Abstract
Proteins perform essential cellular functions as part of protein complexes, often in conjunction with RNA, DNA, metabolites and other small molecules. The genome encodes thousands of proteins but not all of them are expressed in every cell type; and expressed proteins are not active at all times. Such diversity of protein expression and function accounts for the level of biological intricacy seen in nature. Defining protein-protein interactions in protein complexes, and establishing the when, what and where of potential interactions, is therefore crucial to understanding the cellular function of any protein—especially those that have not been well studied by traditional molecular genetic approaches. We generated a large-scale resource of affinity-tagged expression-ready clones and used co-affinity purification combined with tandem mass-spectrometry to identify protein partners of nearly 5,000 Drosophila melanogaster proteins. The resulting protein complex “map” provided a blueprint of metazoan protein complex organization. Here we describe how the map has provided valuable insights into protein function in addition to generating hundreds of testable hypotheses. We also discuss recent technological advancements that will be critical in addressing the next generation of questions arising from the map.
Keywords: mass spectrometry, proteomics, affinity purifications, protein complex map, Drosophila proteome, interaction mapping, high-throughput techniques
At any given point in time, thousands of proteins are interacting with other proteins inside a cell. Protein interactions can be relatively long lasting to form stable protein complexes with defined functions, or transient and short-lived during specific biochemical processes. Thus the function of any protein depends on its ability to participate in such reactions, while mutant or pathogenic conditions may reflect malfunction of normal interactions. Recognizing the fundamental importance of protein interactions and the relationship between protein complexes, much effort has been devoted to approaches that can experimentally define protein interactions within a proteome. Two key techniques have emerged as instrumental in generation of proteome-wide “interactome” maps. The first is based on the yeast two-hybrid (Y2H) system, which determines pair-wise protein interactions between two proteins at a time. Extensive interactome maps for almost all model organisms have been generated using this technique.1-7 The second technique is co-affinity purification combined with mass spectrometric analysis (co-AP/MS), for the isolation of protein complexes using individual affinity-tagged proteins. Using this technique, protein complex maps have been generated for smaller, well-defined proteomic spaces in higher model organisms8-12 and at a proteome-scale for yeast, bacteria13-15 and more recently for Drosophila melanogaster.16 The Drosophila Protein interaction Map (DPiM)16 is the largest unbiased metazoan protein complex map to date.
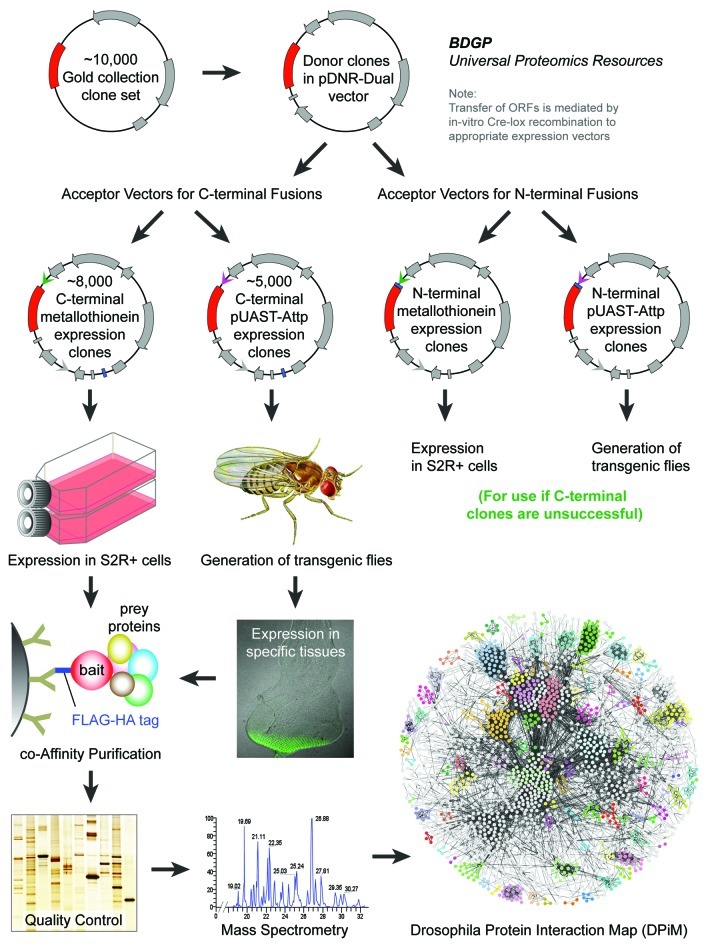
