Abstract
Drosophila melanogaster is widely used as a model system for development and disease. Due to the homology between Drosophila and human genes, as well as the tractable genetics of the fly, its use as a model for neurologic disorders, in particular, has been rising. Locomotive impairment is a commonly used diagnostic for screening and characterization of these models, yet a fast, sensitive and model-free method to compare behavior is lacking. Here, we present a high throughput method to quantify the crawling behavior of larvae. We use the mean squared displacement as well as the direction autocorrelation of the crawling larvae as descriptors of their motion. By tracking larvae from wild-type strains and models of the Fragile X mental retardation as well as Alzheimer disease, we show these mutants exhibit impaired crawling. We further show that the magnitude of impairment correlates with the severity of the mutation, demonstrating the sensitivity and the dynamic range of the method. Finally, we study larvae with altered expression of the shaggy gene, a homolog of Glycogen Synthase Kinase-3 (GSK-3), which has been implicated in Alzheimer disease. Surprisingly, we find that both increased and decreased expression of dGSK-3 lead to similar larval crawling impairment. These findings have implications for the use of GSK-3 inhibitors recently proposed for Alzheimer treatment.
Keywords: Alzheimer disease, FMR1, Fragile X syndrome, GSK-3, Shaggy, behavior, crawling, disease model, larva, neurodegeneration
Introduction
A hallmark of many neurodegenerative diseases is progressive locomotive impairment.1,2 Animal models for movement have previously been used to study the foraging patterns of ants,3 seabirds4 and butterflies5 but are also studied for a plethora of diseases, including Parkinson disease, Alzheimer Disease, Amyotrophic lateral sclerosis and Fragile X syndrome.2 Given the genetic origin of many of these disorders, Drosophila melanogaster has proven to be a powerful model system in many cases.2,6,7 While studying the locomotion of adult flies is feasible,8-10 larval phenotypes are often used to characterize mutations because of the simplicity of their movement patterns. Larvae also have the advantage over adults in that they allow for the study of diseases that are lethal before adulthood.
Different modes of larval movement, ranging from body shape changes to taxis toward food, have been examined depending on the nature of the mutation under study. However, given that many defects in body movement result in defects in larval crawling, the latter has gathered interest since it is amenable to quantification. Attempts to quantify larval crawling have focused on the speed of the larvae and their propensity to change direction, albeit often with arbitrarily set thresholds and restrictive tracks for larval crawling. Nevertheless, comparing larval crawling has been instrumental in studies of models of disease,2,11,12 ion channels,13,14 chemotaxis and foraging15,16 and the interplay between sensory inputs and locomotion.17,18
We describe a high throughput and sensitive method to quantify larval crawling behavior without the need for setting ad hoc thresholds or imposing models. The method uses the Mean Square Displacement (MSD) as well as the Direction AutoCorrelation (DAC) to compare velocities and turning statistics of larvae. We show that our method reproduces the previously analyzed crawling behavior of a fly model of Fragile X syndrome. We use the method to further study a model of Alzheimer disease, a hallmark of which is increased activity of the serine-threonine kinase Glycogen Synthase Kinase 3 (GSK-3).19,20 We used our quantitative method to ask whether overexpression of endogenous GSK-3 leads to crawling phenotypes and whether these phenotypes correlate with gene expression levels.
Results
Fragile X larvae are slow and turn more often
To assess the performance of the methodology we developed, we applied it to a system that has been well characterized and documented in the literature. The Fragile X-related gene in Drosophila (dfmr1) is highly homologous to the human fragile X mental retardation (fmr1) gene. Loss-of-function mutations in the FMR1 gene cause Fragile X syndrome. Previous work has shown that loss-of-function mutations in the Drosophila dfmr1 gene affect the crawling behavior of the larvae by decreasing the duration and percentage of time the larvae spend on linear locomotion.12 This behavior was characterized by counting individual direction switching events using traces of crawling larvae tracked by the DIAS image-analysis system.21,22 In this study as well as others,23,24 a switching event was defined as motion deviating from the previous direction by more than a manually set threshold. Setting thresholds manually could obscure differences if phenotypical traits are not very pronounced. In order to avoid this complication, here we use a parameter-free method to characterize larval locomotion and compare our results with those previously reported.
The details of the method are described in the Methods section. Briefly, paths of multiple larvae at that same developmental stage are tracked from time lapse images. The position data are analyzed to extract the Mean Squared Displacement (MSD) and the Directional Autocorrelation (DAC) as a function of time. Figure 2A compares the MSD for the dfmr14 larvae to the relevant wild type strain (w1118), showing obvious differences. The MSD is a measure of the area explored per unit time averaged over many larvae and is clearly smaller for the dfmr14 larvae. The dependence of the MSD on time portrays the type of underlying motion: for a purely random walk, the MSD is linear with time, while for directed motion along a straight line, the MSD increases quadratically with time. When plot on a log-log plot, random motion gives a line with a slope of 1, while directed ballistic motion gives a slope of 2. As is clear in the inset of Figure 2A, the MSD for both the wild type and mutant larvae have a slope between 1 and 2 which is indicative of super-diffusive motion as is the case for many foraging animals.25 Importantly, the mutant is slower than the wild type larvae as indicated by the smaller intercept with the vertical axis on the log-log plot.
