Abstract
Wolbachia is a genus of parasitic alphaproteobacteria found in arthropods and nematodes, and represents on of the most common, widespread endosymbionts known. Wolbachia affects a variety of reproductive functions in its host (e.g., male killing, cytoplasmic incompatibility, parthenogenesis), which have the potential to dramatically impact host evolution and species formation. Here, we present the first broad-scale study to screen natural populations of native Hawaiian insects for Wolbachia, focusing on the endemic Diptera. Results indicate that Wolbachia infects native Hawaiian taxa, with alleles spanning phylogenetic supergroups, A and B. The overall frequency of Wolbachia incidene in Hawaiian insects was 14%. The incidence of infection in native Hawaiian Diptera was 11% for individuals and 12% for all species screened. Wolbachia was not detected in two large, widespread Hawaiian dipteran families—Dolichopodidae (44 spp screened) and Limoniidae (12 spp screened). Incidence of infection within endemic Hawaiian lineages that carry Wolbachia was 18% in Drosophilidae species, 25% in Caliphoridae species, > 90% in Nesophrosyne species, 20% in Drosophila dasycnemia and 100% in Nesophrosyne craterigena. Twenty unique alleles were recovered in this study, of which 18 are newly recorded. Screening of endemic populations of D. dasycnemia across Hawaii Island revealed 4 unique alleles. Phylogenetic relationships and allele diversity provide evidence for horizontal transfer of Wolbachia among Hawaiian arthropod lineages.
Keywords: Diptera, Hawaii, Wolbachia, horizontal transfer, nesophrosyne, phylogenetics
Introduction
Wolbachia is an endosymbiotic genus of inherited, transovarially transmitted alphaproteobacteria that infects the major arthropod orders and nematodes.1,2 Previous studies have estimated that the incidence of arthropod infections ranges from 20–75% in different systems and up to 65% worldwide.1,3-6 While these estimates are based on the criteria of a positive infection rate of > 1% for individuals of a given species and are corrected for low sample size, they demonstrate that Wolbachia is one of the most abundant and widespread endosymbionts known.1,3-6 Wolbachia is characterized by its varied and potentially dramatic effects on host reproduction, causing cytoplasmic incompatibility, feminization of genetic males and male killing.1 In contrast, the bacterium also provides mutualistic benefits to insect hosts, including wasps, bedbugs and Drosophila.7-9 Due to Wolbachia’s pervasiveness and potential influence on host population demographics, it has been proposed as a possible driver of speciation in highly diverse insects.6
Wolbachia has been a subject of intense interest due to its unusual diversity and potential role in insect diversity. Recent research has investigated the applied science of Wolbachia, by infecting natural populations of Mosquito vectors in an effort to control human disease such as Dengue fever.10,11 However, fundamental questions regarding the mechanisms of Wolbachia’s global persistence and its net effects on speciation remain open questions, with many large geographic areas and insect taxonomic groups remaining to be studied.1 In order to better answer these basic questions, a more complete understanding of Wolbachia occurrence across novel environments and taxonomic groups is required. This will help elucidate the role of this bacterium in host evolution, teasing it apart from other demographic, ecological and geographic factors that may drive local and global insect diversity. This study expands previous work on Wolbachia by conducting the first broad scale screening of environmentally sampled, endemic Hawaiian insects, including multiple lineages of Diptera (Drosophilidae, Dolichopodidae, Limoniidae, Calliphoridae) and Hemiptera (Cicadellidae).
The Hawaiian Islands and their native biota present a unique and simplified system to explore questions regarding Wolbachia persistence and spread between interacting arthropod lineages that are both distantly and closely related. The archipelago is isolated by 4000km of oceanic water, with constituent islands arranged in a linear geologic chronology. As a result, arthropod lineages are taxonomically disharmonic with respect to mainland sources, and are typically high in species diversity.12-14 In order for Wolbachia to persist in Hawaiian arthropods, it must first be present in colonizing individuals, and subsequently persist through repeated colonization of neighboring islands and explosive intra-island host radiations. The pattern of repeated intra-island founder events, and resultant population bottlenecks, may purge Wolbachia infections. Thus, long-term persistence may further require extensive horizontal transfer from infected to uninfected lineages to spread and persist.
Sampling in this study focuses on Hawaiian Diptera, one of the most diverse and dominant components of the native Hawaiian fauna. The endemic genus Drosophila is a classic example of adaptive radiation in nature, comprising over 1000 species and representing 10% of the endemic Hawaiian insect fauna.15 To contrast potential results from Hawaiian Diptera, we further include a number of non-native lineages found on Hawaii and other Pacific locations (Australia and South Pacific Islands), and several species of the large, native leafhopper genus Nesophrosyne (Hemiptera: Cicadellidae). This strategy offers both a broad taxonomic and ecological sampling to compare distantly related lineages that share similar geographic constraints (Drosophila vs. Nesophrosyne, which co-occur in similar habitats and share host plants), span a wide variety of habitat types (mesic forest, rainforest, cloud forest, semi-aquatic habitats, invaded ecosystems), encompass different degrees of ecological specialization (generalists and specialists), and occupy different ecological niches (predators, saprophagous taxa, plant-feeding insects).
Results from this study demonstrate that Wolbachia is present in native Hawaiian insects, a novel result since previous screens in Hawaiian arthropods were based on either non-native flies, produced negative results, or were limited in taxonomic diversity and based on long-standing laboratory cultures.16,17 The incidence of Wolbachia in native Hawaiian insects was 14%, but was lower for native Hawaiian Diptera (11%). Several large groups including the dipteran families Dolichopodidae (Hawaii), Limoniidae (Hawaii) and Mycodrosophila (Oceania Region) showed no evidence of infection. Overall, Wolbachia incidence was generally low (< 20%). Sequenced Wolbachia alleles are placed in both the A and B supergroups, with most alleles from endemic hosts placed in the B supergroup. Twenty alleles were recovered in this screen, including 18 newly discovered, and evidently known only from Hawaii. Phylogenetic relationship of Wolbachia wsp alleles from endemic insects form clades with other Hawaiian taxa or infections from Japan. Concordant with previous results, Hawaiian Wolbachia show evidence of horizontal transfer.4,18
Results
Incidence of Wolbachia infection and allelic diversity
Table 1 lists the positive results for taxa screened in this study, including Wolbachia supergroup placement and allelic designations (Table 3 provides a full breakdown of all taxa screened). Table 2 lists the percent frequency of Wolbachia occurrence in the different categories of insects screened (e.g., all taxa, Native Diptera, Drosophila dasycnemia, etc.). The incidence of Wolbachia in all individuals and species screened was 11%. Native Hawaiian taxa showed a 14% incidence for both species and individuals, with endemic Diptera showing lower individual and species level incidences of 11% and 12%, respectively. Of the four major dipteran families screened, Drosophilidae (18%) and Calliphoridae (25%) yielded positive infection results. In contrast, Dolichopodidae and Limoniidae yielded no Wolbachia despite the relatively large number of taxa screened (n = 120; Table 3). The native Hawaiian leafhoppers (Nesophrosyne) gave positive results for 12 of the 13 individuals screened (92% infection rate), and 100% infection incidence for all species screened.
