Abstract
Background
Toxin-antitoxin (TA) systems are commonly found in bacteria and Archaea, and it is the most common mechanism involved in bacterial programmed cell death or apoptosis. Recently, MazF, the toxin component of the toxin-antitoxin module, has been categorized as an endoribonuclease, or it may have a function similar to that of a RNA interference enzyme.
Results
In this paper, with comparative data and phylogenetic analyses, we are able to identify several potential MazF-conserved motifs in limited subsets of foodborne pathogens and probiotic strains and further provide a molecular basis for the development of engineered/synthetic probiotic strains for the mitigation of foodborne illnesses. Our findings also show that some probiotic strains, as fit as many bacterial foodborne pathogens, can be genetically categorized into three major groups based on phylogenetic analysis of MazF. In each group, potential functional motifs are conserved in phylogenetically distant species, including foodborne pathogens and probiotic strains.
Conclusion
These data provide important knowledge for the identification and computational prediction of functional motifs related to programmed cell death. Potential implications of these findings include the use of engineered probiotic interventions in food or use of a natural probiotic cocktail with specificity for controlling targeted foodborne pathogens.
Keywords: toxin-antitoxin module, probiotic cocktail, engineered probiotics, foodborne pathogens
Background
Foodborne illnesses continue to be an important public health concern in developing, as well as in developed countries, thus prevention of foodborne illness outbreaks through effective and novel interventions should be given priority. The U.S. Public Health Service has identified ten important foodborne pathogens causing human illnesses, including pathogenic strains of Escherichia coli, Salmonella, Listeria, Clostridium botulinum, Shigella, and Campylobacter, which are associated with more than 250 known foodborne diseases (http://www3.niaid.nih.gov/topics/foodborne/default.htm).
In addition, according to the World Health Organization (WHO), antibiotic overuse in food animal production is a major contributor to the emergence of antibiotic resistant foodborne pathogens [1]. The use of antibiotics in food animals for growth promotion and treatment disrupts the normal beneficial commensal bacterial microflora in the animal intestinal tract [2-6]. Recently, the human and chicken gut microbiome projects [7-12] have shed new light on the existence of a bacterial ‘phylogenetic core’ [13] consisting of a wide diversity of gastrointestinal bacteria by using new technologies such as next generation sequencing, 16S rRNA screens, metagenomics, and metaproteomics. Healthful, commensal bacteria found in the GI tract might be key members of known or potential probiotic strains revealed in this ‘phylogenetic core’, which may include Bacillus clausii, Bacillus pumilus, Lactobacillus acidophilus, Lactobacillus reuteri, Lactobacillus rhamnosus GG (ATCC 53103), Bifidobacterium infantis, Saccharomyces boulardii, Lactobacillus ruminis, Lactobacillus johnsonii str. NCC 533, and many others. These known probiotic strains have been used as dietary supplements, as treatments for illnesses caused by foodborne pathogens, and for disease prevention. Use of probiotic strains not only reduce invasion by bacterial pathogens, but also restore and maintain an optimal balance of healthy commensal bacteria in the human gut via production of antimicrobials [14-23].
One of the major mechanisms recognized as being responsible for apoptosis, or programmed cell death, and production of toxic metabolites in bacteria is through the regulation of a wide variety of bacterial toxin-antitoxin modules [24-26], such as MazE/MazF, a chromosomal toxin-antitoxin module [27-30], plasmid-encoded parD [31-33], chpBIK[26,34], relBE[35,36], and the PhoQ-PhoP system [37-40]. MazEF is one of the most well-studied toxin–antitoxin (TA) systems and has been found on the chromosomes of many bacteria. The MazF protein has been recently categorized as an endoribonuclease [41,42] or as a type of RNA interference enzyme [43]. The link between this TA system and quorum sensing has also been explored recently through a small pentapeptide (NNWNN) called the Extracellular Death Factor (EDF) [44]. This small peptide motif (such as NNWNN) is known to be an extra-cellular death factor in E. coli and other bacterial species. The necessity of an “extracellular death factor” (EDF) or “cell death factor” (CDF) via MazEF-mediated cell death is a population phenomenon requiring the activation of quorum-sensing (cell-to-cell signaling) peptides in bacteria. High cell density was found to be associated with elevated concentrations of EDF, and the presence of EDF resulted in MazF-induced cell death [44]. From a food safety and public health perspective, use of EDF or a similar strategy may be used in place of antibiotics, resulting in less usage of antibiotics. We also noticed in one very interesting study that the induction of toxin MazF and the use of antibiotic share a similar mechanism by inhibiting the transcription and/or translation of the MazE antitoxin [45].