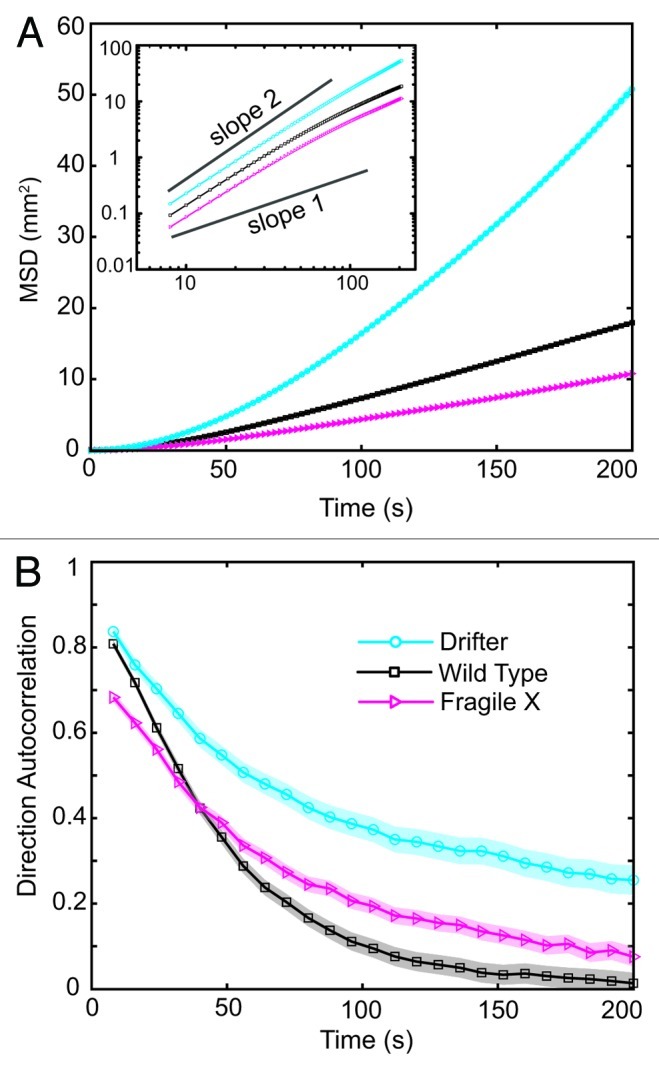
Figure 2. Fragile X larvae crawl more slowly and turn more often. (A) The Mean Squared Displacement (MSD) for crawling larvae from the Fragile X model (dfmr14) is lower than that for wild type larvae w[1118] indicating crawling defects. The smaller y-intercept, inset in (A), is indicative of a smaller mean velocity. Larvae from the Drifter strain (Df), which have decreased levels of the ion channel pickpocket1 that is over abundant in Fragile X individuals, show an opposite trend demonstrating the dynamic range of the method. (B) The Direction Autocorrelation (DAC) shows that the dfmr14 larvae turn more often than wild type larvae at small times and that the Df larvae turn significantly less. Error is standard error of the mean and is shown as a shadowed region around the trace for clarity. MSD error is smaller than the data point symbols and not shown on the logarithmic plot for clarity. Symbols and colors: cyan circles are Drifters, black squares are wild type and magenta triangles are Fragile X larvae, arranged in this order from top to bottom in the MSD plot in (A). (Number of larvae tracked: WT (25), dfmr14 (24), Drifter (17))
The smaller MSD for the mutant larvae could result from either more sluggish crawling or more frequent direction switching, or both. To separate the effect of direction switching, we calculated the directional autocorrelation function shown in Figure 2B. For each time point, the DAC is the cosine of the angle between two segments separated by that amount of time, averaged over all segments during that interval. Larvae crawling in a straight line over the course of a specified time interval will exhibit an autocorrelation value of 1. The dfmr14 larvae are more likely to switch direction very quickly as portrayed by the lower directional correlation at small times. This is also consistent with the findings in ref.12
To test the dynamic range of the method, we decided to analyze the crawling behavior for larvae, named Drifter (Df), which have previously been shown to move in straight trajectories for extended periods of times. Drifter larvae were also reported to be faster than the wild type.13 Loss of the protein Pickpocket1 (ppk1), a Na+ epithelial channel, has been shown to exhibit this behavior. The phenotype being opposite of the dfmr14 mutant is consistent with functional dFMR1 acting to decrease the levels of ppk1 mRNA in vivo.26,27 We used the Gal4 109(2)80 line, driving expression in multiple dendritic neurons, to decrease Pickpocket1 levels through a ppk1RNAi transgene obtained from the Transgenic RNAi Project.12 As shown in Figure 2A, the MSD for the Drifter larvae is much higher than both wild type and dfmr14 larvae. As previously reported,13 the Drifter mutant is faster than the wild type as seen by the increase of the y-intercept in the log-log plot. Moreover, the directional autocorrelation in Figure 2B indicates that Drifter larvae are significantly less likely to switch direction. This analysis on two separate mutants shows that our method reproduces previously published results, with the advantage of providing a high throughput and model-free assessment of the crawling phenotype.