In order to generate DPiM, first we created the expression ready clone set, the “Universal Proteomics Resource” using a subset of the Berkeley Drosophila Genome Project (BDGP) cDNA Gold Collection.17 This comprehensive resource would allow affinity purifications of a large proportion of the Drosophila genome with the ability to add different tags at both the C- and the N-terminus of the cloned cDNAs (Fig. 1). Initially, we tested a range of affinity tags, and the FLAG-HA tag was chosen for its small size, availability of reagents and adaptability to a high-throughput purification platform in addition to better bait and prey recovery for a panel of test clones. The FLAG-HA epitope-tagged clone library was generated by transferring ORFs from the BDGP expression-ready clone set18 to the pMK33-C-FLAG-HA-BDGP Acceptor Vector, which is designed for the expression of FLAG-HA fusion proteins.
Figure 1. The Drosophila Protein interaction Map (DPiM) project’s protein complex purification pipeline. All the sequence verified Drosophila Gold Collection cDNA clones were transferred to a pDNR-Dual vector that allows donor clones to be transferred via Cre-lox recombination into acceptor vectors as distinct C-terminal or N-terminal tagged expression constructs. A collection of different acceptor vectors has been created with choice of affinity tags (FLAG-HA tag, TAP-tag, His-tag etc.) to be used as expression clones in cell culture (as metallothionein-inducible constructs) or to generate transgenic flies (as UAS expression constructs). Nearly 5,000 individual FLAG-HA tagged proteins were expressed in S2R+ cells followed by co-affinity purification and mass spectrometry to identity protein partners and construction of DPiM. The same set of clones has been used to generate a complementary set of stable transgenic lines for tissue and stage specific expression using the UAS-Gal4 system.
Traditionally tandem affinity purifications are used to improve protein sample purity to analyze by mass spectrometry. However in the recent years, availability of better reagents for affinity purifications combined with advancements in the sensitivity and interpretability of mass spectrometry methodologies has rendered single-step purification capable of generating reliable results. Now, with several thousand individual affinity purifications, it has become relatively easy to identify and segregate many non-specific proteins/contaminants using bioinformatics tools. It should be noted that tandem purification using two tags and use of additional controls is still advisable for small-scale experiments. We relied on competitive elution using synthetic HA peptides which is a relatively gentle method compared with chemical methods of displacement such as glycine or SDS elution. The eluted protein samples from the DPiM pipeline were precipitated using trichloroacetic acid and repeated cold acetone washes to remove free HA peptide and buffer components. By multiplexing most of the processes, we developed a high-throughput strategy to process up to 400 individual affinity purifications per month using the HA-affinity purification of protein complexes from Drosophila S2R+ cell lysates.