Table 1. Taxonomic information for Hawaiian insect hosts sequenced Wolbachia wsp.
| Family | Genus | Species | Wolbachia supergroup | Allele | GenBank accession (Study barcode) |
|---|---|---|---|---|---|
| Drosophilidae |
Drosophila |
dasycnemia |
B |
wDasA |
JX134910 (85w) |
| |
|
|
B |
wDasA |
JX134911 (135w) |
| |
|
|
B |
wDasB |
JX134912 (136w) |
| |
|
|
B |
wDasA |
JX134913 (137w) |
| |
|
|
B |
wDasA |
JX134914 (138w) |
| |
|
|
B |
wDasD |
JX134921 (139w) |
| |
|
|
B |
wDasA |
JX134916 (140w) |
| |
|
|
B |
wDasB |
JX134920 (141w) |
| |
|
|
B |
wDasA |
JX134918 (201w) |
| |
|
|
B |
wDasA |
JX134917 (202w) |
| |
|
|
B |
wDasA |
JX134919 (268w) |
| |
|
|
B |
wDasC |
JX134915 (269w) |
| |
|
kikkawai |
B |
wKik |
JX134922 (277w) |
| |
|
apicipuncta |
B |
wApi |
JX134926 (276w) |
| |
|
nr.fundita |
B |
wGin |
JX134924 (200w) |
| |
|
ancyla |
B |
wGin |
JX134925 (87w) |
| |
|
fundita |
B |
wFun |
JX134929 (86w) |
| |
|
eurypeza |
B |
wEur |
JX134927 (142w) |
| |
|
sp1nr.basimacula |
B |
wBas |
JX134934 (81w) |
| |
|
sp2.nr.basimacula |
B |
wBas |
JX134933 (82w) |
| |
|
redunca |
B |
wDasA |
JX134932 (83w) |
| |
|
prodita |
B |
wDasA |
JX134931 (84w) |
| |
|
forficata |
B |
wFor |
JX134928 (149w) |
| |
|
nr.dorsigera |
B |
wFor |
JX134935 (150w) |
| |
|
tetraspilota |
B |
wTet |
JX134936 (281w) |
| |
|
nigrocirrus |
A |
wEla |
JX134930 (42w) |
| |
|
suzukii |
A |
wRi |
JX134952 (70w) |
| |
|
|
A |
wRi |
JX134953 (71w) |
| |
|
|
A |
wRi |
JX134954 (72w) |
| |
Scaptomyza |
flava |
A |
wFla |
JX134938 (15w) |
| |
|
nr.semiflava |
A |
wEla |
JX134939 (43w) |
| |
|
longiestosa |
A |
wEla |
JX134923 (58w) |
| Calliphoridae |
Dyscritomyia |
obscura |
B |
wObs |
JX134937 (199w) |
| Cicadellidae |
Nesophrosyne |
n.sp9 |
B |
wGin |
JX134949 (143w) |
| |
|
n.sp9 |
B |
wGiff |
JX134945 (283w) |
| |
|
giffardi interrupta |
B |
wGin |
JX134950 (270w) |
| |
|
giffardi |
B |
wGiff |
JX134946 (284w) |
| |
|
giffardi |
B |
wGiff |
JX134947 (285w) |
| |
|
giffardi |
B |
wGiff |
JX134948 (286w) |
| |
|
silvicola |
B |
wSil |
JX134944 (282w) |
| |
|
craterigena |
B |
wCraA |
JX134942 (154w) |
| |
|
craterigena |
B |
wCraB |
JX134951 (151w) |
| |
|
craterigena |
B |
wCraA |
JX134943 (155w) |
| |
|
craterigena |
B |
wCraB |
JX134940 (153w) |
| craterigena | B | wCraB | JX134941 (152w) |
Table 3. Summary of taxon sampling arranged by taxonomic groups and geographic locality.
|
Order |
Family |
Genus |
Group |
Species |
Locality |
SamplessScreened |
Individuals infected |
Species infected |
| Diptera |
Calliphoridae |
Dyscritomyia |
|
4 spp |
Hawaiian Islands, USA |
4 |
1 |
1 |
| |
Dolichopodidae |
Adachia |
|
1 sp |
Hawaiian Islands, USA |
1 |
0 |
0 |
| |
|
Arciellia |
|
4 spp |
Hawaiian Islands, USA |
4 |
0 |
0 |
| |
|
Campsicnemus |
|
26 spp |
Hawaiian Islands, USA |
26 |
0 |
0 |
| |
|
Chrysotus |
|
2 spp |
Hawaiian Islands, USA |
2 |
0 |
0 |
| |
|
Dolichopus |
|
2 spp |
Hawaiian Islands, USA |
2 |
0 |
0 |
| |
|
Eurynogaster |
|
8 spp |
Hawaiian Islands, USA |
8 |
0 |
0 |
| |
|
Sweziellia |
|
tergoprolixa |
Hawaiian Islands, USA |
1 |
0 |
0 |
| |
|
Tachytrechus |
|
angustipennis |
Hawaiian Islands, USA |
1 |
0 |
0 |
| |
|
Uropachys |
|
fusticercus |
Hawaiian Islands, USA |
1 |
0 |
0 |
| |
Drosophilidae |
Dichaetophora |
|
plana |
Australia |
1 |
0 |
0 |
| |
|
Drosophila |
dispar |
dispar |
Australia |
1 |
0 |
0 |
| |
|
|
melanogaster |
kikkawai |
Pacific Region |
22 |
1 |
1 |
| |
|
|
|
suzukii |
Pacific Region |
24 |
3 |
1 |
| |
|
|
immigrans |
immigrans |
Pacific Region |
15 |
0 |
0 |
| |
|
|
|
nasuta |
Pacific Region |
3 |
0 |
0 |
| |
|
|
|
nasutoides |
Pacific Region |
2 |
0 |
0 |
| |
|
|
Hawaiians, spoon tarsus |
dasycnemia |
Hawaiian Islands, USA |
56 |
12 |
1 |
| |
|
|
Hawaiians, Drosophila |
81 spp |
Hawaiian Islands, USA |
81 |
13 |
13 |
| |
|
|
Hawaiians, Scaptomyza |
12 spp |
Hawaiian Islands, USA |
12 |
3 |
3 |
| |
|
Hirtodrosophila |
|
4 spp |
Pacific Region |
4 |
0 |
0 |
| |
|
Leucophenga |
|
scutellata |
Australia |
1 |
0 |
0 |
| |
|
Mycodrosophila |
|
47 spp |
Pacific Region |
47 |
0 |
0 |
| |
|
Paramycodrosophila |
|
pictifrons |
French Polynesia |
1 |
0 |
0 |
| |
|
Samoaia |
|
2 spp |
Samoa |
2 |
0 |
0 |
| |
|
Scaptodrosophila |
|
albifrontata |
Australia |
1 |
0 |
0 |
| |
|
Sphaerogastrella |
|
novaguinenensis |
New Guinea |
2 |
0 |
0 |
| |
|
Tambourella |
|
endiandrae |
Australia |
1 |
0 |
0 |
| |
|
Zygothrica |
|
4 spp |
Pacific |
4 |
0 |
0 |
| |
Limoniidae |
Dycranoymia |
|
12 spp |
Hawaiian Islands, USA |
76 |
0 |
0 |
| Hemiptera |
Cicadellidae |
Nesophrosyne |
|
5 spp |
Hawaiian Islands, USA |
13 |
12 |
5 |
| Totals | 230 spp | 419 | 45 | 25 spp |
Table 2. Incidence of Wolbachia for taxonomic groups screened for the wsp loci (individual/species).