GSK-3 overexpression leads to crawling defects
After showing the quantitative methodology accurately describes well documented crawling phenotypes, we decided to study a fly model of Alzheimer disease. Glycogen Synthase Kinase 3 (GSK-3) has been proposed to be a key player in Alzheimer pathology and other neurological disorders.19,28-32 The Shaggy (Sgg) gene in Drosophila encodes for the sole homolog of GSK-3α/β (dGSK-3).33 We used the GAL4-UAS system34 to induce panneuronal overexpression of wild-type dGSK-335 and compared the crawling of these larvae to larvae with wild-type expression of dGSK-3. Figure 3A shows that the MSD is lower for the increased dGSK-3 larvae indicating a slower crawling phenotype similar to that of dfmr14 larvae. Moreover, the directional correlation is also lower indicating more frequent direction switching.
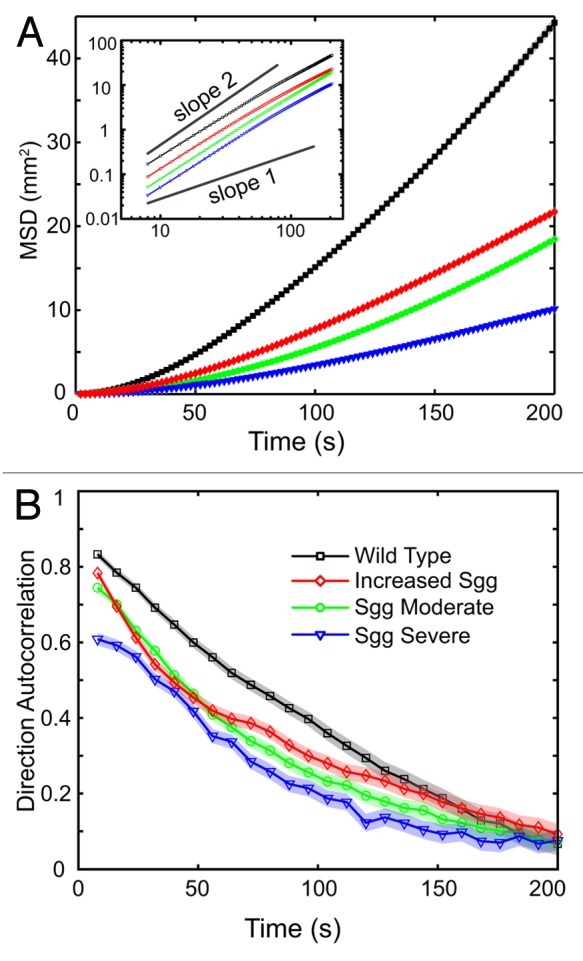
Figure 3. Both GSK-3 overexpression and reduction lead to dosage-dependent impairment of the crawling behavior. The MSD and DAC plots show that overexpression of dGSK-3 leads to a smaller velocity (A, inset) and more frequent turns (B). Two classes of larvae with different degrees of dGSK-3 reduction lead to decrease in velocity and increase in turning frequency. The severity of the phenotype correlates with the expression levels of dGSK-3. The ’SGG severe’ express less dGSK-3 than both wild type and ‘SGG moderate’ larvae and show a smaller velocity (A) as well as a larger turning frequency (B). Error is standard error of the mean and is shown as a shadowed region around the trace for clarity. MSD error is smaller than the data point symbols and not shown on the logarithmic plot for clarity. Symbols and colors: black squares are wild type, red diamonds are increased Sgg, green circles are Sgg moderate and purple triangles are Sgg severe larvae, arranged in this order from top to bottom in the MSD plot in (A). [Number of larvae tracked: WT (22), Increased SGG (19), SGG Moderate (24), SGG Severe (17)]
GSK-3 reduction leads to dosage-dependent crawling defects similar to those of GSK-3 overexpression
We next asked whether the severity of the crawling impairment correlates with gene expression levels. To answer that question, we opted to use flies with reduced dGSK-3 expression. Given that Sgg-null flies die before reaching adulthood, we used the Shaggy mutants to generate a mixed population of larvae with varying expression levels of dGSK-3 (see Methods).35 These larvae fell in two groups based on dGSK-3 expression levels: ‘Sgg moderate’ contains two separate types of larvae, those entirely wild type for Sgg and those heterozygous for a null copy of Sgg. The ‘Sgg severe’ population, however, contains either heterozygous mutants or mutants containing two null copies (homozygous null) of the Sgg gene. We found that both these groups show crawling impairment compared with the wild type (Fig. 3). The impairment was reminiscent of that observed for dGSK-3 overexpression for the ‘Sgg moderate’ larvae and even more severe for the ‘Sgg severe’ larvae. Thus, reduction as well as increase of dGSK-3 expression leads to larvae that are slower and turn more frequently. These findings demonstrate the sensitivity of the method and show that our quantitative measure of the behavioral phenotype correlates with gene expression. Moreover, our observations have implications on the use of GSK-3 inhibitory drugs proposed for Alzheimer treatment as discussed below.
