of the first AUG downstream of the annAUG of all genes, not just those identified in this MS/MS analysis, revealed translation initiation ribosome profiles that are particularly pronounced for genes with poorer Kozak context at their annAUG (Fig. 4C,D). Ribosome tag densities were examined for 81 of the 320 detected downPeptide genes that had tag densities > 0.1 tags/nt in the first 200 nt of their annORFs, and pronounced tag densities (≥ 10 tags/30 nt) in the 30-nt window starting at their dnAUG. All but five of these genes had > 3 ribosome tags in the 30-nt window starting at their annAUG, consistent with translation initiation at the annAUG in addition to the detected dnAUG initiation (Supplementary Table S5). This suggests that most downPeptide genes are under-annotated rather than mis-annotated, with translation initiation occurring at both the annotated AUG and the implicated dnAUG.
Figure 4. Ribosome profiles, gene expression, and TRII score analysis.
(A, B) The ribosome profiles (3) of annAUGs of annPeptide genes have prominent signals at nt positions 2, 3 and 15, and characteristic depression surrounding position 15. Aligned dnAUGs of downPeptide genes show elevated signal at nt 2 and 15 (arrowheads), and depression surrounding nt 15 (arrow). X-axis values show the 14th nt of ribosome tags corresponding to the middle of the ribosome; position 1 is the A of the AUG start codon. Y-axis values show mean relative tag densities for profiles of 259 annPeptide genes, and 17 downPeptide genes with annAUG TRII scores < 8 and >15 nt between the annAUG and dnAUG. Tag densities for each gene were computed relative to their mean tags per nt in the annORF (limited to genes with mean density > 0.1 tags per nt, 5% false identification rate for glutaraldehyde-treatment data, and 1% for PeptideAtlas data). (C, D) Ribosome profiles of all rank 1 dnAUGs with annAUG TRII < 8 (1267 genes) show elevated tag densities at nt positions 2 and 15 suggesting that many of these genes initiate translation at their dnAUG. Elevations at nt 2 and 15 are less pronounced for annAUG TRII ≥ 10 (1372 genes).
(E–G) DownPeptides genes have depressed mRNA expression (E), protein expression (F) and ribosome densities (G) of their annotated ORFs (solid lines) compared to genes with amino-peptides that map to the annAUG (annPeptide genes; broken lines). mRNA expression and ribosome densities are from deep sequence analysis of Ingolia et al. (3). Protein expression is of C-terminal TAP-tagged proteins as reported by Ghaemmaghami et al. (30). The downPeptide and annPeptide distributions are significantly different in (E–G) (chi-square p < 0.01).
(H) DownPeptide genes (solid line) have poorer sequence context surrounding their annAUG, measured by Translation Relative Individual Information (TRII score (17, 26)), compared to annPeptide genes (broken line). The distributions of TRII scores plotted for the two gene sets are significantly different (chi-square p < 0.01).
The unexpected prevalence of downPeptides, and parallel ribosome profile data, suggests that translation initiation at dnAUGs is common. As summarized in Fig. 1D, mechanisms that would lead to dnAUG initiation include:
expression of 5′-truncated mRNAs or spliced mRNAs that do not include the annAUG (27, 28);
leaky scanning where the ribosome sometimes skips the annAUG and instead initiates at a dnAUG (5–8);
translation reinitiation at a dnAUG following translation of an ORF that starts in the 5′UTR (9–11);
translation initiation at internal ribosome entry sites (IRES (29)).
Truncated mRNAs (Supplementary Table S6) may account for 16% of the detected downPeptides. These genes have high-confidence capped mRNAs (27, 28) with 5′ ends that map downstream of the annAUG or within 20 nt upstream; 5′UTRs less than 20 nt in length are reported to reduce translation initiation efficiency (13). Many of these genes also express longer mRNAs from more upstream transcription initiation that could allow translation initiation at their annAUG or their dnAUGs.
Expression levels of mRNAs (27, 28) and proteins (30) are lower in downPeptide genes compared to annPeptide (Fig. 4E–G). The lower mRNA levels may be explained in part by nonsense mediated decay (31) due to frame 2 or 3 translation initiation at dnAUGs or upstream AUGs. The lower protein levels, measured using frame-1 encoded C-terminal epitope tags, may also be explained in part by inefficiency of translation reinitiation (11) or lower protein half lives. Consistent with their lower expression, fewer internal trypsin fragments were detected for downPeptide genes (average of 5 internal peptides) compared to the annPeptides genes (9 internal peptides) DownPeptide detection might also be biased towards genes with lower mRNA and protein expression due to under-annotation, for example, of undetected truncated mRNAs
The Kozak sequence contexts surrounding the annAUGs of downPeptide genes are of poorer quality compared to those of annPeptides genes (Fig. 4H), consistent with the expected underutilization of annAUGs in downPeptide genes. This is emphasized by the prevalence of U at the third nucleotide upstream of the annAUG in downPeptide genes (19%) compared to the strong selection against U at this position in annPeptide genes (10%; chi-square p < 0.0002); the identity of the -3 nucleotide is key to translation initiation (24, 25). The downPeptide genes with poorer annAUG Kozak context also have poorer protein sequence conservation coded near the annAUG compared to the dnAUG (Supplementary Fig. S3). The downPeptide genes also exhibit elevated Codon Adaptation Indices (CAI (32)) immediately downstream of their implicated dnAUG, suggesting selection for more commonly used codons in this region (Supplementary Fig. S4).