| Geographic area | Groups | Positives | Total screened | % Infected |
|---|---|---|---|---|
| Pacific |
|
|
|
|
| |
Total |
45/25 |
419/230 |
11%/11% |
| |
Diptera |
33/20 |
406/220 |
8/9% |
| Hawaii |
|
|
|
|
| |
Native Hawaiian Insects |
41/23 |
288/161 |
14%/14% |
| |
Native Diptera |
29/18 |
275/156 |
11%/12% |
| |
Native Drosophilidae |
27/17 |
149/94 |
18%/18% |
| |
Calliphoridae |
1/1 |
4/4 |
25% |
| |
Dolichopodidae |
0 |
46/50 |
0% |
| |
Limoniidae |
0 |
12/76 |
0% |
| |
Nesophrosyne |
13/5 |
12/5 |
92%/100% |
| |
Drosophila dasycnemiea |
12 |
56 |
20% |
| |
Drosophila suzukii |
3 |
24 |
13% |
| |
Drosophila kikkawai |
1 |
22 |
5% |
| Nesophrosyne craterigena | 5 | 5 | 100% |
Wolbachia incidence within species varied. Of the 58 Drosophila dasycnemia taxa screened, 12 (21%) individuals were infected. The widespread, non-native Drosophila suzukii and D. kikkawai demonstrated relatively low within-species Wolbachia incidence of 13% and 5%, respectively. In Nesophrosyne species, a 100% infection incidence was observed for the five individuals of N. craterigena, three individuals N. giffardi and two individuals of N. n.sp9.
Table 1 lists the Wolbachia alleles discovered in this study. Twenty unique alleles were sequenced, of which 18 are novel and sequenced from native Hawaiian insects. Several species share alleles, including Drosophila dasycnemia, D. prodita and D. redunca (wDasA); D. forficata and D. dorsigera (wFor); D. ancyla, D. nr.fundita, Nesophrosyne giffardi interrupta and N. n.sp9 (wGin); D. nigrocirrus, D. nr.semiflava and D. longiestosa (wEla); Nesophrosyne n.sp9 and N. giffardi (wGiff). Individuals of the species, Nesophrosyne craterigena, have either allele wCraA or wCraB. Of the multiple individuals screened for D. dasycnemia, four unique alleles were recovered (wDasA–wDasD).
Phylogenetic relationships
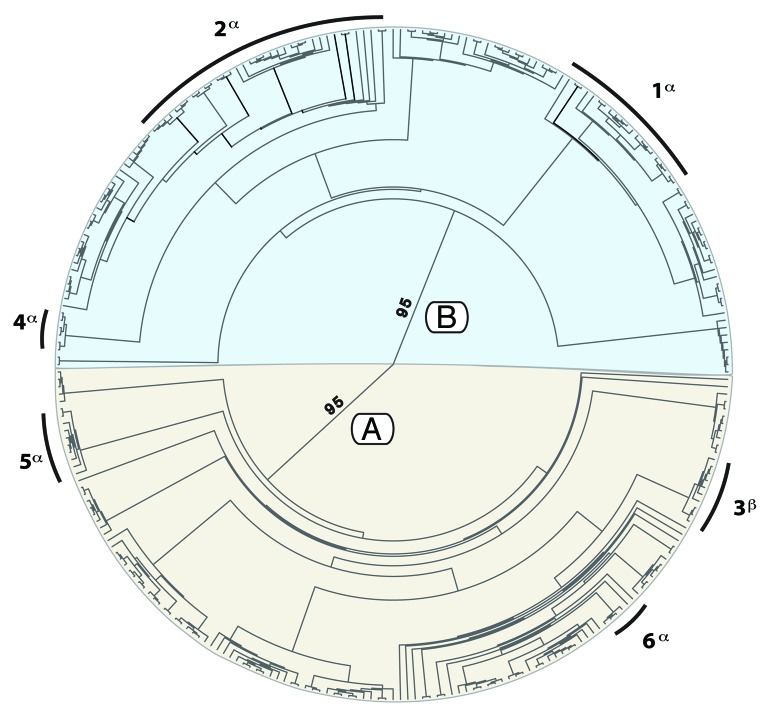
Global Maximum Likelihood (ML) results for the 284 Wolbachia wsp sequences included in this study are shown in Figure 1 (–lnL = 8396.93). Sequences were placed in the Wolbachia phylogenetic supergroup core clades, A and B, with high bootstrap support (BS = 95). Hawaiian alleles are distributed between the supergroups A and B, but placed predominantly in the B supergroup (38 of 45 sequenced results). The global tree is split into six different sections for ease of discussion; hereafter referred to as Hawaiian subgroups 1–6 (Figs. 1–4). Support for relationships range from moderate to high support (Fig. 2, subgroup 2; BS = 60–97) to poor or no support (subgroup 1; BS < 50), and are summarized below:
Figure 1. Maximum-likelihood phylogeny of 284 Wolbachia isolates for surface protein gene wsp conducted using RAxML-HPC2 v7.2.7.51,52 Phylogeny is mid-point rooted, and color-coded according to the (A) (red) and (B) (blue) supergroup systematic classifications. Inset numbers correspond to bootstrap support values. Numbers 1–6 demarcate subsections of the phylogeny containing Wolbachia sequenced from Hawaiian insects in this study. Symbols α and β indicate groups that contain endemic Hawaiian taxa and those that do not, respectively. Hawaiian subsections are enlarged to show relationships and endemicity of constituent taxa in Figures 2–4.