Discussion
The methodology we developed in this study enabled a quantitative description of locomotion for a population of Drosophila larvae. Larval velocities and direction switching were readily compared using the Mean Squared Displacement (MSD) and directional autocorrelation (DAC) calculations which were used to highlight differences between wild type larvae and models of neurodegenerative diseases. These parameters had previously been shown to correlate with some neurological disorders in larval models of disease, yet the low throughput and ad hoc choice of parameter threshold needed limited their applicability.
Our method uses a low-cost and easily replicable setup that generates highly contrasted image sequences of the millimeter-sized first-instar larvae. We showed that our trajectory tracking method can be used with six larvae crawling at once on the large Petri dish; the larvae seldom interfere. Given that as few as 15 larvae are needed to measure a statistically significant crawling phenotype, this batch imaging allows for high throughput data collection compared with methods requiring collection, conditioning and recording of one larva at-a-time.12,22,23 Throughput can be further increased by using a wide-field lens, allowing simultaneous recording of 15 or more larvae.
Previous studies have focused on the movement of third-instar larvae, a 72 h-long stage, which leads to uncertainty in age that could affect the behavior if gene expression varies over that time for the trait investigated. Our ability to use the small first-instar larvae enabled us to standardize for age when looking at larva populations by allowing the larvae to hatch from embryos on the Petri dish while imaging and analyzing their crawling a fixed amount of time post-hatching. Previous works, as well as our Fragile X results, showed that first instar larvae show similar behavior to later stage larvae in many cases.18,36,37 Being able to use first-instar larvae has an additional advantage in cases where the mutation is lethal at later stages. However, the same methodology could be used for later stage larvae if the trait investigated requires that.
The larval tracks are statistically analyzed to calculate the MSD and the DAC. The shape of the MSD allows comparing larval speeds as well as distinguishing between different types of larval motion (e.g., ballistic vs. diffusive). Close inspection of the MSD in Figures 2A and 3A shows that its slope is slightly lower than 2 at early times and drops to a smaller value for later times. This indicates that the motion of the larvae is super-diffusive. Such super-diffusive motion was observed for many foraging animals25,38 and more recently for T-cell migration.39 The DAC gives a detailed readout of the direction switching over different timescales that is independent of speed. These statistical descriptors of the locomotion are average quantities that do not require manually setting any threshold to compare two populations. Moreover, since we are comparing populations rather than individuals, our method is less prone to variability arising from occasional coincidence of behavior over some time segment of individuals from the two populations. When averaged over many larvae (15–25), the characteristic traits of that genotype can be immediately seen in the MSD and DAC plots. One must caution, however, when using running averages to increase data precision in MSD calculations, as typically done with statistical analyses in physical systems, since larvae can change their behavior as they age. Our results show that three-minute tracks are sufficient to discern small changes in population behavior and that the trends are continuous as demonstrated by the analysis of mixed populations in Figure 3. This illustrates the sensitivity of the method.
We tested our methodology on the previously studied model of Fragile X syndrome (dfmr14). We find that the dfmr14 mutation leads to a decrease in crawling velocity along with more frequent turns. On the other hand, the Drifter mutation, one which leads to decreased levels of the ion channel pickpocket1 which is profuse in Fragile X individuals, exhibits increases in larval crawling velocity and less frequent turns. These findings are in qualitative agreement with previous results.12,13
We studied a Drosophila model of Alzheimer disease: mutants in the shaggy gene (Sgg) which encodes for the fly homolog of Glycogen Synthase Kinase-3 (GSK-3).33 Elevated levels or altered activation of GSK-3 have been implicated in Alzheimer disease, amyotrophic lateral sclerosis (ALS), bipolar disorder and schizophrenia.19,28-32 Increasing dGSK-3 expression in first-instar larvae led to a decrease in crawling velocity and increased turning frequency compared with the wild type larvae, similar to what was seen in the Fragile X Drosophila model. Importantly, we also found that populations of larvae with reduced dGSK-3 expression had both a lower velocity and higher turning frequency. We further found that this trend is Sgg copy number dependent and changes continuously with the reduction of dGSK-3.
