Abstract
Support for molecular biology researchers has been limited to traditional library resources and services in most academic health sciences libraries. The University of Washington Health Sciences Libraries have been providing specialized services to this user community since 1995. The library recruited a Ph.D. biologist to assess the molecular biological information needs of researchers and design strategies to enhance library resources and services. A survey of laboratory research groups identified areas of greatest need and led to the development of a three-pronged program: consultation, education, and resource development. Outcomes of this program include bioinformatics consultation services, library-based and graduate level courses, networking of sequence analysis tools, and a biological research Web site. Bioinformatics clients are drawn from diverse departments and include clinical researchers in need of tools that are not readily available outside of basic sciences laboratories. Evaluation and usage statistics indicate that researchers, regardless of departmental affiliation or position, require support to access molecular biology and genetics resources. Centralizing such services in the library is a natural synergy of interests and enhances the provision of traditional library resources. Successful implementation of a library-based bioinformatics program requires both subject-specific and library and information technology expertise.
INTRODUCTION
Biomedical research now takes place in the context of an exploding information environment. For example, GenBank [1], the public repository for DNA sequence information maintained by the National Center for Biotechnology Information (NCBI) of the National Library of Medicine (NLM), has been doubling in size every fifteen to eighteen months, growing from 5,700 entries in 1985 to over three million sequence records with over two billion base pairs by 1998. This growth has occurred in the context of the parallel expansion of the Internet and the nearly universal adoption of the Web as the medium for connectivity and database access. GenBank is only one of many hundreds of diverse molecular biological and genetics information resources available to researchers over the Internet. These resources include primary DNA and protein sequence databases, human and model organism genome sites, genetic mutation and variation databases, protein structural databases, visualization tools, molecular sequence database similarity search systems, gene and protein structure prediction servers, and pattern and motif identification tools [2–7]. The 1998 annual database issue of Nucleic Acids Research contains descriptions of 102 databases, an increase of 36% from 1997 [8]. Factors driving the expansion of molecular biological and genetic information resources include the establishment and funding of national and international coordinating agencies [9, 10], technological advances [11, 12], and large-scale collaborative genetic mapping and sequencing programs [13, 14].
The explosive growth of molecular sequence databases and other biological information resources are having major impacts on how biomedical research is performed. As DNA and protein sequence databases have grown, questions that a few years ago would have been addressed at the laboratory bench are now being answered on the information desktop. Biomedical researchers are replacing “wet” (in vitro or in vivo) bench science with “dry” (in silico) computer activities [15–17]. As the size and complexity of biological information resources has grown, the ability to find and use the information in molecular sequence databases has become increasingly important to researchers. The techniques for gene cloning, DNA manipulation, and DNA sequencing that have been developed by molecular biologists and virologists have now become ubiquitous in biomedical research laboratories. As the applications of gene and protein analysis have expanded, so have the needs of diverse researchers for access to molecular sequence information resources. The term bioinformatics has come to be used broadly to describe the use of computer and information technology in the life sciences and specifically to refer to the development and application of molecular sequence databases and software [18, 19].
Grefsheim and Cunningham [20, 21] conducted studies of the biotechnology-related information needs of researchers in academic health sciences centers in 1988. The scientists surveyed obtained their information from three basic sources: most importantly, their own research; secondarily, from personal communication with other scientists; and, thirdly, from printed and electronic media. Major conclusions of the study were that health sciences libraries were perceived as becoming increasingly tangential to the scientific process and that the role of the library and librarians should be to assist and train researchers in accessing and using information in diverse distributed sources more effectively.
Medical libraries at several institutions have developed bioinformatics support programs. The Houston Academy of Medicine-Texas Medical Center Library had a prototype liaison program to serve the information needs of biotechnology clients [22]. The responsibilities of the liaison included providing education and training in biotechnology to other library personnel, representing the interests of biotechnology clients in collection development, and exploring the use of databases to improve access to research results. The Medical Library at the Washington University School of Medicine participated as a test site for the NCBI GenInfo program [23], developed a networked system to provide access to molecular sequence and mapping databases and analysis software, and established a support program to coordinate access to both bibliographic and nonbibliographic databases. The University of California at San Francisco included an introductory class in genome informatics in its library bibliographic instruction program to familiarize students with the major genetic and molecular sequence databases, Internet tools for database access, and effective use of these resources [24].
The University of Washington (UW) Health Sciences Libraries (HSL) have developed a bioinformatics services program to enhance access for researchers to molecular sequence databases and other biological information resources. The UW HSL serves the Health Sciences Center, which consists of schools of medicine, dentistry, nursing, pharmacy, social work, and public health and community medicine. The diverse user population includes 1,800 faculty, 3,000 graduate and professional students, and several thousand research staff. UW is one of the nation's leading research centers and has ranked among the top five federally funded research institutions for the past twenty years. In recent years, it has ranked second overall and first among public universities in receipt of federal research funds. In 1998, UW received $557 million in contract and grant awards, with approximately 60% going to researchers in the biomedical sciences. Several major genomic mapping and sequencing programs are based at the UW, as are a number of Howard Hughes Medical Institute (HHMI) funded investigators and research groups. The university serves as an incubator for the vibrant and growing biotechnology industry in the Seattle area.
THE HSL BIOINFORMATICS PROGRAM
In 1994, HHMI began awarding grants to host institutions to support the work of medical libraries. Discussions between HSL administrators and UW HHMI researchers resulted in the development of an innovative program to enhance access to molecular biology information resources. The major components of the program included consultation services, education and training, bioinformatics Web pages, and networked biological information resources. These services were the primary responsibility of a new bioinformatics consultant position, which has been held by an individual with a doctoral degree in molecular and cellular biology. Bioinformatics services have been provided at HSL since October 1995.