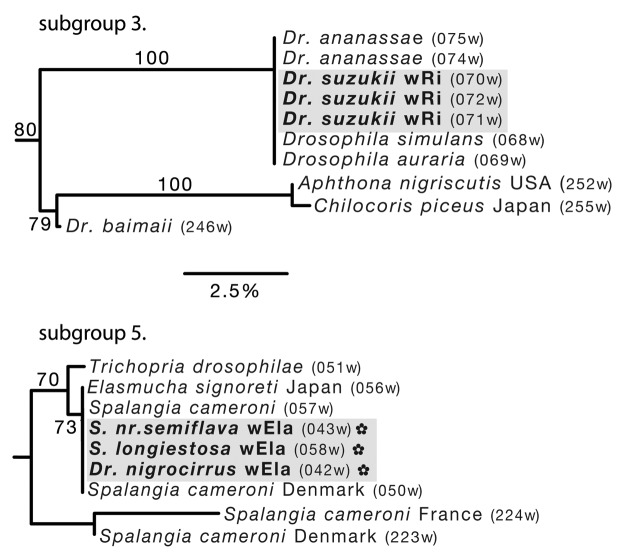
Figure 4. Hawaiian Wolbachia subgroups 3 and 5 (supergroup A). See Figure 2 legend for explanation and interpretation of species abbreviations, branch support, DNA barcodes and other symbols.
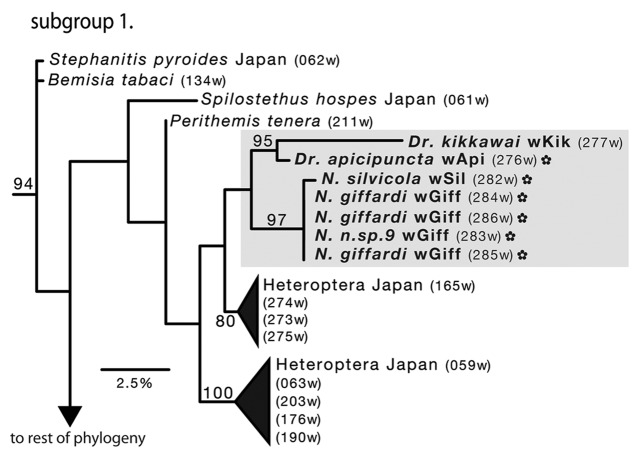
Figure 2. Hawaiian Wolbachia subgroup 1 (supergroup B) from the maximum-likelihood phylogenetic reconstruction presented in Figure 1. The gray box delineates Hawaiian sequences, with endemic taxa starred. Hawaiian genera abbreviations are as follow: Dr. = Drosophila (Diptera: Drosophilidae), Dy. = Dyscritomyia (Diptera: Calliphoridae), S. = Scaptomyza (Diptera: Drosophilidae), and N. = Nesophrosyne (Hemiptera Cicadellidae). Branch numbers correspond to bootstrap values (BS < 50 not shown). Names correspond to the taxonomic placement of the host species and unique allele name (e.g., wGiff); parenthetical three-digit barcode corresponds to Table S1.
Hawaiian subgroups 1 and 2 are placed within the Wolbachia B supergroup (Figs. 1–3: BS = 95). The closest relatives of these alleles are primarily other Hawaiian sequences, and Lepidoptera and Heteroptera from Japan and Africa. Subgroup 1 (Fig. 2) includes infections sequenced from two Drosophila species, D. kikkawai and D. apicipuncta (BS = 95) and three Nesophrosyne species. Relationships of Wolbachia sequences in this clade are generally poorly supported, however Nesophrosyne infections represent a highly supported clade (BS = 97). Wolbachia alleles from Drosophila are placed sister with high support (BS = 95). The closest alleles to the Hawaiian infections are Heteroptera from Japan, although support is low (BS < 50)
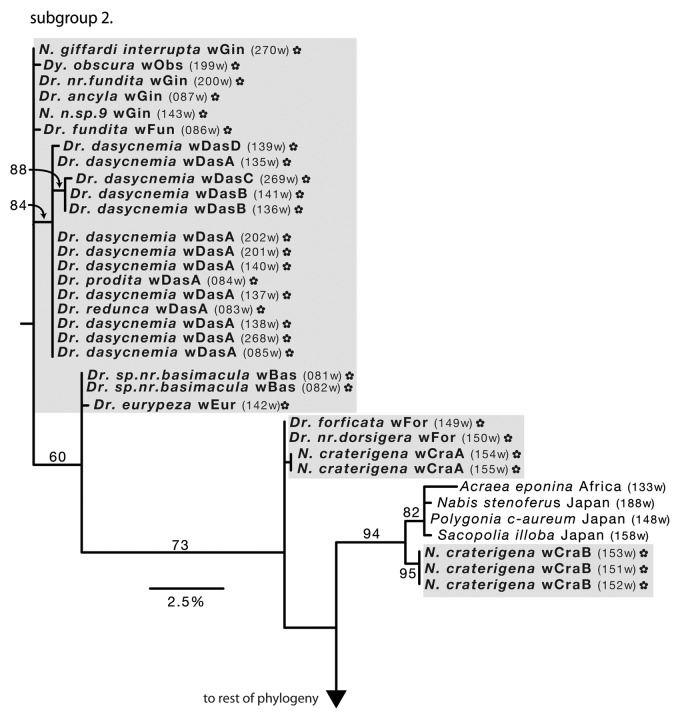
Figure 3. Hawaiian Wolbachia subgroup 2 (supergroup B). See Figure 2 legend for explanation and interpretation of species abbreviations, branch support, DNA barcodes and other symbols.