Given that a hallmark of the Alzheimer pathology is an increase in GSK-3 activity,20 our finding that reduced dGSK-3 leads to impaired larval locomotion reminiscent of the Fragile X model might seem surprising. However, we have recently shown that both reducing dGSK-3 as well as increasing its expression leads to increased axonal accumulations in larval segmental nerves.35 Our behavioral assay suggests that such accumulations could be the cause of impaired locomotion. With GSK-3 inhibitors being investigated as potential Alzheimer therapies40,41 our findings suggest that care must be taken in evaluating such therapies.
In summary, we presented a quantitative and sensitive methodology to record and analyze locomotion of Drosophila larvae with a high throughput. The framework described in this work is not limited to fly larvae and can be used with other genetic systems, albeit with imaging adapted to the specific system. While we studied models of neuronal disorders, the method lends itself to characterizing locomotion in other diseases, especially in cases where the mutation leads to lethality at the pupal or adult stages, as well as locomotion in the presence of external cues. Further, the high throughput and sensitivity of the method we described make it suitable for drug as well as genetic screens.42
Materials and Methods
Fly stocks
Flies were grown on standard yeast-agar media at room-temperature. All stocks were obtained from the Bloomington Drosophila Stock Center, unless otherwise specified. Wild type controls were either w[1118] (FBid: 6326) or YW. The dfmr14 stock we used as our Fragile X model has been described previously24,27 and was provided to us by Fen-Biao Gao (UCSF). The Drifter (Df) mutation was created by crossing Gal4 109(2)80 (FBid: 8768) virgins to UAS-ppk1-RNAi (TRiP, Harvard Medical School, FBid: 29571) males, achieving overexpression of ppk1-RNAi in all multiple dendritic neurons. This stock has been described extensively.13 Overexpression of dGSK-3 was accomplished by crossing elav Gal4 (FBid: 458) virgins with UAS-SggS9A (FBid: 5255) males. For dGSK-3 reduction sgg1/FM7, Actin-GFP females were crossed to sgg1/Dp(1;2;Y)w+ (FBid: 4095) males that survive due to a small X duplication containing the Sgg gene on their Y chromosome.35 The first-instar larvae expressing GFP as seen under a dissection fluorescence microscope were termed the ‘Sgg moderate’ population while the GFP-negative larvae were termed the ‘Sgg severe’ population, based on the corresponding dGSK-3 expression levels.
Camera mount and stand
Larval crawling time-lapse images were recorded on a computer hard drive using a CCD camera (The Imaging Source) fitted with a 1:1.4/16mm lens (Fuji) and mounted to an aluminum stand (Fig. 1, inset). The camera was positioned 45 cm above the crawling surface of the agar plate. All recordings were taken at 0.5 fps and re-sampled for analysis as described below.
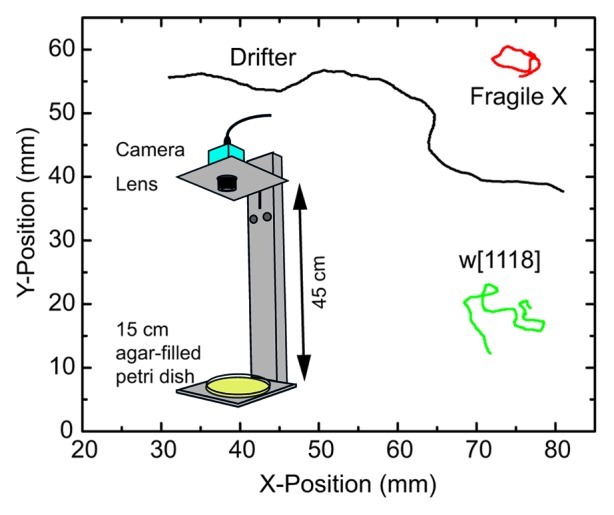
Figure 1. Representative larva tracks alongside a schematic representation of the experimental setup. The inset shows the experimental setup which consists of a camera and lens fixed on an aluminum stand 45 cm away from the Petri dish on which the larvae crawl. Larval tracks for the Fragile X model (dfmr14), Drifters (Df), and their corresponding wild-type strain (w[1118]) show that differences cannot always be visible in the tracks unless the phenotype is extreme as in the case of the Drifters. All tracks represent 20 min of crawling.
Larval crawling assay
Drosophila embryos were placed onto a 15 cm diameter plastic Petri dish with a layer of 2.5% agarose gel at room temperature. To prevent interactions between larvae, no more than six embryos were deposited onto the agar simultaneously. The typical separation between embryos was about 1 cm. Video recording began after all embryos were deposited onto the agarose gel and ended a few hours after the last hatching event (total recording time ~3–4 h). Short 200 sec segments at the beginning of the tracks were used for all analysis. To standardize for developmental age and potential initial transient effects, position tracks began 10 sec after hatching. The absence of food prevented any potential directional bias on the locomotive pattern of larvae. Phototactic and geotactic cues were eliminated by uniform lighting and flat agar surfaces.