Needs analysis
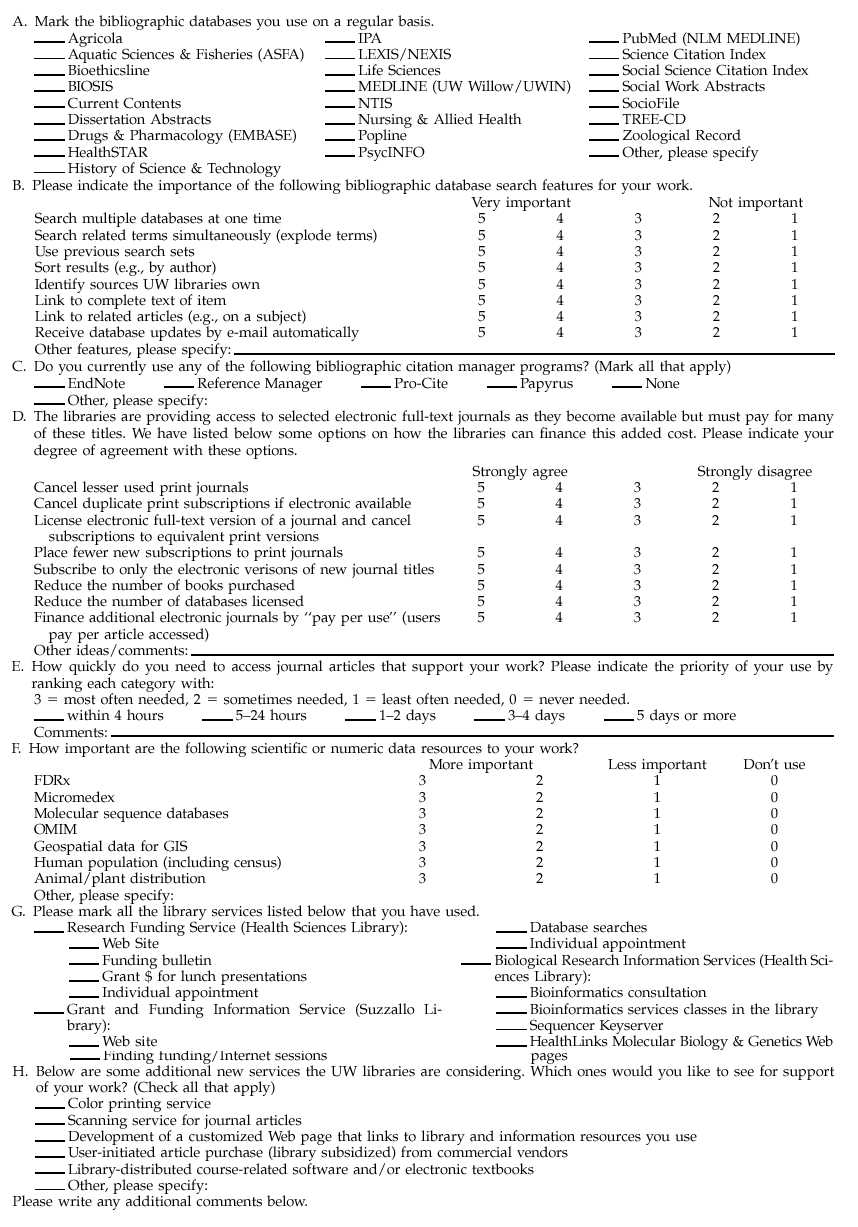
As a first step in the process of establishing this program, the bioinformatics consultant attended laboratory meetings of nine of the eleven HHMI research groups in the fall of 1995 and winter of 1996. Participants included eight laboratory group heads and research faculty, twenty-three postdoctoral researchers, twelve research technicians, and fifteen graduate students. These sessions consisted of live demonstrations of biological information resources on the Web and discussions about researchers' information needs and library services. At the beginning of each session, the researchers were asked to complete a survey about their use of both library services and biological information resources (Appendix A). Approximately 45% of the 127 UW HHMI researchers participated in these meetings and completed the surveys.
A large majority of the respondents reported utilizing DNA or protein sequence databases (85%) or sequence analysis tools (73%) in their research, most commonly on a weekly or monthly basis (Figure 1). The most widely used software for these purposes were the commercial Genetics Computer Group (GCG) package accessible over the campus network and publicly available programs on the Internet and Web. The primary sources of information about these resources were personal contacts with other researchers. Few of the respondents reported a high level of awareness of library services. A large majority described themselves as somewhat or barely aware of specific services. The most widely used services reported were self-searching of bibliographic databases from home or laboratory computers and reading or copying of journal articles (Figure 2).
Figure 1.
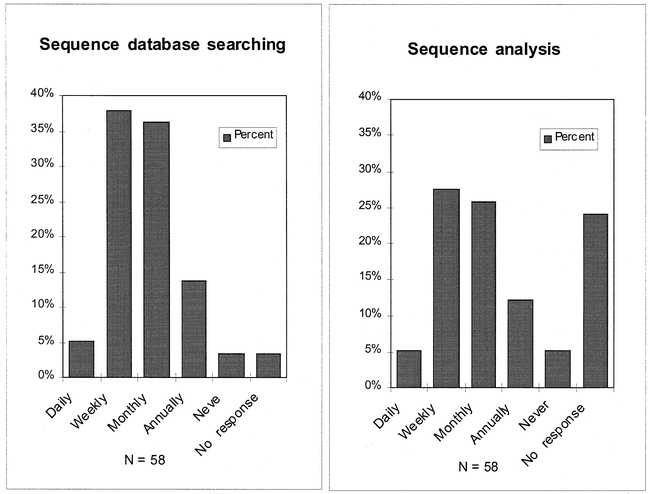
Frequency of molecular sequence activities by HHMI researchers
Figure 2.