The S2R+ cells that hosted the expressed protein complexes, while offering the most tractable experimental system to establish the first protein complex map for Drosophila, necessarily allowed only a partial, static picture of the proteome. For example, this cell line naturally expresses less than half of the predicted Drosophila proteome (as extrapolated from gene modeling, RNA expression and RNA-seq19 data), limiting the repertoire of proteome members that can associate with the tagged proteins we used as baits. Nevertheless, the analysis we performed, while not fully saturated, allowed us to visualize 60% of the predicted S2R+ proteome and found the distribution of analyzed protein classes20 to be similar to that of the entire Drosophila proteome.16 The expression of nearly one thousand proteins as baits that are not expressed in this cell type improved our coverage. The raw mass spectrometry data that passed our quality control regime have been regularly made available, prior to publication, through FlyBase (http://flybase.org/) where they have been widely used by the community. While the bait protein was recovered in most affinity purifications, the ability to recover bait was maximized when the proteins were naturally expressed in this cell type. In addition to nearly 5,000 proteins identified from the FlyBase annotation of the Drosophila melanogaster genome, a significant proportion of the mass spectral data has not been assigned to any predicted protein sequence. Some of these may correspond to peptides with distinct post-translational modifications that have not been examined or predicted yet. Additionally, it also remains to be determined that if the spectral data from thousands of mass spectrometry runs might be mined to discover novel exons, protein isoforms and translated regions of the genome.
Generally, non-specific/contaminant proteins are present in a large number of data sets independent of the bait used, as opposed to genuine interactors that tend to “co-occur” with their partners across experiments. There are currently two dominant methods for scoring such co-occurrence of protein-protein interactions from co-AP/MS experiments, the spoke model and the matrix model. In the spoke model, all the interactions are evaluated as bait-prey interactions, while in the matrix model, interactions are scored irrespective of the bait used, accounting for both bait-prey as well as prey-prey interactions. In other words, regardless of the actual topology of physical interactions in a protein complex, the spoke model assigns all interactions to the bait used, whereas the matrix model assumes potential interactions between all members.21
We performed co-occurrence analysis for the resulting protein-protein interactions in a matrix model using a modified version of the hypergeometric error model22 by taking peptide spectral counts into consideration for improved prediction of co-complex members. In this novel scoring model called “HGSCore,” the spectral count indeed adds a quasi-quantitative dimension to the scoring strategy. A total of 4,927 individual proteins were identified by mass spectrometric analysis, with 209,912 interactions being observed and scored statistically among them. We also showed that, compared with other published methods (including several spoke-model based algorithms), our HGSCore method performed significantly better at recovering previously documented interactions listed in the DroID reference database.23 To generate the high quality map, we used a stringent statistical cutoff by restricting the analysis to the top 5% of the interactions observed. We used the Markov clustering algorithm24 to assess co-complex membership interactions and thus define putative protein complexes. The DPiM includes 556 protein clusters, providing the basis to address the “second generation” questions we discuss here. Expanding the S2R+ based map with additional baits is expected to add more proteins and complexes while reinforcing the current version of the map. On the other hand, the same analysis in different cell lines will create a wealth of additional information, as has been found from analyses of the full transcriptomes of these cell lines by microarray and deep-sequencing studies. Since different cell lines comprise overlapping but distinct proteomes, they will permit us to test the universality of the map. The panoply of existing cell lines has been isolated from discrete developmental contexts and they will accentuate the developmental variations of this map.
Several existing data sets were very useful in assessing the overall quality of DPiM. The raw DPiM data set was analyzed using HGSCore and other published algorithms and the resulting interactions were used to calculate the recovery of known interactions from DroID23 (including interolog data). It was clear that the integration of spectral count data for all peptides with the HGSCore algorithm helped improve its accuracy and predictive ability. Time-course and tissue-specific transcription profiling data19,25,26 were used to show significant correlation of relative and absolute levels of gene expression among the interacting proteins in clusters compared with rest of the genome. Such comparisons and integrations will be possible in innovative ways as numerous unique data sets are being generated and continually updated as part of the Drosophila modENCODE (model organism Encyclopedia of DNA Elements) project. This large consortium effort aims to comprehensively map transcripts, histone modifications, chromosomal proteins, transcription factors, replication components and nucleosome properties, across a developmental time course and in multiple cell lines. So far, this large-scale effort has generated more than 700 data sets and discovered protein coding, non-coding, RNA regulatory, replication and chromatin elements, which have more than tripled the annotated portion of the Drosophila genome.27 The availability of such enormous genomic and proteomic data sets necessitates concomitant advances in bioinformatics approaches to effectively mine the wealth of biological information and gain insights that are not possible from traditional small-scale focused studies. At the same time, these findings have to be integrated with all the genetic, physical and biochemical interactions defined in thousands of individual, detailed, small-scale studies in Drosophila and different model organisms.