Hawaiian subgroup 2 (Fig. 3) contains four well-supported clades (BS = 71–97). The largest clade is composed of Wolbachia from Drosophila dasycnemia mixed with D. redunca and D. prodita from across Hawaii Island (BS = 84). Wolbachia from two distantly related species, D. spnr.basimacula and D. eurypeza, are placed sister with moderate support (BS = 71). Wolbachia sequenced from Nesophrosyne species are placed in two separate clades: a monophyletic N. craterigena sister to Japanese and African Lepidoptera (BS = 94), and a Drosophila–Nesophrosyne mixed clade (BS = 97). Several Wolbachia alleles are unplaced with no supported relationships, including infections from Dyscritomyia obscura (wObs, Calliphoridae), Drosophila ancyla (wGin), D. nr.fundita (wGin), D. fundita (wFun), Nesophrosyne giffardi interrupta (wGin) and N. n.sp9 (wGin).
Hawaiian subgroups 3 (BS = 80) and 5 (BS = 70: both shown in Fig. 4) and two native drosophilids are placed in the taxonomic supergroup A. Subgroup 3 comprises the widespread Drosophila species D. suzukii from Hawaii, D. ananassae, D. simulans and D.auria (BS = 100). Subgroup 5 is moderately supported (BS = 70), containing three Hawaiian sequences from both native Hawaiian drosophilid genera Scaptomyza (S. nr.semiflava and S. longiestosa) and Drosophila (D. nigrocirrus). Hawaiian sequences are placed within a moderately supported clade (BS = 73), comprising infections from a Japanese Heteroptera and Hymenoptera parasitoides (Spalangia cameroni).
Finally, Wolbachia from Drosophila tetraspilota (subgroup 4; BS > 97) and Scaptomyza flava (subgroup 6: BS = 100, results not shown, but see Fig. 1) are placed individually in supergroup B and A, respectively. Neither shares alleles with other Hawaiian Drosophila.
Discussion
Incidence of Wolbachia on Hawaii
This study is the first to conduct broad-scale screening for Wolbachia in naturally occurring populations of native Hawaiian insects. Our results demonstrate that Wolbachia is present in the native Hawaiian Drosophilidae (> 1000 species), Calliphoridae (25 species), and Cicadellidae (> 200 species). This result is significant, as previous studies have failed to find Wolbachia in lab reared native and non-native Drosophila from Hawaii,16,17 and it is the first to show Wolbachia infecting other Hawaiian families. Eighteen of the recovered haplotypes are known only from native Hawaiian taxa. This suggests the intriguing possibility that Hawaii may host endemic Wolbachia lineages, but further Pacific-wide screening is required to confirm this. An equally interesting outcome of this study is the apparent absence of Wolbachia from the endemic fly families Dolichopodidae (> 120 species) and Limoniidae (12 species), despite the large numbers of individuals screened (see Table 3). Several of these groups represent some of the largest Hawaiian insect radiations, and they are all common components of the Hawaiian entomofauna, sharing habitats throughout the islands.
Previous studies have estimated that local and global species level incidence of Wolbachia ranges from 20–75%, however these estimates are potentially biased due to incomplete taxon sampling and the inconsistent number of individuals screened per species.4,6,20,21 Thus, direct comparisons of Wolbachia incidence between geographic localities, or to worldwide estimates, are difficult to interpret directly. Among the Hawaiian insects screened in this study, the overall incidence of Wolbachia at the species level is 14%, with the endemic Hawaiian Diptera demonstrating an even lower incidence of 12% for species screened (see Table 2). Contrary to this finding, the native Nesophrosyne Hawaiian leafhoppers exhibited a high incidence of Wolbachia of 100% for species screened. The exceptional Wolbachia occurrence in Nesophrosyne needs to be further examined, since sampling is taxonomically limited and all specimens were acquired from the same geographic location—Kipuka Puaulu, Hawaii Island. This result may represent a localized infection with an unusually high level of infection. A closer examination of Wolbachia in individual groups demonstrate a higher infection incidence of ~20% at the family level (18% for drosophilids, including the genera Drosophila and Scaptomyza) and the species level (21% within Drosophila dasycnemia).6 Further, the non-native D. kikkawai and D. suzukii show an incidences of 5% and 13%, respectively.
The depressed incidence and complete absence of Wolbachia in some Hawaiian lineages suggests that the dynamics of Wolbachia may be different than for other geographic regions.6,19,22 A plausible hypothesis for the observed or general absence of Wolbachia from some groups may be due to the remote location of the Hawaiian Islands: Hawaii is isolated from the nearest continent by over 4000 km of water, with an estimated successful arthropod colonization every 175,000 y.23 The rarity of these events, combined with the global finding that few if any lineages have a 100% Wolbachia incidence,4-6,19,20 suggests that infections may simply be left behind as uninfected colonizers disperse to Hawaii. Similarly, after establishment on the archipelago, lineages typically disperse to neighboring islands, speciate in allopatry, and experience resultant reductions in effective population size and genetic diversity.24,25 As lineages colonize new islands within the Archipelago, infection may be further reduced or purged through populations bottlenecks and sorting events. The interplay between these processes could explain the low incidence and complete absence of Wolbachia in some Hawaiian groups (e.g., Dolichopodidae and Limoniidae). Thus, Wolbachia, in order to persist, must survive multiple rounds of colonization or find alternative routes to spread, such as horizontal transfer.
Relationships of Hawaiian Wolbachia
Native Hawaiian Wolbachia alleles were placed predominantly in the B supergroup (~75% of the species screened), with only four isolates placed in the A supergroup (Drosophila flava, D. nigrocirrus, Scaptomyza. nr.semiflava and S. longiestosa). The non-native Hawaiian Drosophila, D. suzukii and D. kikkawai, were also placed in the A supergroup along with other widespread species (Fig. 4), indicating a potentially shared infection for these species. While the A and B supergroups are known to primarily infect insect hosts, little is known about the phylogenetic split between them or basic biological roles of either group.
Phylogenetic reconstructions tend to cluster Hawaiian Wolbachia together, along with the Japanese and African Heteroptera and Lepidoptera infections (Figs. 3 and 4). The close association of Hawaiian alleles suggests that Wolbachia is maintained through speciation or potentially transferred between related hosts (e.g., horizontal transfer). Several of the infected Drosophila species are closely related and share the same or closely related Wolbachia alleles. For example, D. ancyla and D. nr.fundita share the wGin alleles, and both are placed in the split tarsus species subgroup along with D. fundita, which carries the closely related Wolbachia allele, wFun (Fig. 3).26 Similarly, sister species D. redunca and D. prodita both share the wDasA allele. (Fig. 3) The close relationships between these species suggest that Wolbachia infections may persist through cladogenesis. On the other hand, closely related species share distantly related alleles and vice versa. The wGin allele shared among split tarsus species, is also shared with native Hawaiian leafhoppers (Fig. 3). The shared allele between D. redunca and D. prodita, is also carried by the distantly related D. dasycnemia. Finally, Nesophrosyne species in subgroup 1, which share the wGiff and closely related wSil alleles, are not closely related (Fig. 2). Rather, these species only occur in sympatry, but occupy different host plants and are members of unrelated species groups (Bennett, unpublished). This result provides evidence for other mechanisms of Wolbachia spread and persistence, such as horizontal transfer, which is described in more detail below.