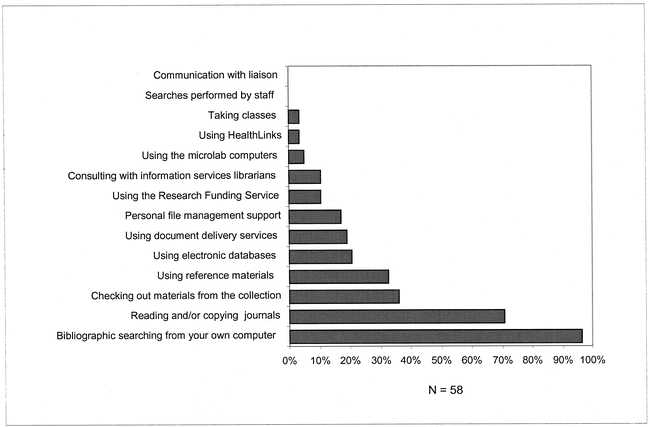
Use of HSL services by HHMI researchers
Strong interest was expressed in additional services, including desktop access to networked reference resources, molecular biological consultation services, classes in molecular sequence databases and analysis, and access to sequence analysis software. In a follow-up focus group, researchers recommended expanded access to electronic journals, training and support for basic computer and network skills, and establishment of an e-mail update list for communication. The discussions and survey results clearly showed that these scientists had broad ranges of computer, Internet, and molecular sequence analysis experience. There was strong interest for the library to become an active agent in providing bioinformatics consultation and training services for different levels of sophistication and skill.
Molecular biology consultation services
The consultation services developed to meet these needs included molecular sequence database similarity searching, interpretation of database search results, and DNA and protein sequence analysis. Approximately 340 clients have been served over the first three years of the program. Many of the initial users of this service were researchers who had attended the needs analysis sessions. Over time, the clientele has been drawn from diverse departments and laboratories throughout the Health Sciences Center. Of the 131 consultation clients for whom departmental status was identified, 55% were associated with basic sciences departments and 45% with clinical departments. The three departments with the largest number of users were biochemistry (20%), pathology (17%), and pharmacology (7%). The largest percentage of clients have been faculty and clinical fellows (37%), followed by postdoctoral research fellows (28%), research technicians (22%), and graduate students (13%).
Many client interactions entailed multiple contacts between the consultant and the client, with an average of 3.2 contacts per client. The largest percentage of these contacts took place by e-mail (81%), followed by inperson meetings (13%) and telephone conversations (6%). The preference for e-mail reflected both the predominant fashion in which the clients contacted the consultant, as well as follow-up messages to e-mail, phone conversations, and meetings. Most of the inperson consultation sessions were with individual researchers, but sometimes meetings were held with groups of two to ten persons. The format, number of sessions, and time required to resolve a particular problem depended on multiple factors including the complexity of the question, the client's background knowledge, and the consultant's familiarity with the topic.
Questions have been answered in such areas as programs for identifying patterns in proteins, performance and analysis of Basic Local Alignment Search Tool (BLAST) searches, access to GenBank, software for scientific plotting and drawing, computer hardware, Internet connectivity, and sources for specific biological reagents. These questions could be classified into three categories based on an estimation of the degree of knowledge required on the part of the consultant to answer adequately:
1. Basic questions required the least amount of specialized molecular biological subject-specific expertise to answer and were similar to the types of questions asked of health sciences or biomedical reference librarians, such as how to locate biological information resources, how to find a particular program or database, and whether a manual or help file was available. An example of a basic question was: “How do I find the Seqpup program for sequence assembly” This type of question could be expected to be answered by a biomedical librarian with some bioinformatics training. Many basic questions were answered by directing the client to the HealthLinks Molecular Biologist Toolkit, a component of the HSL Web gateway. On the basis of analysis of the questions asked of the consultant in year two of the program, about 15% of the inquiries were of this basic type.
2. Technical questions that required significant subject-specific knowledge to answer were the most common. Approximately 75% of the inquires were of this type. Many of these questions concerned the identification of the most appropriate database or program for a specific task, use of a search tool or sequence analysis program, or interpretation of database search results. An example of a technical questions was: “Is there a program to design primers from a peptide with known amino acid sequence?” Many of the resource identification questions were addressed by directing the client to resources listed in the HealthLinks Molecular Biologist Toolkit, but required more knowledge of the specific attributes of the resources than would be expected from a biomedical librarian without advanced training.
3. Analytical questions required the most time and expertise on the part of the consultant, including providing indepth assistance with data analysis or help in planning experimental strategies. Approximately 10% of the inquiries were of this type, and these inquiries usually entailed multiple inperson meetings between the consultant and client. One example of this type of question involved the following iterative process: assistance with BLAST searching of protein and expressed sequence tag (EST) databases with a newly discovered DNA sequence, instruction in how to perform multiple alignments of this protein to related sequences, and demonstration of various tools to identify conserved motifs associated with different functional activities within the protein family.
EDUCATIONAL PROGRAM
Skills training
Bioinformatics classes were added to the HSL education program in January 1996. In the first year, two classes were offered: Netsurfing for Biologists, a survey of Web resources for basic biomedical sciences, and Molecular Biology Database Searching, a more advanced class on Web tools for molecular sequence database searching and analysis. Each class consisted of a two-hour session held in the library microcomputer laboratory. The class format for netsurfing was an instructor guided hands-on exploration of selected sites using a Web browser. A Web page with links and annotation was developed and used to structure the class. The molecular biology database class consisted of an introduction to various databases and search tools, and brief hands-on demonstrations. Attendees included faculty, staff, and students from throughout the Health Sciences Center. Approximately fifty individuals attended classes during the first year of the program.