DPiM also provides material for several lines of additional investigation that generate several straightforward testable hypotheses. For example: mapping different genetic screens targeting the same signaling or biochemical pathways onto the proteomic maps to identify protein complexes that are significantly enriched for hits/modifiers that might point to an important but overlooked function.28 A similar recent application of DPiM was in understanding the mediation of Drosophila autosomal dosage effects and compensation by network interactions using deficiency strains.29 The authors examined the expression changes in the Df/+ lines for genes encoding members of 23 protein complexes from DPiM and found 37 cases in which a change in expression in one member was associated with expression change in another gene encoding a member of the same complex. These results provide convincing evidence that one member of a DPiM complex can preferentially affect expression of genes encoding other members of the same protein complex.
The extensive genetic tools available to analyze gene function render Drosophila an ideal organism for such studies, as it offers in vivo assays that can investigate molecular relationships implied by the map. Remarkably, despite the success of the extensive genetic, molecular and genomic studies in Drosophila, approximately one-third of Drosophila gene products still lack detailed functional annotation and another third are annotated only through indirect evidence or sequence homology.30 Therefore the protein complex map will serve as a powerful tool to generate a wealth of testable biological hypotheses and will provide us a systematic way for defining functional association and characterization for poorly annotated genes. Given that the level of gene annotation is also incomplete in mammals, we expected that the Drosophila map would be useful in annotation of mammalian proteomes. To evaluate the degree to which DPiM can be paradigmatic for other metazoan proteomes, we tested a subset of 94 proteins and their DPiM interactions in human-derived HEK-293F cells. Using orthologous proteins and our existing purification strategy, we found that 51% of the interactions were valid across species, suggesting conservation of a large proportion of protein interactions. Among these human-validated DPiM interactions, a total of 268 were novel. Recently, an independent study31 describing protein complexes from soluble protein extracts isolated from human HeLa S3 and HEK293 cells using extensive biochemical fractionation and quantitative mass spectrometry also found excellent overlap with the fly protein interactions and complexes from DPiM.
The map also provides strong experimental evidence and potential mechanistic insights for novel members of well-defined protein complexes. All the co-complex memberships lead to a strong set of testable hypotheses that indicate physical interactions between the members. It is important to note that high-scoring interactions in the map and membership in complexes does not mean that the proteins in question are interacting most of the time, or present in most biochemical contexts. Additional experimentation is necessary to test how the interactions are functionally relevant and to determine which of those interactions are common across cell types and which are found in only a small subset of contexts/conditions. However, it is worth noting that many protein complexes that were originally defined painstakingly over many years from independent studies were immediately apparent in their entirety in DPiM. Definition of entire protein complexes in such an unbiased manner is a testament to the quality of biological information encompassed in the protein complex map. The existence of this map allows us, for the first time, to ask questions regarding how intra-complex protein interactions as well as inter-complex interactions affect the overall architecture of the network. Such relationships will provide insights into how functional cellular units, defined by the existence of specific protein complexes, are integrated within the cell.
A fundamental property of protein networks is their interconnectedness. The giant component of the DPiM map is composed of 357 complexes, which shows a high degree of interconnectivity.16 The accumulated evidence, emphasized by genetic modifier analyses, indicates that a given protein complex with a particular function almost never acts alone but is modulated by other cellular elements (e.g., refs. 32–35). While mechanistic characterization of such connections is very challenging, the map we have generated offers the opportunity to examine how, and if, the disruption of a complex can affect its mechanistic “surroundings” and hence its functionality. Additionally, some proteins in the map have multiple connections with other proteins and help to connect different protein clusters with distinct functions. By disrupting the expression or function of such “hubs” we can test the dynamics of intra- or inter-complex relationships. The significance of complex interconnectedness is in our view profound, as it will open the way to explore protein interaction dynamics in different genetic backgrounds, physiological conditions, metabolic states etc., at a system-wide level. In order to do this we do not need to re-establish proteome maps for specific conditions but rather, based on the existing map, we can obtain a better picture by probing the presence, distribution and composition of a select subset of complexes when confronted with diverse developmental contexts in vivo. Such an analysis has both basic and translational value, given that it will open the way to probe complex, system-wide interactions under different physiological conditions imposed by genetic mutation or pharmacological manipulation.