Locomotion tracking and data analysis
Calibrated position tracks were obtained using a centroid-tracking program written in LabView.43 Mean squared displacement and directional autocorrelation analyses were performed using programs written in Matlab. The programs are made available at http://chaos.utexas.edu/people/faculty/george-t-shubeita. Each genotype was analyzed using 17–25 individuals.
The Mean Squared Displacement (MSD) was calculated using:
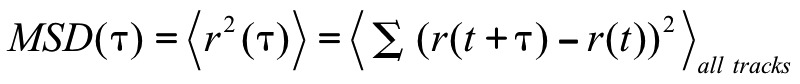 |
Where r(t) is the position at time t. The MSD is proportional to the average area covered in a given amount of time, τ, by a large number of particles starting from the same spatiotemporal origin and having the same statistical behavior. Thus, this function starts from zero and increases over time as more area is explored. When plotted on a logarithmic scale, the MSD enables measuring physical parameters of the locomotive behavior. The slope indicates to what degree the motion is directed or is a random walk. The MSD as a function of time for a directed, or ballistic, movement will have a slope of 2 on the logarithmic plot, while a purely random walk will have a slope of 1. The transition between the two regimes indicates the average time the larva moves in a straight line before switching direction. When represented on a linear scale, these regimes translate into a parabolic or linear relation, respectively. A slope between 1 and 2 is indicative of a super-diffusive motion.
The value of the y-intercept on the logarithmic plot is related to a generalized particle velocity. In particular, when the slope has a value of 2, the y-intercept is proportional to the logarithm of the particle’s velocity. This relationship does not hold, however, if the slope does not equal 2.
When comparing the MSD for larvae of different genotypes, lower MSD values can be due to either a smaller velocity or more frequent turning events, or both. The MSD would, in principle, enable differentiating between these two parameters by measuring the y-intercept on a logarithmic plot, and estimating the transition time between a slope 2 and slope 1. Practically, however, the transition time is not always well defined especially when the ballistic motion is not resolved or when motion is super-diffusive.
To get a more detailed description of turning behavior of the larvae we used the Direction AutoCorrelation (DAC) function given by:
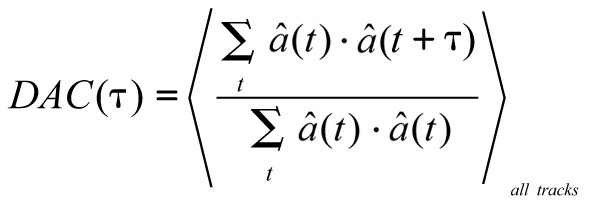 |
where 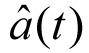 is a unit vector tangential to the path of the larvae at time t, and τ is the time interval at which the correlation is calculated. Given that our method describes the long range crawling of the larvae rather than changes in the body shape, we calculated the autocorrelation after reducing the frame rate to 0.125 fps. This value was chosen because it is the typical time it takes a larva to move twice its body length. This also had the effect of reducing the noise resulting from any inaccuracy in determining the centroid position of the larva.
is a unit vector tangential to the path of the larvae at time t, and τ is the time interval at which the correlation is calculated. Given that our method describes the long range crawling of the larvae rather than changes in the body shape, we calculated the autocorrelation after reducing the frame rate to 0.125 fps. This value was chosen because it is the typical time it takes a larva to move twice its body length. This also had the effect of reducing the noise resulting from any inaccuracy in determining the centroid position of the larva.
A direction autocorrelation of 1 indicates that, for the time interval considered, the direction did not change. The autocorrelation would be -1 if the larvae reversed direction and 0 if their directions at the beginning and end of the time interval are completely uncorrelated on average. For a crawling larva, the DAC would start close to 1 and drop as a function of time. The rate of decline of the DAC is a measure of the “straightness” of the track. One important feature of the direction autocorrelation function is that it is only sensitive to turning events and not to velocity. This allows for a direct comparison of the turning characteristics between different genotypes regardless of whether they move at the same velocity.