Postclass forms and e-mail surveys were used to evaluate the classes. Based on this feedback, the classes were restructured in the second year. The Web survey class was renamed Web Resources for Molecular Biology and focused more narrowly on identifying and locating tools for molecular biological research. Rather than trying to cover diverse molecular sequence database and analysis tools in a single class, Molecular Biology Database Searching was divided into separate classes on the NCBI Entrez system, BLAST sequence similarity searching, and protein motif identification. An additional class in using the Sequencher program for DNA sequence assembling and editing was added. The classes were presented and advertised as a Bioinformatics Series and taught over a two to three–week period one or two times each academic quarter. In the second year, 189 participants attended twenty-four classes and, in the third year, 259 participants attended twenty-six sessions.
The format for each two-hour class consisted of three components: (1) PowerPoint background presentation, (2) hands-on guided demonstration of resources and program features, and (3) Web-based tutorial. Hypertext markup language (HTML) versions of the background presentations and tutorials were linked to the bioinformatics consultant's Web page [25]. Classes took place in a laboratory with an assistant to guide students as they followed along during the demonstrations and exercises, and handouts included background slide show outlines and Web page printouts.
Classes were publicized in the HSL educational program brochures, newsletter, Web site, posted announcements, and targeted e-mail. E-mail messages with schedules and registration instructions were sent to HSL lists and to medical school administrators for forwarding to departmental lists. E-mail was the most effective publicity method; about 75% of recent participants specified it as their source of information about class availability.
A follow-up evaluation survey was sent to Bioinformatics Series participants in May 1998 (Appendix B). The survey form and an anonymous reply envelope were mailed to all researchers who had attended one or more of the classes in the previous twelve months. Responses were received from forty out of eighty individuals. Participants were asked how often they had used the relevant resources before and after taking each class and how they evaluated the overall impact of the class on their ability to use these resources in their research. Scoring was based on Likert scales of 1 to 5, with 1 corresponding to “never used” or “no impact” and 5 corresponding to “very often” or “very high impact.”
Survey results indicated that the Bioinformatics Series classes were considered to be of value to participants. For each of the five different classes, the majority of respondents reported that they were using the resources covered more frequently after taking the class (Figure 3). The increase in self-estimated use was greatest for Using the Entrez System (Likert median 1 before to 3 after, P = 0.0002)†, followed by Web Resources for Molecular Biologists (Likert median 2 before to 3 after, P = 0.001)† and BLAST Sequence Database Similarity Searching (Likert median 1 before to 3 after, P = 0.003)†. The predominant reason cited for not using the covered resources after training was lack of need rather than insufficient skills. Median self-estimated impact scores for the classes ranged from 3 for DNA Sequence Assembly and Editing Using Sequencher to 4 for Using the Entrez System (Figure 4).
Figure 3.
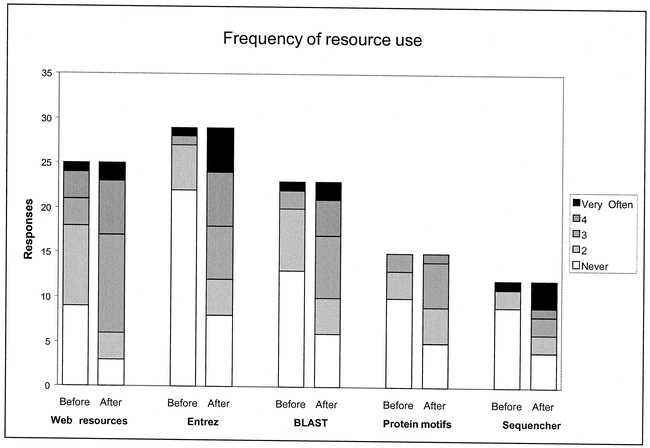
Changes in frequency of resource use reported by participants in Bioinformatics Series classes
Figure 4.
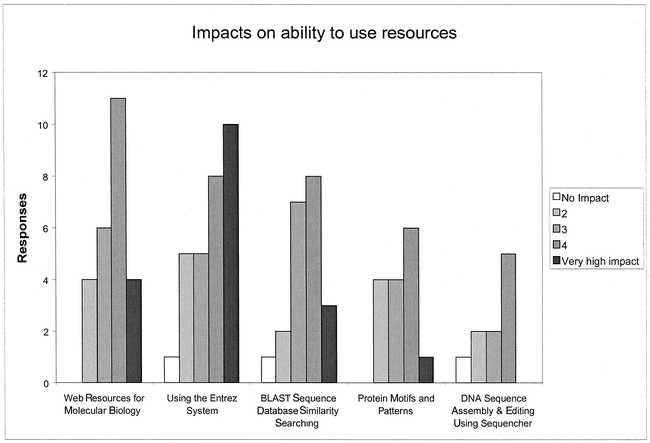
Impacts on ability to use resources reported by participants in Bioinformatics Series classes
Graduate curriculum courses
The bioinformatics consultant has been teaching a graduate level course entitled “Bioinformatics and Gene Sequence Analysis.” This course is offered jointly by the departments of pathobiology and medical education, and is sponsored by the HSL. It is organized and coordinated by the bioinformatics consultant in collaboration with a pathobiology faculty member. The course is part of the curriculum for the doctoral degree program in pathobiology and is open to students and postgraduate trainees in the biomedical, computer, and information sciences. Covered topics include the generation and manipulation of sequence information, protein and DNA sequence analysis, database searching, multiple sequence alignments, phylogenetic analysis, genome projects, and protein structure modeling. The course format is a one-hour weekly lecture presented either by one of the course coordinators or a guest lecturer and a two-hour computer laboratory session. The bioinformatics consultant also regularly presents lectures about biological information resources to graduate classes in the Department of Medical Education and the School of Library and Information Sciences.