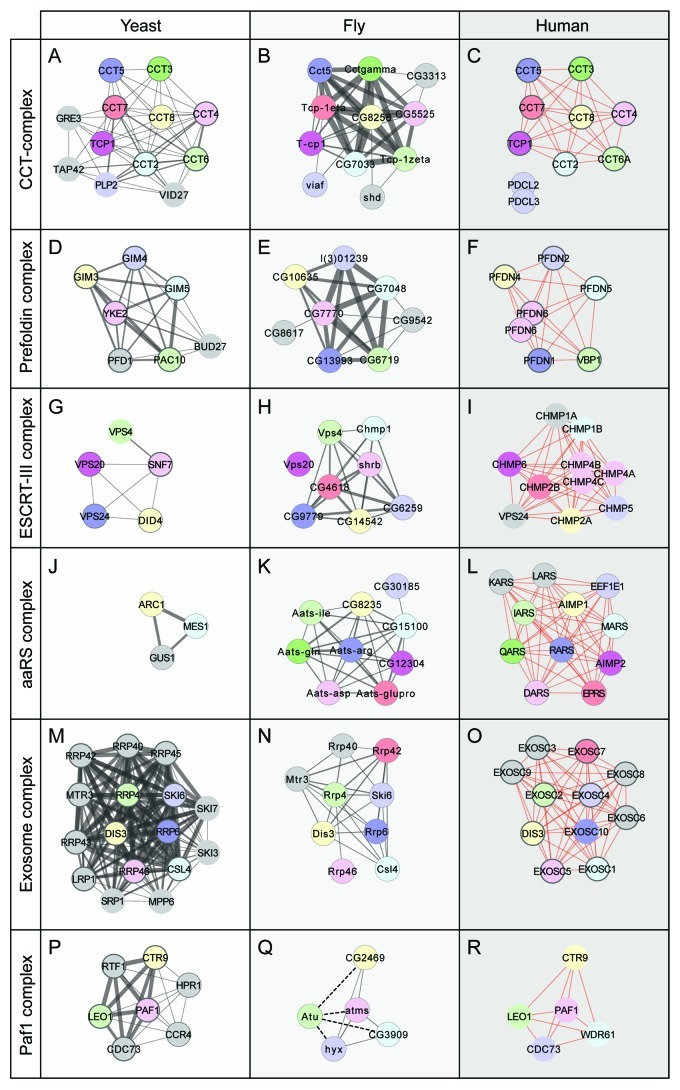
Examining the evolution of protein complex organization across species can provide important insights by establishing conserved and orthologous relationships and help improve the annotation of uncharacterized genes. To gain evolutionary insights, individual protein complexes defined in DPiM were compared with orthologous protein complexes from yeast and humans. We used the most extensive available manually curated annotations of protein complexes for the yeast (MIPS, CYC2008) and human (REACTOME, CORUM) proteomes. In most cases where the complexes were well defined, the relationships could be established directly due to linear conservation. For example, the CCT and Prefoldin complexes (Fig. 2A-F) have not changed significantly during evolutionary time reflecting the fundamental function of protein folding that has remained relatively unaltered. On the other hand, in the cases of ESCRT-III and aminoacyl tRNA synthetases, there seems to have been a linear expansion of complex members (Fig. 2G–L). In several cases in which the relationships were not immediately clear, we were able to show distant functional or sequence similarities,16 though there are exceptions (see Exosome complex, Fig. 2M–O). In certain cases, interactions known in other species were only identified as sub-threshold interactions in DPiM (see Paf1 complex, Fig. 2P–R), highlighting the utility of analyzing all the available interaction data to deduce the structure of the complexes.