It has been theorized that there is “one toxin for one antitoxin” and interestingly MazF, in some bacteria, exhibits a selective inhibition of ribosomes and mRNAs [43,46]. Numerous strains of probiotic bacteria, such as Lactobacillus spp., have been reported to produce antimicrobial agents [47], such as bacteriocins, that inhibit or kill closely-related species, or even different strains of the same species through the inhibition of transcription and translation by receptor binding. The antimicrobial activities of bacteriocins are due to a heterologous subgroup of ribosomally synthesized cationic peptides [48]. Nisin, a polycyclic antibacterial peptide 34 amino acid residues long, is one of the most studied bacteriocins and is produced by many strains of Lactococcus lactis. It has been approved by the FDA for use as a food preservative, and certain probiotic Lactobacillus strains have been thoroughly studied and evaluated in vitro and in vivo. For example, L. reuteri controls diarrhea in children and suppresses the growth and pathogenicity of harmful foodborne pathogens such as Salmonella, E. coli, Staphylococcus, and Listeria[49,50]. L. casei GG has been used to treat Clostridium difficile infections and to reduce intestinal permeability [51-54]. L. reuteri is known to produce a broad-spectrum antimicrobial agent, reuterin, composed of the natural metabolic compound 3-hydroxypropionaldehyde, which has been used on the surface of sausages to inhibit growth of harmful bacteria and fungi [15,16]. However, the molecular mechanisms underlying the effectiveness of individual probiotic strains have not been systematically studied and characterized. Several potential mechanisms of action, including their ability to generate diverse natural toxic metabolites, lactic acid, and other organic acids, enzymes, vitamins, and hydrogen peroxide, as well as antimicrobial peptides such as nisin, have been well-described [20].
The work reported herein explores the experimental antimicrobial possibilities and/or procedures for (1) expression of an engineered, stress-induced recombinant secreted fusion gene encoded by MazF and a small extracellular cell death factor (CDF) on the surface or extracellular space of recombinant probiotic bacteria or (2) for potential application of a cocktail of natural probiotic strains via experimental in vitro selection to control foodborne pathogens for improving the safety and quality of foods, as well as improving human health. The use of engineered probiotic strains or the natural probiotic cocktail consisting of mixed probiotic strain populations for targeting foodborne pathogens will potentially be able to selectively inactivate these pathogens, even in complex food matrices. Through gene/motif reshuffling [55,56] and computational molecular modeling, this engineered and secreted bacterial fusion protein, MazF-CDF, which contains an enterokinase (EK) cleavage site [57-60] for releasing active CDF and MazF after protein secretion, should have a narrow range of applicability, limited to inactivating only specific foodborne pathogens such as E. coli O157, Salmonella, Listeria, Campylobacter, and potentially other human pathogens. This pathogen specificity is due to the fact that the genetically-engineered MazF-CDF fusion protein could be modified by the inclusion of specific genomic DNA sequences from various commensal, as well as pathogenic bacterial strains, identified through DNA/protein structure and functional comparisons. Additionally, this engineered antimicrobial polypeptide and MazF will be non-toxic to beneficial (healthful/probiotic) bacteria, as well as to its host that expresses the protein/peptide. Moreover, these engineered and secreted CDFs and MazF proteins/peptides can easily pass through infectious foodborne pathogen cell barriers mediating cell death, and thus could potentially reduce the use of deleterious compounds such as antibiotics or other harmful chemicals in the feed and food industry.