It is not possible to propose biogeographic hypotheses regarding the origins of Hawaiian Wolbachia at this time. Geographic and taxonomic sampling of Wolbachia is biased toward certain taxonomic groups and geographic areas. While the associations with Japanese Heteroptera and Lepidoptera Wolbachia alleles are intriguing,19,22 significant regions of the Western Pacific basin have not been sampled, nor have many of the dipteran groups. It is possible that the shared alleles found in this study include widely dispersed, pan-pacific Wolbachia lineages. In this case, the Wolbachia lineages recovered here may have arrived to Hawaii from other sources not yet screened. Or, the sister relationships between Japanese and Hawaiian Wolbachia is merely an artifact of incomplete taxon sampling. In order to robustly test the potential origins of Wolbachia infection on Hawaii, broad scale taxonomic sampling across the Pacific region that targets known outgroups and related genera is required.
Horizontal transmission of Wolbachia among Hawaiian lineages
Wolbachia in lineages with relatively low incidence rates (< 20%) may rely on other mechanisms to sustain infection, including mutualistic benefits to hosts, interactions with other endosymbionts, or widespread horizontal transfer.27-29 Our results provide several lines of evidence for extensive horizontal transmission of Wolbachia between Hawaiian taxa at multiple taxonomic scales, which is congruent with other studies.1,30-32 At the ordinal level, nearly identical Wolbachia alleles are shared between Diptera species (e.g., Drosophila forficata, and D. spnr.dorsigera) and Hemiptera (Nesophrosyne craterigena: Fig. 3). A plausible mechanism for this scenario is explained by the reliance of both Drosophila and Nesophrosyne species on shared host plants across their ranges (e.g., both feed from and lay their eggs directly into their host plants).33-35 These results corroborate the findings of Sintupachee et al.36 that hosts spanning multiple insect orders, which rely on the same host plants, tend to share closely related Wolbachia alleles. It is notable that in contrast to this result, the uninfected dolichopodid and limoniid flies do not rely on host plants, as they are predatory and semi-aquatic or aquatic, respectively.37,38 This difference in habitat use and life strategy may eliminate potential mechanisms for horizontal Wolbachia transmission.
Within the Drosophilidae family, there is evidence for horizontal transmission of Wolbachia between species. Shared alleles emerge between distantly related species (Drosophila eurypeza and D. nr.basimacula), and between the widely sampled D. dasycnemia, D. redunca and D. prodita. The shared alleles between D. dasycnemia and D. redunca, which are endemic to different islands (Hawaii Island and Molokai, respectively), suggests that horizontal transmission has occurred somewhat recently and potentially through a network of interacting species. The native and non-native drosophilid lineages placed in the A supergroup also share similar alleles (D suzukii, D. ananassae, D. simulans and D. auria; and, D. nigrocirrus, Scaptomyza. nr.semiflava and S. longiestosa: Fig. 4) both with other widespread taxa and parasitoid wasps from other studies (see Table S1 for references). This close association suggests parasitoids as a second potential mechanism for Wolbachia horizontal transmission between native and non-native Hawaiian Drosophila. There is substantial evidence that wasps transfer Wolbachia among the hosts they provision themselves with, and especially Drosophila, which are susceptible to parasitism.32,39,40 Parasitoid relationships within D. dasycnemia, D. redunca and D. redunca are currently unknown. However, Hawaii is home to a large diversity of endemic and introduced parasitoid wasps known to attack both native and non-native insects.41 This mechanism may allow for newly colonizing lineages to introduce and spread novel Wolbachia alleles to previously uninfected Hawaiian arthropods.
Materials and Methods
Taxon sampling and DNA extraction
Taxonomic sampling included 419 specimens, focusing on Diptera in four families (Drosophilidae, Limoniidae, Calliphoridae, Dolichopodidae) and 22 genera (see Table 3 for the taxonomic placement of all species screened for this study). Hawaiian Diptera sampling consisted primarily of native flies, but included several introduced Drosophila in the D. melanogaster and D. immigrans species groups, and Pacific-wide species not known to occur in Hawaii (e.g., Drosophila dispar species group and Mycodrosophila). Thirteen native Hawaiian leafhopper (Nesophrosyne) individuals in five species were also included from an ongoing project examining endosymbiont associations in this genus. To examine the potential rate of infection within a single population, 56 individuals of native Drosophila dasycnemia from Hawaii Island were screened for Wolbachia. Field collected material was placed in 95% ethanol for preservation and then identified to species. DNA was extracted from whole insects specimens using the DNeasy Tissue Kit (QIAGEN) following the manufacturer's protocol.
Wsp screening, PCR and DNA sequencing
Presence of Wolbachia was determined by screening potential dipteran and hemipteran hosts, using primers specific to the wsp gene. Despite some criticisms of the single use of the wsp gene,43 few studies have screened Pacific islands for Wolbachia infections, and the available wsp sequence data provides the best opportunity for geographic contextualization of potential infections in Hawaiian taxa. The wsp gene has been one of the most widely used in Wolbachia identification and systematics (~2200 worldwide GenBank sequences), including extensive sampling of Japanese Lepidoptera and Heteroptera.19,22 As well, the wsp gene provides adequate terminal resolution to assess relationships of Wolbachia alleles among Hawaiian taxa. The wsp locus was sequenced for the forward and reverse directions using Polymerase Chain Reaction (PCR) primers: wspF 5′-TGGTCCAATAAGTGATGAAGAAACTAGCTAG-3′ and wspR 5′-AAAAATTAAACGCTACTCCAGCTTCTGCAC-3′.3,17 A general touchdown PCR protocol was implemented: 3 min at 94°C; 5 cycles of 30 sec at 94°C, 30 sec at 65°C and 90 sec at 72°C; 5 cycles of 30 sec at 94°C, 30 sec at 60°C and 90 sec at 72°C; 25 cycles of 30 sec at 94°C, 30 sec at 55°C and 90 sec at 72°C; 5 min at 72°C. Positive infections were determined from successful PCR amplifications with the wsp primers, which were then cleaned using ExoSap (Strategene) following the manufacturer's protocol and sequenced at the U.C. Berkeley, Barker Sequencing Center. Sequenced specimens gave clean wave profiles, indicating single Wolbachia infections of screened Hawaiian Insects. New sequences derived from this study were submitted to GenBank (see Table 1 for accession numbers).