Figure 2. Patterns of Protein complex evolution. Comparison of protein complexes defined in DPiM (center panels, fly) with the same functional complexes from yeast (left panels, using MIPS, CYC2008) and human cells (right panels, using REACTOME, CORUM). Protein partners found in one database have thinner outlines while proteins found in both databases have thicker outlines. Proteins with obvious sequence similarity are colored identically and arranged in similar location for all three species. In case of multiple paralogs, the protein with highest sequence homology is considered as the corresponding homolog. Proteins shown as gray circles are members that have no clear sequence similarity with others, although they may have similar names. The thickness of gray lines connecting the proteins is proportional to the score/weight of the interaction, while red lines connecting human proteins are unweighted interactions in the databases. (A–C) The CCT (chaperonin-containing T-complex) comprises nine conserved proteins conserved in all three species, while in yeast and fly, a small number of other proteins are also found to be associated. (D–F) The Prefoldin complex does not show significant variation from yeast to human, with only minor addition or subtraction of new complex members (two Ensembl gene IDs for PFDN6). (G–I) The ESCRTIII complex is a multiprotein assembly with a conserved role in endocytosis. In yeast, it contains five members, which are retained in both fly and human; although the VPS24 components of yeast and human do not share noticeable sequence similarity. In DPiM, the complex has also been found to be associated with the Flotillin complex (not shown) in fly cells. (J–L) The aminoacyl-tRNA synthetases consist of both conserved and variant subunits, due to their derivation by both paralogous evolution and horizontal gene transfer. Comparison shows that fly and human complexes are very similar, while in yeast, only a subset of these is found. (M–O) The Exosome (RNase) complex in all three species has six conserved core members. The other proteins, despite similar names, do not appear to be closely related in sequence, exemplifying how the same functional complex can have different protein components across species. (P–R) The Paf1 complex is involved in RNA Polymerase II function, and one member, Atu has sub-threshold interaction scores (dotted lines, Q) with other subunits in DPiM.
We were able to compare a handful of complexes in our analysis, but at this point in the development of bioinformatics methods, doing so on a large scale is quite challenging. A number of tools have recently been developed for such analyses (e.g., refs. 36, 37 and others). While quality of the tools varies, the more relevant concern is the origin and quality of the individual input data sets. For instance, comparing conservation between organisms where one data set was generated using AP/MS and the other from Y2H (as in human studies) may show divergence of complex members that, is merely a consequence of the methods used. Generally, the fact that complexes are not defined at the same level of detail in all organisms makes it difficult to carry out reliable large-scale evolutionary analysis, and there are as yet no commonly accepted standards of quality or statistical significance.
Given the conservation of protein sequence and function between species, it is becoming clear that the key to understanding the diversity of protein interactions is not just a question of which proteins are involved but how much of each is present. We have also shown that members of a protein complex generally have similar expression profiles in tissues and in development and have comparable absolute levels of expression.16 This suggests stoichiometry may determine the extent of interaction between any two proteins and therefore account for the compositional differences of a given complex in different developmental contexts. Quantitative mass spectrometry techniques such as isobaric labeling will permit the determination of protein sequence and the relative amounts of peptides (based on the reporter tags’ abundances). Moreover, tagging relies only on chemical labeling, as opposed to approaches that necessitate propagation of cells in different conditions (such as SILAC, for example see refs 38, 39). The availability of tandem mass tags (TMT reagents),40,41 which define isobaric tags that are now available in multiplex combinations, can allow examination and comparison of up to six samples, i.e., experimental conditions, simultaneously.
Not all proteins that are part of a protein complex are physically attached to each other. Most proteins interact with each other in a defined manner with specific domains coming into direct contact. Such interactions are determined by the tertiary structure and surface charge of the protein domains in their particular cellular milieu. It will be interesting to see which of the proteins that the map defined as strong interactors have any proteins domains or structures (known or novel) that are complementary to protein domains in partner proteins. Such domain interaction analyses will help delineate the topology of interactions within members of a protein complex.