Bioinformatics Web pages
Another component of the program has been the development of pathfinder Web pages to aid researchers in accessing biological information resources. These pages are integrated into HealthLinks [26], a joint project of HSL and the Integrated Advanced Information Management Systems (IAIMS) Program. HealthLinks is an electronic gateway or portal to local UW and remote information resources for the biomedical sciences and includes various “toolkits” that bring together information based on role (e.g., Grantseeker, Care Provider) and format (e.g., databases, texts, journals). The HealthLinks Molecular Biologist Toolkit is organized as a subject-specific hierarchical menu of links to molecular biology and genetics resources along with descriptive annotation (Figure 5). These pages receive an average of approximately 2,000 hits per month and are employed by HSL staff to direct clients to appropriate resources.
Figure 5.
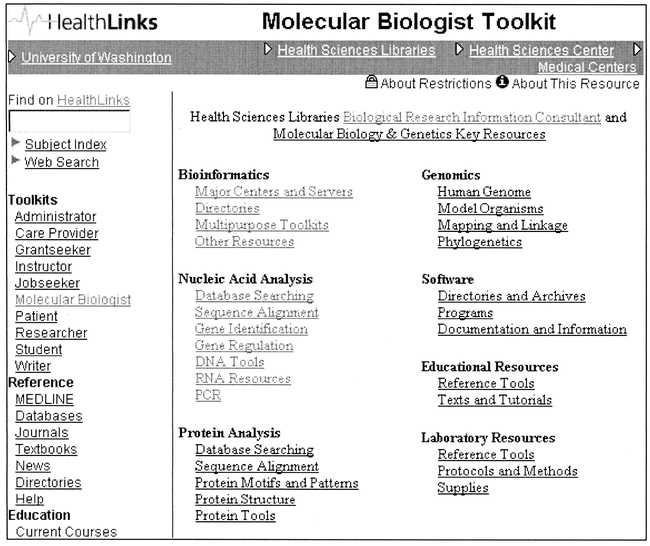
HealthLinks Molecular Biologist Toolkit
Statistics from a spring 1998 survey of 2,100 UW biological and health sciences faculty (31% return) and a sample of 400 graduate and professional students (35% return) indicated the value of this service to our users (Appendix C). Molecular sequence databases were considered to be of high importance by 15% of faculty respondents and 8% of students, while 9% of faculty and 19% of students reported use of biological research information services. HealthLinks molecular biology and genetics Web pages were used by 11% of faculty and 16% of students.
Networked biological information resources
The bioinformatics consultant has actively participated in the librarian liaison group and taken part in information resource decision making. Interest in new products and services obtained from the survey of HHMI researchers and subsequent queries of potential users through the BioList e-mail list has provided a qualitative increase in researcher input. Based on survey priorities, Current Contents became a campus network database service and two electronic full-text reference titles, Methods in Enzymology: Molecular Biology and Encyclopedia of Virology, were made available on the library CD-ROM network.
A pilot project to provide networked access to molecular biology software has been in place since 1997. The goal is to address identified needs for specific resources not available via the Web. These include DNA sequence assembly or editing software and integrated, multi-purpose sequence analysis packages for Windows- and Macintosh-based computers. Most laboratory groups do not purchase such programs because of high cost, irregular use, or lack of awareness. There has been little or no departmental level or university-wide support for this type of software. The pilot project has included a three-user license for Sequencher (Gene Codes Corporation), a Macintosh-based program for DNA sequence assembling and editing, and a ten-user license for KeyServer (Sassafras Software), an access control and monitoring program. Network connection to the KeyServer server is via transmission-control protocol/Internet protocol (TCP/IP); access is restricted to UW users by IP address control and the number of simultaneous connections controlled by a hardware device and the KeyServer program.
The Sequencher program has been installed on over 120 Macintosh computers for sixty laboratory groups from fifteen departments. The bioinformatics consultant has installed the program and necessary system components on laboratory computers, demonstrated program features to contact people, and collected laboratory, contact, and equipment information. Sequencher training classes have been offered as part of the Bioinformatics Series, and copies of the program manual and tutorial have been available in the library. Most users have required little follow-up support because of the well-designed interface. Usage statistics for Sequencher were analyzed for the seven-month period from September 1997 through March 1998. The program was used 2,400 times, on 100 different computers, with an average connect time of sixty minutes per session. The three-user limit denied access to potential users 765 times, and there was an average wait time of twenty-two minutes for those unable to obtain a key.
Following up on the success of the Macintosh Sequencher pilot project, network licenses for the new Windows version of the program and a general-purpose sequence analysis software package have recently been purchased with funds from the HSL collection budget.
Communication and outreach
Introductory letters were sent to the departmental chairs in the basic sciences departments during the first year of the program to inform the potential user community about the library's bioinformatics services. As a result of these contacts, the bioinformatics consultant was invited to make presentations about biological information resources and HSL services to a number of basic science departments.
An electronic mailing list of researchers interested in bioinformatics, the BioList, has been an important component of outreach efforts. During the first year of the program, e-mail messages describing the bioinformatics consultation, education services, and Web pages were sent to more than 1,000 basic sciences researchers (faculty, research technicians, postdoctoral fellows, graduate students). About 10% of those queried responded that they were interested in receiving e-mail updates, and the list has subsequently grown to 400 recipients from basic sciences and clinical departments.
Messages have been sent to the BioList every few months. The content has been less than two screens and has included Bioinformatics Series schedules, Web sites for polymerase chain reaction (PCR) primer selection, new library databases, and changes in NCBI Entrez and BLAST servers. The list has served as a reminder about the program, as evidenced by an increase in client-initiated contacts following each update. Messages about topics of more limited interest and surveys about specific services have been sent to subsets of the list.