Results and discussion
General genetic analysis of probiotics and foodborne pathogen genomes
The genomic information from selected probiotic strains and foodborne pathogens is shown in Table 1. It revealed little diversity in genomic GC-content in the Bifidobacterium genus, while showing an astonishing diversity in the GC-content among Lactobacillus species, ranging from 33 to 51.5%. It has been demonstrated that genomic GC-content is correlated with a number of factors [61], including genome size [62] from species such as Lactobacillus, which ranges from 1.8 to 3.4 Mb in length. This demonstrates that the GC-content and genome size of Lactobacillus genomes may have implications related to the biological complexity and adaptation of this genus, and could be due to the rate of recombination that has been extensively studied in the E. coli genome [63]. Knowledge of genetic diversity is fundamental to development of synthetic probiotic genomes and/or nucleic acid sequence reshuffling strategies. In this paper, we demonstrate that the MazF protein is a suitable candidate for the determination of genetic relationships within sets of MazF proteins in combination with with functional motif analysis.
Table 1.
Genomic information of selected microorganisms considered as potential probiotic strains and some major food-borne pathogens used in this study
| Organism | Strain | Pathotype /or others | GenBank Accession /Assembly | GC (%) | Genome Size (~ Mb) |
|---|---|---|---|---|---|
|
Campylobacter upsaliensis |
JV21 |
pathogenic |
ASM18534v1 |
NA |
1.6 |
|
Lactobacillus coleohominis |
101-4-CHN |
probiotic |
ASM16193v1 |
41.3 |
1.7 |
|
Campylobacter jejuni |
RM1221 |
pathogenic |
NC_003912.7 |
30.3 |
1.8 |
|
Lactobacillus johnsonii |
FI9785 |
probiotic |
NC_013504.1 |
34.4 |
1.8 |
|
Pediococcus pentosaceus |
ATCC 25745 |
probiotic |
CP000422.1 |
37.4 |
1.8 |
|
Streptococcus thermophilus LMG |
LMG 18311 |
probiotic |
NC_006448.1 |
39.1 |
1.8 |
|
Lactobacillus gasseri |
ATCC 33323 |
probiotic |
NC_008530.1 |
35.3 |
1.9 |
|
Lactobacillus sakei subsp. sakei |
23K |
probiotic |
NC_007576.1 |
41.3 |
1.9 |
|
Lactobacillus delbrueckii subsp. bulgaricus |
ATCC 11842 |
probiotic |
NC_008054.1 |
49.7 |
1.9 |
|
Lactobacillus delbrueckii subsp. bulgaricus |
ATCC BAA-365 |
probiotic |
NC_008529.1 |
49.7 |
1.9 |
|
Leuconostoc citreum |
KM20 |
probiotic |
ASM2640v1 |
38.9 |
1.9 |
|
Bifidobacterium animalis subsp. Lactis |
Bl-04 |
probiotic |
NC_012814.1 |
60.5 |
1.9 |
|
Bifidobacterium animalis subsp. Lactis |