The N-terminal peptides that map to annAUGs and dnAUGs are most readily explained by translation initiation at the implicated AUGs. However, it is formally possible that alternative mechanisms, such as protease activities, could account for some instances of these N-terminal peptides, even though they map to AUG codons. For example, Vogtle et al. (33) characterized the amino-termini of 615 proteins following cleavage of mitochondrial signal peptides, and one of these proteins coincidentally has the same amino terminus as one of the detected downPeptides (for the SDH2 Gene). However, the vast majority of our reported downPeptides are most likely explained by translation initiation at dnAUGs, especially given the supporting data from ribosome profiling, TRII score analysis, and CAI assessment.
Translation initiation at dnAUGs expands the proteome’s repertoire of protein functions as well as targeting and localization within the cell (Supplementary Tables S7, S8). A large number (25) of the detected frame-1 downPeptide genes encode a predicted signal sequence between the annAUG and dnAUG, which in some cases may target the full-length and truncated versions of the proteins to different cellular compartments (Fig. 1B). Predicted motifs for several enzyme activities are also encoded between the annAUG and dnAUG (Supplementary Tables S7, S8). In addition, the amino acid composition coded between the annAUG and dnAUG of downPeptide genes has a greater tendency towards predicted disorder than the equivalent regions of randomly selected genes (Supplementary Fig. S5), which can facilitate association with regulatory cofactors (34). Although translation initiation at annAUGs and dnAUGs may result in proteins with different functions or localizations, it is also likely that for many genes, the alternative frame-1 products are not significantly different in their behaviors. Indeed, over evolutionary time, the diversification of initially similar products could underlie the emergence of new gene functions. Global assessment of the Genome Ontology terms (GO terms; Supplementary Table S9) did not reveal significant differences between the terms associated with annotated frame-1 proteins of the downPeptide genes compared to the terms associated with the detected annPeptide genes. However, proteins initiating at frame-2 and -3 dnAUGs add to the yeast proteome complexity and potentially provide significant new functions.
These results suggest that the yeast proteome is more complex than currently annotated. The proteomes of yeast, and by extension other species, are likely under-annotated. In some cases, the amino-termini of proteins may be misannotated, but in many cases, genes probably utilize multiple translation start sites, as also suggested by recent ribosome profile results (3, 4), raising the interesting possibility that translation initiation can be a regulated or probabilistic event. Translation of alternative protein products with different functions and cellular localizations or targeting, provides a versatile mechanism to regulate and evolve gene expression behaviors in response to environmental and developmental cues (35). Moreover, analyses of individual gene products do not generally assume that proteins have a diversity of amino termini, and this knowledge could have profound implications for the design and interpretation of these studies.
Supplementary Material
Acknowledgments
Funding Sources:
This work was supported in part by funds from the Howard Hughes Medical Institute to support undergraduate initiatives in the life sciences, NSF grant CNS-0959856 and NIH grant 1R15GM096228.
We thank David Han, Mark Flory, Robert Lane, Michael Rice, and Will Gladstone for discussions.
Abbreviations
- annAUG
annotated AUG
- dnAUG
downstream AUG
Footnotes
Author Contributions:
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Lander ES, Linton LM, Birren, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res. 2002;30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kochetov AV. AUG codons at the beginning of protein coding sequences are frequent in eukaryotic mRNAs with a suboptimal start codon context. Bioinformatics. 2005;21:837–840. doi: 10.1093/bioinformatics/bti136. [DOI] [PubMed] [Google Scholar]
- 7.Kochetov AV. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays. 2008;30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 8.Wang XQ, Rothnagel JA. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res. 2004;32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porras P, Padilla CA, Krayl M, Voos W, Barcena JA. One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J Biol Chem. 2006;281:16551–16562. doi: 10.1074/jbc.M600790200. [DOI] [PubMed] [Google Scholar]
- 10.Kochetov AV, Sarai A, Rogozin IB, Shumny VK, Kolchanov NA. The role of alternative translation start sites in the generation of human protein diversity. Mol Genet Genomics. 2005;273:491–496. doi: 10.1007/s00438-005-1152-7. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 14.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546, 563–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 15.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 16.Russo A, Chandramouli N, Zhang L, Deng H. Reductive glutaraldehydation of amine groups for identification of protein N-termini. J Proteome Res. 2008;7:4178–4182. doi: 10.1021/pr800224v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins-Pianka A, Rice MD, Weir MP. The mRNA landscape at yeast translation initiation sites. Bioinformatics. 2010;26:2651–2655. doi: 10.1093/bioinformatics/btq509. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, Vetro JA, Chang YH. The specificity in vivo of two distinct methionine aminopeptidases in Saccharomyces cerevisiae. Arch Biochem Biophys. 2002;398:87–93. doi: 10.1006/abbi.2001.2675. [DOI] [PubMed] [Google Scholar]
- 19.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgibbon M, Li Q, McIntosh M. Modes of inference for evaluating the confidence of peptide identifications. J Proteome Res. 2008;7:35–39. doi: 10.1021/pr7007303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 22.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Lundgren DH, Han DK, Eng JK. Protein identification using TurboSEQUEST. Curr Protoc Bioinformatics. 2005;Chapter 13(Unit 13):13. doi: 10.1002/0471250953.bi1303s10. [DOI] [PubMed] [Google Scholar]
- 24.Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 26.Weir MP, Rice MD. TRII: A Probabilistic Scoring of Drosophila melanogaster Translation Initiation Sites. EURASIP Journal on Bioinformatics and Systems Biology. 2010;2010 doi: 10.1155/2010/814127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci U S A. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez CI, Bhattacharya A, Wang W, Peltz SW. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene. 2001;274:15–25. doi: 10.1016/s0378-1119(01)00552-2. [DOI] [PubMed] [Google Scholar]
- 32.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 34.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Thomas D, Plant LD, Wilkens CM, McCrossan ZA, Goldstein SA. Alternative translation initiation in rat brain yields K2P2.1 potassium channels permeable to sodium. Neuron. 2008;58:859–870. doi: 10.1016/j.neuron.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.