Sequence editing and alignment
Sequenced Hawaiian taxa were imported into Geneious Pro v5.0.4,44 which was used to build and edit contigs for forward and reverse sequence fragments. Nucleotide BLAST searches were conducted with Hawaiian sequences to identify the most closely related wsp sequences, which were then imported into a large alignment. All individual sequences were given a four-digit barcode (e.g., 001w–286w) for reference, which corresponds to host taxonomic information, geographic collection locality and references (see Table S1).
A total data matrix containing 284 wsp sequences (45 are new to this study) was aligned using Muscle V.3.5.45 Two taxa obtained from GenBank (006w and 007w: see Table S1) were removed from further analyses due to difficulty in aligning. The total alignment was then exported to MacClade v4.08,46 translated into amino acid sequences, and checked against the amino acid conceptual translation for the wsp gene acquired from GenBank. Two ambiguously aligned regions between 239–250 and 572–589 (30 base pairs) were removed from analyses, yielding a total gene length of 605 base pairs. The edited alignment is available from the O’Grady lab website (http://www.drosophilaevolution.com/index.html). All sequence data generated for this study has been deposited to the GenBank database and assigned sequence accession number JX134910–JX134954 (see Table 1).
Phylogenetic analyses
Phylogenetic analyses were performed using Maximum Likelihood methods. A likelihood model of nucleotide substitution for the wsp gene data set was estimated using Modeltest v3.7 run in PAUP* and determined via Akaike Information Criterion,47-49 which approximated GTR+inv+gamma as the best fit model. In order to successfully reconstruct a statistical phylogenetic inference of Wolbachia relationships for all 284 taxa (Parsimony and Bayesian analyses failed to finish), RAxML-HPC2 v7.2.7 was employed through the CIPRES portal on the ABE server.50-52 RAxML was run with the rapid bootstrapping option for 1000 bootstrap iterations with a GTRCAT model of nucleotide substitution. A best-scoring maximum likelihood tree was then estimated under a GTRGAMMA model, with the suggested default of 25 rate categories. Resultant trees were exported into FigTree v1.3.1 for viewing and editing, and rooted at its mid-point.53
Conclusion
Results from this study provide the foundation for understanding Wolbachia infections on the remote Hawaiian Archipelago. The local Wolbachia incidence for Hawaiian groups is low (generally less than 20%), suggesting that the direct effects on the sexual reproduction and population dynamics of endemic lineages may be reduced or even absent.28,42 Thus, the overall role of Wolbachia in speciation of native Hawaiian insects may not be significant, however this remains to be directly tested. While horizontal transfer appears to be common in native Hawaiian taxa, broad scale screening of different native lineages is required to confirm its overall role in the persistence of Wolbachia among endemic Hawaiian arthropods. Phylogenetic evidence indicates two potential, testable mechanisms may contribute to the observed horizontal transmission of Wolbachia: shared host plants and parasitoid interactions. Future studies should screen not only insect lineages sharing particular host plants and habitats, but also the shared host plant tissues and parasitoids known to occur or provision themselves with geographically overlapping species. The Hawaiian drosophilids and cidadellids present tractable systems to examine these and other mechanisms in future studies, as sister species in these groups are geographically isolated between islands and volcanoes, species are members of large intra-island radiations within discreet island boundaries, and much is known about the age and ecology of their species.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank three anonymous reviewers for their insightful comments that helped improve the quality of this manuscript. We also thank Michael Turelli, David Hembryand Brian Ort for thoughtful comments and discussion on drafts of this manuscript. We thank Rick Lapoint and Karl Magnacca for fly samples, and Therese Markow and her lab-group for Wolbachia wsp primers and PCR protocol suggestions. This research was partially funded by the U.C. Berkeley Walker Grant. Some work was partially funded by NSF DEB-0842348 to PM O’Grady and NL Evenhuis.
Supplemental Material
Supplemental material may be downloaded here:
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/21161
References
- 1.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–51. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 2.Zhou WF, Rousset F, O’Neil S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265:509–15. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 4.Jiggins FM, Bentley JK, Majerus MEN, Hurst GDD. How many species are infected with Wolbachia? Cryptic sex ratio distorters revealed to be common by intensive sampling. Proc Biol Sci. 2001;268:1123–6. doi: 10.1098/rspb.2001.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ros VI, Fleming VM, Feil EJ, Breeuwer JAJ. How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae) Appl Environ Microbiol. 2009;75:1036–43. doi: 10.1128/AEM.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–85. doi: 10.1098/rspb.2000.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dedeine F, Boulétreau M, Vavre F. Wolbachia requirement for oogenesis: occurrence within the genus Asobara (Hymenoptera, Braconidae) and evidence for intraspecific variation in A. tabida. Heredity (Edinb) 2005;95:394–400. doi: 10.1038/sj.hdy.6800739. [DOI] [PubMed] [Google Scholar]
- 8.Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 2010;107:769–74. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starr DJ, Cline TW. A host parasite interaction rescues Drosophila oogenesis defects. Nature 2002; 418:76-79. [DOI] [PubMed]
- 10.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–7. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 11.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–3. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie RG, Roderick GK. Arthropods on islands: colonization, speciation, and conservation. Annu Rev Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. [DOI] [PubMed] [Google Scholar]