DSM 10140 |
probiotic |
NC_012815.1 |
60.5 |
1.9 |
|
Bifidobacterium animalis subsp. Lactis |
AD011 |
probiotic |
NC_011835.1 |
60.5 |
1.9 |
|
Lactobacillus amylovorus |
GRL1118 |
probiotics |
ASM19411v1 |
38.0 |
2.0 |
|
Lactobacillus johnsonii |
NCC 533 |
probiotic |
NC_005362.1 |
34.6 |
2.0 |
|
Lactobacillus acidophilus |
NCFM |
probiotic |
NC_006814.3 |
34.7 |
2.0 |
|
Lactobacillus reuteri |
DSM 20016 |
probiotic |
NC_009513.1 |
38.9 |
2.0 |
|
Lactobacillus reuteri |
JCM 1112 |
probiotic |
NC_010609.1 |
38.9 |
2.0 |
|
Listeria monocytogenes |
FSL J1-208 |
pathogenic |
ASM16843v1 |
37.7 |
2.0 |
|
Lactobacillus salivarius |
UCC118 |
probiotic |
NC_007929.1 |
33 |
2.1 |
|
Lactobacillus helveticus |
DPC 4571 |
probiotic |
NC_010080.1 |
37.1 |
2.1 |
|
Leuconostoc mesenteroides subsp. mesenteroides |
ATCC 8293 |
probiotic |
NC_008531.1 |
37.7 |
2.1 |
|
Campylobacter concisus |
13826 |
pathogenic |
NC_009802.1 |
39.3 |
2.1 |
|
Lactobacillus fermentum |
IFO 3956 |
probiotic |
NC_010610.1 |
51.5 |
2.1 |
|
Bifidobacterium adolescentis |
ATCC 15703 |
probiotic |
NC_008618.1 |
59.2 |
2.1 |
|
Bifidobacterium bifidum |
PRL2010 |
probiotic |
NC_014638.1 |
Na |
2.2 |
|
Bifidobacterium bifidum |
S17 |
probiotic |
NC_014616.1 |
Na |
2.2 |
|
Lactobacillus amylovorus |
GRL 1112 |
probiotic |
NC_014724.1 |
Na |
2.2 |
|
Lactobacillus brevis |
ATCC 367 |
probiotic |
NC_008497.1 |
46.1 |
2.3 |
|
Lactobacillus crispatus |
CTV-05 |
probiotic |
ASM16588v1 |
37.1 |
2.3 |
|
Bifidobacterium longum |
NCC2705 |
probiotic |
NC_004307.2 |
60.1 |
2.3 |
|
Bifidobacterium longum subsp. longum |
BBMN68 |
probiotic |
NC_014656.1 |
Na |
2.3 |
|
Bifidobacterium longum subsp. infantis |
157F |
probiotic |
NC_015052.1 |
Na |
2.4 |
|
Bifidobacterium longum subsp. longum |
JCM 1217 |
probiotic |
NC_015067.1 |
Na |
2.4 |
|
Lactococcus lactis subsp. lactis |
Il1403 |
probiotic |
NC_002662.1 |
35.3 |
2.4 |
|
Bifidobacterium longum |
DJO10A |
probiotic |
NC_010816.1 |
60.2 |
2.4 |
|
Bifidobacterium longum subsp. longum |
JDM301 |
probiotic |
NC_014169.1 |
Na |
2.5 |
|
Lactococcus lactis subsp. cremoris |
MG1363 |
probiotic |
NC_009004.1 |
35.7 |
2.5 |
|
Listeria grayi |
DSM 20601 |
pathogenic |
ASM14899v1* |
41.6 |
2.6 |
|
Propionibacterium freudenreichii subsp. shermanii |
CIRM-BIA1 |
probiotic |
NC_014215.1 |
Na |
2.6 |
|
Lactococcus lactis subsp. lactis |
KF147 |
probiotic |
NC_013656.1 |
34.9 |
2.6 |
|
Lactococcus lactis subsp. cremoris |
SK11 |
probiotic |
NC_008527.1 |
35.8 |
2.6 |
|
Bifidobacterium dentium |
Bd1 |
probiotic |