DISCUSSION
The response by researchers to the HSL bioinformatics program demonstrates that an extensive range of biological information services can be successfully provided by a health sciences library. Use of the educational and consultational components of the program by researchers from clinical and basic sciences departments indicates a widespread need for bioinformatics training and support. In addition, the extensive use of the Sequencher program illustrates the demand for access to sophisticated molecular sequence analysis software. The HSL Bioinformatics program has enhanced the ability of biomedical researchers to use biological information resources effectively. In the follow-up survey of the bioinformatics classes, participants have reported increases in frequency of use of the resources covered, and most have reported that the classes have a high impact on their ability to use these resources.
Academic biomedical researchers are increasingly becoming at a competitive disadvantage relative to those working in the biotechnology and pharmaceutical industries, where there is abundant corporate recognition and support for bioinformatics [27]. Although the University of Washington is one of the major biomedical research facilities in the world, there has been little or no institutional recognition of bioinformatics and little in the way of resources provided for nonbibliographic information infrastructure. Many companies have bioinformatics service groups, often comprised of systems personnel, programmers, librarians, and research scientists [28]. With the appropriate allocation of institutional resources, biomedical and health sciences libraries can help serve to meet the broad informational needs of researchers and students. Biomedical libraries should consider nonbibliographic biological information resources, including software for molecular sequence analysis, as part of collection development.
CONCLUSION
As the primary role of the library has shifted from information repository to portal, the need for specialized subject expertise has both broadened and intensified. The HSL bioinformatics program illustrates the changing roles of professionals in the new information-based basic research environment. The type and scope of bioinformatics services that can be provided by a biomedical library will greatly depend on the level of available subject-specific expertise. In the experience at HSL, less than 20% of the bioinformatics consultation questions could be expected to be answered by a librarian who does not have a strong background in molecular biology or specialized training in this area. Few biomedical librarians have the training or experience that would enable them to provide a full range of bioinformatics services including such activities as advising about the interpretation of molecular sequence database search results. The great majority of the questions addressed by the bioinformatics consultant require a significant level of technical expertise to answer competently. However, many of these questions still fall within a range of subject-specific knowledge similar to that exhibited by expert reference librarians in relation to clinical medicine. The need for expanded support services may be met either by educating biomedical librarians about relevant biological information resources or by adding bioinformatics professionals to the library staff.
The Medical Library Association Molecular Biology and Genomics Special Interest Group (MBGSIG) offers an opportunity for professional development in these areas. Its goals are to provide a forum for exchange of knowledge about molecular biology, genome projects, and clinical genetics information resources; to facilitate communication and collaboration in the development and delivery of relevant information services; and to promote the professional development of librarians. The Web site provides information about membership, projects, and links to various genetic and molecular biological educational resources [29].
The MBGSIG organized a symposium entitled the “Impact of Genomics on Twenty-first Century Medicine,” which was held at the 1998 Medical Library Association meeting. A major area of discussion was the need for increased bioinformatics professional development opportunities for librarians and library and information sciences students. Participants suggested that the NLM and NCBI should play a more active role in providing these opportunities.
Acknowledgments
The authors thank Catherine Burroughs for assistance with the Bioinformatics Series evaluation survey and helpful comments on the manuscript, and Doug Schaad for help with the analysis of survey results. The authors would like to recognize the important contributions of Emily Hull, Leilani A. St. Anna, Steve Rauch, Kevin Ibrahim, and Joanne West in HealthLinks Web site development and maintenance. The authors also would like to acknowledge the significant role that Joanne West has played in the development of training materials for the Bioinformatics Series classes and thank her for her dedication and skillful work.
APPENDIX A
Survey for University of Washington HSLIC bioinformatics services
Name
Phone
E-mail address
Today's date
University status:
Department:
Lab group:
Years at UW:
——Faculty
——Postdoctoral fellow
——Staff
——Grad student
——Undergrad student
——Other (specify)
Computer systems employed:
——Macintosh
——DOS
——Windows
——Unix
——Other (specify)
Do you utilize DNA or protein sequence databases or analysis tools in your work?
Databases:——Yes
——No
Analysis:——Yes
——No
What types of software do you use for database access and sequence analysis?
——Shareware/freeware on lab microcomputer (i.e., DNA Strider)
——Commercial software package on lab microcomputer (i.e., Geneworks)
——Shareware/freeware on networked computer (i.e., ClustalV)
——Commercial software package on networked computer (i.e., GCG, IG).
——Web and other Internet resources (i.e., BLAST, Retrieve).
——Other (specify)
List any packages or programs you find useful:
How often do you perform:
database searching? sequence analysis?
——Daily ——Daily
——Weekly ——Weekly
——Monthly ——Monthly
——Annually ——Annually
——Never ——Never
How do you find out about the availability and use of these resources?
——Other people in the lab
——Other UW researchers
——Other personal contacts
——Reading the literature
——UW classes
——Commercial advertising
——Meetings or workshops
——Internet searching
——Health Sciences Library
——Other (Specify)
Are you interested in participating in graduate level courses in molecular biology database searching and sequence analysis?
——Yes (teaching)
——No
——Yes (student)
——No
How do you use the services of the Health Sciences Library and Information Center (HSLIC)?; check any that apply.