Acknowledgments
We thank Fen-Biao Gao for providing us with the dfmr14 fly line and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study. We also thank Emma Yee, Erika Park and Priya Tembekhar for building the camera mount. This work was supported in part by the National Science Foundation grant PHY- 0957811 to G.T.S.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/fly/article/21582
References
- 1.Hardy J, Gwinn-Hardy K. Genetic classification of primary neurodegenerative disease. Science. 1998;282:1075–9. doi: 10.1126/science.282.5391.1075. [DOI] [PubMed] [Google Scholar]
- 2.LeDoux M, ed. Movement Disorders: Genetics and Models: Academic Press, 2005. [Google Scholar]
- 3.Crist TO, MacMahon JA. Individual foraging components of harvester ants: movement patterns and seed patch fidelity. Insectes Soc. 1991;38:379–96. doi: 10.1007/BF01241873. [DOI] [Google Scholar]
- 4.Fritz H, Said S, Weimerskirch H. Scale-dependent hierarchical adjustments of movement patterns in a long-range foraging seabird. Proc Biol Sci. 2003;270:1143–8. doi: 10.1098/rspb.2003.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Root RB, Kareiva PM. The Search for Resources by Cabbage Butterflies (Pieris Rapae): Ecological Consequences and Adaptive Significance of Markovian Movements in a Patchy Environment. Ecology. 1984;65:147–65. doi: 10.2307/1939467. [DOI] [Google Scholar]
- 6.Ambegaokar SS, Roy B, Jackson GR. Neurodegenerative models in Drosophila: polyglutamine disorders, Parkinson disease, and amyotrophic lateral sclerosis. Neurobiol Dis. 2010;40:29–39. doi: 10.1016/j.nbd.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sang T-K, Jackson GR. Drosophila models of neurodegenerative disease. NeuroRx. 2005;2:438–46. doi: 10.1602/neurorx.2.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feiguin F, Godena VK, Romano G, D’Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–92. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Jahn TR, Kohlhoff KJ, Scott M, Tartaglia GG, Lomas DA, Dobson CM, et al. Detection of early locomotor abnormalities in a Drosophila model of Alzheimer’s disease. J Neurosci Methods. 2011;197:186–9. doi: 10.1016/j.jneumeth.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slawson JB, Kim EZ, Griffith LC. High-resolution video tracking of locomotion in adult Drosophila melanogaster. J Vis Exp. 2009 doi: 10.3791/1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mudher A, Shepherd D, Newman TA, Mildren P, Jukes JP, Squire A, et al. GSK-3beta inhibition reverses axonal transport defects and behavioural phenotypes in Drosophila. Mol Psychiatry. 2004;9:522–30. doi: 10.1038/sj.mp.4001483. [DOI] [PubMed] [Google Scholar]
- 12.Xu K, Bogert BA, Li W, Su K, Lee A, Gao F-B. The fragile X-related gene affects the crawling behavior of Drosophila larvae by regulating the mRNA level of the DEG/ENaC protein pickpocket1. Curr Biol. 2004;14:1025–34. doi: 10.1016/j.cub.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 13.Ainsley JA, Pettus JM, Bosenko D, Gerstein CE, Zinkevich N, Anderson MG, et al. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr Biol. 2003;13:1557–63. doi: 10.1016/S0960-9822(03)00596-7. [DOI] [PubMed] [Google Scholar]
- 14.Wang JW, Sylwester AW, Reed D, Wu DA, Soll DR, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: aberrant locomotion in Na+ and K+ channel mutants revealed by computer-assisted motion analysis. J Neurogenet. 1997;11:231–54. doi: 10.3109/01677069709115098. [DOI] [PubMed] [Google Scholar]
- 15.Gomez-Marin A, Stephens GJ, Louis M. Active sampling and decision making in Drosophila chemotaxis. Nat Commun. 2011;2 doi: 10.1038/ncomms1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sokolowski MB. Foraging strategies of Drosophila melanogaster: a chromosomal analysis. Behav Genet. 1980;10:291–302. doi: 10.1007/BF01067774. [DOI] [PubMed] [Google Scholar]
- 17.Louis M, Huber T, Benton R, Sakmar TP, Vosshall LB. Bilateral olfactory sensory input enhances chemotaxis behavior. Nat Neurosci. 2008;11:187–99. doi: 10.1038/nn2031. [DOI] [PubMed] [Google Scholar]
- 18.Suster ML, Bate M. Embryonic assembly of a central pattern generator without sensory input. Nature. 2002;416:174–8. doi: 10.1038/416174a. [DOI] [PubMed] [Google Scholar]
- 19.Hooper C, Killick R, Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. J Neurochem. 2008;104:1433–9. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takashima A. GSK-3 is essential in the pathogenesis of Alzheimer’s disease. J Alzheimers Dis. 2006;9(Suppl):309–17. doi: 10.3233/jad-2006-9s335. [DOI] [PubMed] [Google Scholar]