NC_013714.1 |
58.5 |
2.6 |
|
Bifidobacterium longum subsp. infantis |
ATCC 15697 |
probiotic |
NC_011593.1 |
59.9 |
2.8 |
|
Listeria monocytogenes |
EGD-e |
pathogenic |
NC_003210.1 |
38 |
2.9 |
|
Lactobacillus casei str. |
Zhang |
probiotic |
NC_014334.1 |
40.1 |
2.9 |
|
Lactobacillus casei |
ATCC 334 |
probiotic |
NC_008526.1 |
46.6 |
2.9 |
|
Lactobacillus paracasei subsp. paracasei |
ATCC 25302 |
probiotic |
ASM15949v1 |
46.5 |
2.9 |
|
Lactobacillus rhamnosus |
GG |
probiotic |
NC_013198.1 |
46.7 |
3.0 |
|
Lactobacillus rhamnosus |
Lc 705 |
probiotic |
NC_013199.1 |
46.7 |
3.0 |
|
Lactobacillus casei |
BL23 |
probiotic |
NC_010999.1 |
46.3 |
3.1 |
|
Lactobacillus plantarum |
JDM1 |
probiotic |
NC_012984.1 |
44.7 |
3.2 |
|
Enterococcus faecalis |
V583 |
probiotic |
NC_004668.1 |
37.4 |
3.3 |
|
Lactobacillus plantarum |
WCFS1 |
probiotic |
NC_004567.1 |
44.4 |
3.3 |
|
Lactobacillus plantarum subsp. plantarum |
ST-III |
probiotic |
NC_014554.1 |
Na |
3.4 |
|
Bacillus selenitireducens |
MLS10 |
Bio agent |
ASM9308v1 |
48.7 |
3.6 |
|
Bacillus pumilus |
SAFR-032 |
probiotic |
NC_009848.1 |
41.3 |
3.7 |
|
Coprobacillus sp. |
29_1 |
Non-pathogenic |
Coprobacillus_sp_29_1_V1 |
NA |
3.8 |
|
Vibrio cholerae |
M66-2 |
pathogenic |
NC_012578.1 |
47.6 |
3.9 |
|
Bacillus halodurans |
C-125 |
Non-pathogenic |
ASM1114v1 |
43.7 |
4.2 |
|
Bacillus amyloliquefaciens |
Y2 |
probiotic |
ASM28439v1 |
45.9 |
4.2 |
|
Clostridium difficile |
630 |
pathogenic |
NC_009089.1 |
29.1 |
4.3 |
|
Bacillus clausii |
KSM-K16 |
probiotic |
NC_006582.1 |
44.8 |
4.3 |
|
Shigella dysenteriae |
Sd197 |
pathogenic |
ASM1200v1] |
50.9 |
4.6 |
|
Shigella flexneri |
8401 |
pathogenic |
NC_008258.1 |
50.9 |
4.6 |
|
Salmonella enterica subsp. arizonae serovar |
RSK2980 |
pathogenic |
NC_010067.1 |
51.4 |
4.6 |
|
Enterobacter sp. |
638 |
pathogenic |
NC_009436.1 |
52.9 |
4.7 |
|
Vibrio vulnificus |
CMCP6 |
pathogenic |
NC_004459.2 |
46.7 |
5.1 |
|
Escherichia coli |
E24377A |
pathogenic |
NC_009801.1 |
50.6 |
5.2 |
|
Citrobacter rodentium |
ICC168 |
pathogenic |
NC_013716.1 |
54.6 |
5.4 |
|
Klebsiella variicola |
At-22 |
pathogenic |
NC_013850.1 |
57.6 |
5.5 |
|
Bacillus megaterium |
QM B1551 |
probiotic |
ASM2582v1 |
38.0 |
5.5 |
|
Escherichia coli O157:H7 |
Sakai |
pathogenic |
NC_002695.1 |
50.5 |
5.6 |
|
Bacillus thuringiensis serovar israelensis |
ATCC 35646 |
Bio agents |
ASM16769v1 |
35.0 |
5.9 |
| Lachnospiraceae bacterium | 3_1_57FAA_CT1 | Non-pathogenic | Lach_bact_3_1_57FAA_CT1_V1 | NA | 7.7 |
*: assembly accession.