——Bibliographic searching from your own computer (UWIN or Willow)
——Searches performed by HSLIC staff
——Reading and/or copying HSLIC journals
——Using reference materials at the HSLIC (i.e., Current Protocols in Immunology)
——Using HSLIC electronic databases (i.e., Current Contents, CARL)
——Consulting with information services librarians
——Using the microlab computers or other TLC resources
——Using interlibrary loan or other document delivery services
——Checking out materials from the collection
——Using the Research Funding Service
——Taking HSLIC classes
——Using HealthLinks
——Personal file management support
——Reference Manager
——Endnote
——Communication with HSLIC liaison to your department
——Other (specify)
Rate your awareness of HSLIC services.
Oblivious
Barely aware
Somewhat aware
Very aware
Highly aware
How useful are HSLIC services to you?
Don't use
Of little use
Somewhat useful
Very useful
Extremely useful
What additional services would you like the HSLIC to provide?
——Provide networked access to Current Contents
——Provide networked access to Methods in Enzymology
——Provide networked access to current protocols
——Provide online access to other databases and reference sources (specify)
——Consult/advise about molecular biology information resources
——Provide access to commercial sequence analysis software for microcomputers
——Perform sequence database searches and analysis
——Increase holdings of laboratory manuals, handbooks, and other procedure references
——Establish a collection of research laboratory templates
——Provide access to more full-text journals online (list three)
1.——————
2.——————
3.——————
——Other (specify)
Would you take HSLIC sponsored mini-classes in such areas as molecular biology database searching and sequence analysis?
——Yes ——No
Are you interested in participating in a focus group to strengthen HSLIC services to basic science researchers?
——Yes ——No
Suggestions?comments?questions:————
APPENDIX B
Survey for Bioinformatics Series classes
According to our records, you attended one or more of the Health Sciences Libraries and Information Center's Bioinformatics Series classes in the past year. Please fill out this brief survey to help us to evaluate the impact of these classes. Use the accompanying reply envelope to ensure confidentiality. Thank you very much for your assistance.
1. Web Resources for Molecular Biology
Did you take this class?
——Yes ——No ——Don't remember
If you took this class answer the following, otherwise skip to question #2:
How often did you use molecular biological databases and analysis resources on the Web before taking this class?
——Scale of 1 to 5 (never to very often)
How often have you used these resources since taking this class?
Scale of 1 to 5 (never to very often)
If you have not used these resources, why not?
——Lack of need
——Insufficient skills
——Other (specify)
Do you use the Web pages introduced in this class for connecting to molecular biological databases and analysis resources?
——Scale of 1 to 5 (never to very often)
Rate the overall impact of this class on your ability to use molecular biological databases and analysis resources.
——Scale of 1 to 5 (no impact to very high impact)
What additional topics would you like to see covered in this class?
2. Using the Entrez System
Did you take this class?
——Yes ——No ——Don't remember
If you took this class answer the following, otherwise skip to question #3:
How often had you used Entrez before taking this class?
——Scale of 1 to 5 (never to very often)
How often have you used Entrez since taking this class?
——Scale of 1 to 5 (never to very often)
If you have not used Entrez, why not?
——Lack of need
——Insufficient skills
——Other (specify)
Rate the overall impact of this class on your ability to access the information in the Entrez databases.
——Scale of 1 to 5 (no impact to very high impact)
What additional topics would you like to see covered in this class?
3. BLAST Sequence Database Similarity Searching
Did you take this class?
——Yes ——No ——Don't remember
If you took this class answer the following, otherwise skip to question #4:
How often did you perform BLAST searches before taking this class?
——Scale of 1 to 5 (never to very often)
How often have you performed BLAST searches since taking this class?
——Scale of 1 to 5 (never to very often)
If you have not performed BLAST searches, why not?
——Lack of need
——Insufficient skills
——Other (specify)
Rate the overall impact of this class on your competence in performing BLAST searches.
——Scale of 1 to 5 (no impact to very high impact)
What additional topics would you like to see covered in this class?
4. Protein Motifs and Patterns
Did you take this class?
——Yes ——No ——Don't remember
If you took this class answer the following, otherwise skip to question #5:
How often did you perform protein pattern searches before taking this class?
——Scale of 1 to 5 (never to very often)
How often have you performed protein pattern searches since taking this class?
——Scale of 1 to 5 (never to very often)
If you have not performed these searches, why not?
——Lack of need
——Insufficient skills
——Other (specify)
Rate the overall impact of this class on your competence in performing protein pattern searches?
——Scale of 1 to 5 (no impact to very high impact)
What additional topics would you like to see covered in this class?
5. DNA Sequence Assembly and Editing Using Sequencher
Did you take this class?
——Yes ——No ——Don't remember
If you took this class answer the following, otherwise skip to question #6:
How often did you use the Sequencher program before taking this class?
——Scale of 1 to 5 (never to very often)
How often have you used the Sequencher program since taking this class?
——Scale of 1 to 5 (never to very often)
If you don't use Sequencher, why not?
——Lack of need
——Insufficient skills
——Other (specify)
Rate the overall impact of this class on your competence in using Sequencher.
——Scale of 1 to 5 (no impact to very high impact)
What features of the Sequencher program are most useful to you? (check any that apply)
——Sequence assembly
——Sequence editing
——Restriction mapping
——Pattern identification
——Formatting of output
——Other (specify)
6. What other classes would you like to be added to the bioinformatics series?
7. Comments:
APPENDIX C
Additional survey for biological and health sciences faculty
Footnotes
*The library-based bioinformatics services program at the University of Washington program is supported in part by a contribution from the Howard Hughes Medical Institute.
†Wilcoxon two-tail probability.