- 13.Mendelson TC, Shaw KL. Sexual behaviour: rapid speciation in an arthropod. Nature. 2005;433:375–6. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- 14.Price JP, Clague DA. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc Biol Sci. 2002;269:2429–35. doi: 10.1098/rspb.2002.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Grady P, Desalle R. Out of Hawaii: the origin and biogeography of the genus Scaptomyza (Diptera: Drosophilidae) Biol Lett. 2008;4:195–9. doi: 10.1098/rsbl.2007.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A. 1992;89:2699–702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. Heritable endosymbionts of Drosophila. Genetics. 2006;174:363–76. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werren JH, Bartos JD. Recombination in Wolbachia. Curr Biol. 2001;11:431–5. doi: 10.1016/S0960-9822(01)00101-4. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi Y, Fukatsu T. Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl Environ Microbiol. 2003;69:6082–90. doi: 10.1128/AEM.69.10.6082-6090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ono M, Braig HR, Munstermann LE, Ferro C, O’Neill SL. Wolbachia infections of phlebotomine sand flies (Diptera: Psychodidae) J Med Entomol. 2001;38:237–41. doi: 10.1603/0022-2585-38.2.237. [DOI] [PubMed] [Google Scholar]
- 21.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?--A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–20. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagami Y, Miura K. Distribution and prevalence of Wolbachia in Japanese populations of Lepidoptera. Insect Mol Biol. 2004;13:359–64. doi: 10.1111/j.0962-1075.2004.00492.x. [DOI] [PubMed] [Google Scholar]
- 23.Howarth FG, Mull WP. Hawaiian insects and their kin. Honolulu, HI: University of Hawaii Press, 1992. [Google Scholar]
- 24.Carson HL, Templeton AR. Genetic revolutions in relation to speciation phenomena: The founding of new populations. Annu Rev Ecol Syst. 1984;15:97–131. doi: 10.1146/annurev.es.15.110184.000525. [DOI] [Google Scholar]
- 25.Wagner WL, Funk VA. Hawaiian Biogeography: Evolution on a Host Spot Archipelago. Washington DC: Smithsonian Institute Press, 1995. [Google Scholar]
- 26.O’Grady PM, Lapoint RT, Bonacum J, Lasola J, Owen E, Wu Y, et al. Phylogenetic and ecological relationships of the Hawaiian Drosophila inferred by mitochondrial DNA analysis. Mol Phylogenet Evol. 2011;58:244–56. doi: 10.1016/j.ympev.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 2008;17:557–69. doi: 10.1111/j.1365-294X.2007.03608.x. [DOI] [PubMed] [Google Scholar]
- 28.Hughes GL, Allsopp PG, Brumbley SM, Woolfit M, McGraw EA, O’Neill SL. Variable infection frequency and high diversity of multiple strains of Wolbachia pipientis in Perkinsiella Planthoppers. Appl Environ Microbiol. 2011;77:2165–8. doi: 10.1128/AEM.02878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel-Salzat A, Cordaux R, Bouchon D. Wolbachia diversity in the Procelloinides pruinosus complex of species (Crustacea: Onsicidea): evidence for host-dependent patterns of infection. Heredity. 2005;87:428–34. doi: 10.1046/j.1365-2540.2001.00920.x. [DOI] [PubMed] [Google Scholar]
- 30.Ahrens ME, Shoemaker D. Evolutionary history of Wolbachia infections in the fire ant Solenopsis invicta. BMC Evol Biol. 2005;5:35. doi: 10.1186/1471-2148-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shoemkaer DD, Machado CA, Molbo D, Werren JH, Windsor DM, Herre EA. The distribution of Wolbachia in fig wasps: correlation with host phylogeny, ecology, and population structure. Proc Biol Sci. 2001;269:2257–67. doi: 10.1098/rspb.2002.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vavre F, Fleury F, Lepetit D, Fouillet P, Boulétreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol Biol Evol. 1999;16:1711–23. doi: 10.1093/oxfordjournals.molbev.a026084. [DOI] [PubMed] [Google Scholar]
- 33.Bennett GM, O’Grady PM. Review of the native Hawaiian leafhopper genus Nesophrosyne (Hemiptera: Cicadellidae: Deltocephalinae) with description of eight new species associated with Broussaisia arguta (Hydrangeaceae) Zootaxa. 2011;2805:1–25. [Google Scholar]
- 34.Magnacca KN, O’Grady PM. Revision of the ‘nudidrosophila’ and ‘ateledrosophila’ species groups of Hawaiian Drosophila (Diptera: Drosophilidae), with descriptions of twenty-two new species. Syst Ent. 2008;33:395–428. doi: 10.1111/j.1365-3113.2007.00400.x. [DOI] [Google Scholar]
- 35.Zimmerman EC. Insects of Hawaii. Volume 4. Homoptera: Auchenorrhyncha. Honolulu, HI: University of Hawaii Press, 1948;37-81. [Google Scholar]
- 36.Sintupachee S, Milne JR, Poonchaisri S, Baimai V, Kittayapong P. Closely related Wolbachia strains within the pumpkin arthropod community and the potential for horizontal transmission via the plant. Microb Ecol. 2006;51:294–301. doi: 10.1007/s00248-006-9036-x. [DOI] [PubMed] [Google Scholar]
- 37.Hardy DE, Kohn MA. Insects of Hawaii. Volume 11. Family Dolichopodidae Latreille. Honolulu, HI: University of Hawaii Press, 1964;13–296. [Google Scholar]
- 38.Nitta JH, O’Grady PM. Mitochondrial phylogeny of the endemic Hawaiian craneflies (Diptera, Limoniidae, Dicranomyia): implications for biogeography and species formation. Mol Phylogenet Evol. 2008;46:1182–90. doi: 10.1016/j.ympev.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–49. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- 40.Cordaux R, Michel-Salzat A, Bouchon D. Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J Evol Biol. 2001;14:237–43. doi: 10.1046/j.1420-9101.2001.00279.x. [DOI] [Google Scholar]
- 41.Henneman ML, Memmott J. Infiltration of a Hawaiian community by introduced biological control agents. Science. 2001;293:1314–6. doi: 10.1126/science.1060788. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda T, Ishikawa H, Sasaki T. Infection density of Wolbachia and level of cytoplasmic incompatibility in the Mediterranean flour moth, Ephestia kuehniella. J Invertebr Pathol. 2003;84:1–5. doi: 10.1016/S0022-2011(03)00106-X. [DOI] [PubMed] [Google Scholar]
- 43.Baldo L, Dunning Hotopp JC, Jolley KA, Bordenstein SR, Biber SA, Choudhury RR, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, et al. Geneious v5.1, 2010. Available from http://www.geneious.com
- 45.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maddison D, Maddison W. MacClade: analysis of phylogeny and character evolution, v. 4.08. 2005. Sunderland MA: Sinauer Associates. [Google Scholar]
- 47.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 48.Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006;34(Web Server issue):W700-3. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swofford DL. PAUP* Phylogenetic analysis using parsimony (*and other methods), Version 4, 2002. Sinauer Associates, Sunder- land, MA. [Google Scholar]
- 50.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceeding of the Gateway Computing Environments Workshop (GCE), 14Nov2010, New Orleans, LA. [Google Scholar]
- 51.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 52.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–71. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 53.Rambaut A. FigTree v.1.3.1. Institute of Evolutionary Biology, University of Edinburgh. 2009.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.