- 21.Soil DR. The use of computers in understanding how animal cells crawl. Int Rev Cytol. 1995;163:43–104. doi: 10.1016/S0074-7696(08)62209-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang JW, Sylwester AW, Reed D, Wu DA, Soll DR, Wu CF. Morphometric description of the wandering behavior in Drosophila larvae: aberrant locomotion in Na+ and K+ channel mutants revealed by computer-assisted motion analysis. J Neurogenet. 1997;11:231–54. doi: 10.3109/01677069709115098. [DOI] [PubMed] [Google Scholar]
- 23.Caldwell JC, Miller MM, Wing S, Soll DR, Eberl DF. Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc Natl Acad Sci U S A. 2003;100:16053–8. doi: 10.1073/pnas.2535546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee A, Li W, Xu K, Bogert BA, Su K, Gao F-B. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–52. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- 25.Viswanathan GM, Buldyrev SV, Havlin S, da Luz MG, Raposo EP, Stanley HE. Optimizing the success of random searches. Nature. 1999;401:911–4. doi: 10.1038/44831. [DOI] [PubMed] [Google Scholar]
- 26.Papadopoulou D, Bianchi MW, Bourouis M. Functional studies of shaggy/glycogen synthase kinase 3 phosphorylation sites in Drosophila melanogaster. Mol Cell Biol. 2004;24:4909–19. doi: 10.1128/MCB.24.11.4909-4919.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–8. doi: 10.1016/0092-8674(93)90420-U. [DOI] [PubMed] [Google Scholar]
- 28.Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- 29.Gould TD, Picchini AM, Einat H, Manji HK. Targeting glycogen synthase kinase-3 in the CNS: implications for the development of new treatments for mood disorders. Curr Drug Targets. 2006;7:1399–409. doi: 10.2174/1389450110607011399. [DOI] [PubMed] [Google Scholar]
- 30.Jope RS, Roh MS. Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Curr Drug Targets. 2006;7:1421–34. doi: 10.2174/1389450110607011421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peineau S, Bradley C, Taghibiglou C, Doherty A, Bortolotto ZA, Wang YT, et al. The role of GSK-3 in synaptic plasticity. Br J Pharmacol. 2008;153(Suppl 1):S428–37. doi: 10.1038/bjp.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Leystra-Lantz C, Strong MJ. Upregulation of GSK3beta expression in frontal and temporal cortex in ALS with cognitive impairment (ALSci) Brain Res. 2008;1196:131–9. doi: 10.1016/j.brainres.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried E, Chou TB, Perrimon N. wingless signaling acts through zeste-white 3, the Drosophila homolog of glycogen synthase kinase-3, to regulate engrailed and establish cell fate. Cell. 1992;71:1167–79. doi: 10.1016/S0092-8674(05)80065-0. [DOI] [PubMed] [Google Scholar]
- 34.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 35.Weaver C, Leidel C, Szpankowski L, Farley N, Shubeita GT, Goldstein LSB. Endogenous GSK-3/Shaggy regulates bidirectional axonal transport of the Amyloid Precursor Protein. doi: 10.1111/tra.12037. submitted. [DOI] [PubMed] [Google Scholar]
- 36.Carhan A, Reeve S, Dee CT, Baines RA, Moffat KG. Mutation in slowmo causes defects in Drosophila larval locomotor behaviour. Invert Neurosci. 2004;5:65–75. doi: 10.1007/s10158-003-0028-y. [DOI] [PubMed] [Google Scholar]
- 37.Godoy-Herrera R, Burnet B, Connolly K, Gogarty J. The development of locomotor activity in Drosophila melanogaster larvae. Heredity. 1984;52:63–75. doi: 10.1038/hdy.1984.7. [DOI] [Google Scholar]
- 38.Humphries NE, Queiroz N, Dyer JR, Pade NG, Musyl MK, Schaefer KM, et al. Environmental context explains Lévy and Brownian movement patterns of marine predators. Nature. 2010;465:1066–9. doi: 10.1038/nature09116. [DOI] [PubMed] [Google Scholar]
- 39.Harris TH, Banigan EJ, Christian DA, Konradt C, Tait Wojno ED, Norose K, et al. Generalized Lévy walks and the role of chemokines in migration of effector CD8+ T cells. Nature. 2012;486:545–8. doi: 10.1038/nature11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang DM, Manji HK. In search of the Holy Grail for the treatment of neurodegenerative disorders: has a simple cation been overlooked? Biol Psychiatry. 2007;62:4–6. doi: 10.1016/j.biopsych.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang HC, Klein PS. Multiple roles for glycogen synthase kinase-3 as a drug target in Alzheimer’s disease. Curr Drug Targets. 2006;7:1389–97. doi: 10.2174/1389450110607011389. [DOI] [PubMed] [Google Scholar]
- 42.Anholt RR, Mackay TF. Quantitative genetic analyses of complex behaviours in Drosophila. Nat Rev Genet. 2004;5:838–49. doi: 10.1038/nrg1472. [DOI] [PubMed] [Google Scholar]
- 43.Carter BC, Shubeita GT, Gross SP. Tracking single particles: a user-friendly quantitative evaluation. Phys Biol. 2005;2:60–72. doi: 10.1088/1478-3967/2/1/008. [DOI] [PubMed] [Google Scholar]