REFERENCES
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BFF. GenBank. Nucleic Acids Res. 1998 Jan 1;26(1):1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop MJ. Guide to human genome computing. 2d ed. San Diego, CA: Academic Press, 1998. [Google Scholar]
- Peruski LF. The Internet and the new biology: tools for genomic and molecular research. Washington, DC: ASM Press, 1997. [Google Scholar]
- Davidson L. Human genetic databases on the Internet: a Webliography. Med Ref Serv Q. 1997;16(2):49–54. doi: 10.1300/J115v16n02_06. [DOI] [PubMed] [Google Scholar]
- Hart JL, Hart GE. Biotechnology resources. C&RL News. 1997 Dec;58(11):759–62. [Google Scholar]
- Jourdan DR. Accessing genetic sequence information on the Net. Database. 1996;19(2):33–41. [Google Scholar]
- Lei PP, Martinez JP. Gateway to bioinformatics: the Internet. Med Ref Serv Q. 1996;15(2):1–22. [Google Scholar]
- Anonymous Editorial. Nucleic Acids Res 1998January1261 i [Google Scholar]
- Woodsmall RM, Benson DA. Information resources at the National Center for Biotechnology Information. Bull Med Libr Assoc. 1993 Jul;81(3):282–4. [PMC free article] [PubMed] [Google Scholar]
- Bains W. Bioinformatics in Europe—the federation strikes back. Trends Biotechnol. 1993 Jun;11(6):217–8. doi: 10.1016/0167-7799(93)90129-W. [DOI] [PubMed] [Google Scholar]
- Hunkapiller MW. Advances in DNA sequencing technology. Curr Opin Genet Dev. 1991 Jun;1(1):88–92. doi: 10.1016/0959-437x(91)80047-p. [DOI] [PubMed] [Google Scholar]
- Venter JC, Smith HO, Hood L. A new strategy for genome sequencing. Nature. 1996 May 30;381(6581):364–6. doi: 10.1038/381364a0. [DOI] [PubMed] [Google Scholar]
- Yager TD, Nickerson DA, Hood LE. The Human Genome Project: creating an infrastructure for biology and medicine. Trends Biochem Sci. 1991 Dec;16(12):454–8. doi: 10.1016/0968-0004(91)90177-w. [DOI] [PubMed] [Google Scholar]
- Guyer MS, Collins FS. How is the Human Genome Project doing, and what have we learned so far? Proc Natl Acad Sci USA. 1995 Nov 21;92(24):10841–8. doi: 10.1073/pnas.92.24.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Jonassen I, Vilo J, Ukkonen E. Predicting gene regulatory elements in silico on a genomic scale. Genome Res. 1998 Nov;8(11):1202–15. doi: 10.1101/gr.8.11.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavorgna G, Boncinelli E, Wagner A, Werner T. Detection of potential target genes in silico? Trends Genet. 1998 Sep;14(9):375–6. doi: 10.1016/s0168-9525(98)01551-0. [DOI] [PubMed] [Google Scholar]
- Wei MH, Karavanova I, Ivanov SV, Popescu NC, Keck CL, Pack S, Eisen JA, Lerman MI. In silico-initiated cloning and molecular characterization of a novel human member of the L1 gene family of neural cell adhesion molecules. Human Genet. 1998 Sep;103(3):355–64. doi: 10.1007/s004390050829. [DOI] [PubMed] [Google Scholar]
- Foo EL, Heden LG, Bioinformatics and the development of biotechnology. In: DaSilva EJ, Dommergues YR, Nyns RJ, and Ratledge C. eds. Microbial technology in the developing world. New York, NY: Oxford University Press. 1987 396–417. [Google Scholar]
- Franklin J. Bioinformatics changing the face of information. Ann NY Acad Sci. 1993 Dec 21;700:145–52. doi: 10.1111/j.1749-6632.1993.tb26314.x. [DOI] [PubMed] [Google Scholar]
- Grefsheim S, Franklin J, Cunningham D. Biotechnology awareness study, part 1: where scientists get their information. Bull Med Libr Assoc. 1991 Jan;79(1):36–44. [PMC free article] [PubMed] [Google Scholar]
- Cunningham D, Grefsheim S, Simon M, Lansing PS. Biotechnology awanreness study, part 2: meeting the information needs of biotechnologists. Bull Med Libr Assoc. 1991 Jan;79(1):45–52. [PMC free article] [PubMed] [Google Scholar]
- Pratt GF. A health sciences library liaison project to support biotechnology research. Bull Med Libr Assoc. 1990 Jul;78(3):302–3. [PMC free article] [PubMed] [Google Scholar]
- Crawford SY, Stucki LH, Halbrook B, Managing information in biomedical research: the human genome project. In: and Broering NC. ed. High-performance medical libraries: advances in information management for the virtual era. Westport, CT: Meckler Publishing. 1993 127–36. [Google Scholar]
- Owen DJ. Library instruction in genome informatics: an introductory library class for retrieving information from molecular genetics databases. Sci Technol Libr. 1995;15(3):3–15. [Google Scholar]
- University of Washington. HealthLinks. bioinformatics consultant. [Web document]. Seattle, WA: University of Washington Health Sciences Libraries, 1999. [rev 24 Jul 1999; cited 3 Sep 1999]. <http://healthlinks.washington.edu/hsl/liaisons/yarfitz/>. [Google Scholar]
- University of Washington. HealthLinks. [Web document]. Seattle, WA: University of Washington Health Sciences Libraries, 1999. [rev 1 Sep 1999; cited 3 Sep 1999]. <http://healthlinks.washington.edu/>. [Google Scholar]
- Cole NJ, Bawden D. Bioinformatics in the pharmaceutical industry. J Doc. 1996 Mar;52:51–68. [Google Scholar]
- Ibid. [Google Scholar]
- Medical Library Association. Molecular Biology and Genomics Special Interest Group. [Web document]. Chicago, IL: The Association, 1999. [rev. 16 Jul 1999; cited 3 Sep 1999]. <http://medicine.wustl.edu/~molbio/>. [Google Scholar]