The analysis of hemocyte-like embryonic cells (S2R+ cell line) revealed a protein interaction network encompassing 556 complexes. Included in these complexes are the products of several hundred previously unannotated genes for which DPiM provides the first empirical evidence and functional insights by associating with other proteins. It is now possible to examine if and how the protein complex interactions derived from S2R+ cells change in different tissues, developmental or genetic backgrounds. To promote such studies, we have produced a total of 4,228 stable transgenic fly lines carrying the same FLAG-HA tagged version of the proteins under the control of a UAS promoter. These lines were generated at the Drosophila Transgenic Fly Facility (http://www.ncbs.res.in/tff) and are listed on the DPiM website (https://interfly.med.harvard.edu/transgenic_info.php). These lines were generated using the Attp site-specific insertion of the constructs on the third chromosome and thus they should be free of local enhancer effects. The expression of tagged proteins can be spatiotemporally regulated by the use of appropriate Gal4 drivers so that specific interactions of interest can be pursued in greater detail.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIH 5RO1HG003616) to S.A.T. Generation of the clone set was supported by a grant from the National Human Genome Research Institute (NHGRI P41HG3487) to our collaborator Susan Celniker.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/22108
References
- 1.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–36. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–3. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–68. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 6.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–8. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 7.Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, et al. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 10.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105:13333–8. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 14.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 15.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–6. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 16.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C, Wan KH, Hammonds AS, Stapleton M, Carlson JW, Celniker SE. Development of expression-ready constructs for generation of proteomic libraries. Methods Mol Biol. 2011;723:257–72. doi: 10.1007/978-1-61779-043-0_17. [DOI] [PubMed] [Google Scholar]
- 19.Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–14. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakes L, Robertson DL, Oliver SG, Lovell SC. Protein interactions from complexes: a structural perspective. Comp Funct Genomics. 2007 doi: 10.1155/2007/49356. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart GT, Lee I, Marcotte ER. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murali T, Pacifico S, Yu J, Guest S, Roberts GG, 3rd, Finley RL., Jr. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39(Database issue):D736–43. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–9. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–20. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–66. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone JH, Cho DY, Mattiuzzo NR, Artieri CG, Jiang L, Dale RK, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13:r28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, et al. FlyBase Consortium FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37(Database issue):D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–81. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kankel MW, Hurlbut GD, Upadhyay G, Yajnik V, Yedvobnick B, Artavanis-Tsakonas S. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics. 2007;177:2493–505. doi: 10.1534/genetics.107.080994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurlbut GD, Kankel MW, Artavanis-Tsakonas S. Nodal points and complexity of Notch-Ras signal integration. Proc Natl Acad Sci U S A. 2009;106:2218–23. doi: 10.1073/pnas.0812024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, et al. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sen A, Yokokura T, Kankel MW, Dimlich DN, Manent J, Sanyal S, et al. Modeling spinal muscular atrophy in Drosophila links Smn to FGF signaling. J Cell Biol. 2011;192:481–95. doi: 10.1083/jcb.201004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–9. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao CS, Lu K, Baym M, Singh R, Berger B. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics. 2009;25:i253–8. doi: 10.1093/bioinformatics/btp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amanchy R, Kalume DE, Pandey A. Stable isotope labeling with amino acids in cell culture (SILAC) for studying dynamics of protein abundance and posttranslational modifications. Sci STKE. 2005;2005:pl2. doi: 10.1126/stke.2672005pl2. [DOI] [PubMed] [Google Scholar]
- 39.Rigbolt KT, Blagoev B. Proteome-wide quantitation by SILAC. Methods Mol Biol. 2010;658:187–204. doi: 10.1007/978-1-60761-780-8_11. [DOI] [PubMed] [Google Scholar]
- 40.Dayon LHA, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–31. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 41.Thompson ASJ, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]