Ubiquitous existence of MazE/toxin, MazF
MazEF is a toxin-antitoxin (TA) module widely distributed among many bacterial species, including both foodborne pathogens and probiotic strains (Table 1, 2), such as Lactobacillus acidophilus, Lactobacillus reuteri, Lactobacillus rhamnosus GG (ATCC 53103), Escherichia coli, Selenomonas sputigena, Enterobacter spp., Campylobacter spp., Citrobacter spp., Thermoanaerobacter tengcongensis, Pelotomaculum thermopropionicum, Lactobacillus casei, Lactobacillus crispatus, Lactobacillus buchneri, Bifidobacterium longum subsp. infantis, Clostridium botulinum, Clostridium difficile, Vibrio spp., Listeria spp., Bacillus spp., Klebsiella variicola, and Salmonella enterica. Recent literature and computational analyses have shown the presence of many different types of TA modules in various localizations, e.g. some TA modules have been found within prophages, prophage-like elements, and other mobile genetic elements, such as plasmids [31-33]. Due to the existence of varying types of toxin-antitoxin modules and possible gene duplication events, in this paper we present the above one-to-one best matches of chromosomal-encoded mazF orthologs and homologs among 75 publically available genome sequences of foodborne pathogens and probiotic strains (Table 1), and the publically-available databases such as NCBI and the Uniprot database (http://www.uniprot.org/) in Table 2.
Table 2.
The genetic characterization of the transcriptional modulator MazF, a chromosomal cell death factor from potential probiotic strains and some major food-borne pathogens used in this study
| Organism | Type of organisms | Strains | Protein accession (GenBank) | Gene annotation |
|---|---|---|---|---|
|
E. coli |
Pathogen |
EDL933 |
NP_289336.1 |
toxin ChpA |
| |
Pathogen |
Sakai |
NP_311669.1 |
|
| |
Antibiotic resistance |
B185 |
ZP_06660634.1* |
|
|
Enterobacter sp. |
pathogen |
638 |
ABP60743.1* |
transcriptional modulator of MazE/toxin, MazF |
|
Shigella dysenteriae |
pathogen |
Sd197 |
YP_405833.1* |
toxin ChpB |
|
Citrobacter rodentium |
pathogen |
ICC168 |
YP_003366767.1* |
Toxin component of the ChpB-ChpS toxin-antitoxin system |
|
Vibrio vulnificus |
pathogen |
CECT4999=R99 |
YP_001393091.1* |
growth inhibitor |
|
Listeria welshimeri serovar 6b str. |
pathogen |
SLCC5334 |
YP_849071.1 |
PemK family transcriptional regulator |
| |
|
FSL J1-208 |
ZP_05296226.1* |
PemK family transcriptional regulator |
|
Listeria monocytogenes |
|
SLCC3954 |
YP_003464028.1 |
transcriptional regulator, PemK family |
| |
|
FSL S4-120 |
ZP_07870171.1 |
toxin-antitoxin system, antitoxin component, MazF family |
|
Listeria seeligeri serovar 1/2b str. |
|
|
|
|
|
Listeria marthii |
|
|
|
|
|
Listeria grayi |
pathogen |
DSM 20601 |
ZP_07054243.1* |
MazF family toxin-antitoxin system protein |
|
Clostridium difficile |
pathogen |
630 |
YP_001089981.1* |
putative regulator of cell growth |
| |
|
QCD-66c26 |
ZP_05273557.1 |
putative regulator of cell growth |
| |
|
CIP 107932 |
ZP_05323891.1 |
putative regulator of cell growth |
|
Klebsiella variicola |
pathogen |
At-22 |
YP_003438577.1* |
transcriptional modulator of MazE/toxin, MazF pemK; protein PemK |
| |
|
1_1_55 |
ZP_06548086.1 |
|
|
Salmonella enterica subsp. arizonae serovar |
pathogen |
RSK2980 |
YP_001573441.1* |
hypothetical protein SARI_04525 |
|
Campylobacter showae |
pathogen |
RM3277 |
ZP_05364729.1 |
addiction module antitoxin, RelB/DinJ family |
|
Campylobacter concisus |
pathogen |
13826 |
YP_001466677.1 |
addiction module antitoxin |
|
Campylobacter jejuni subsp. |
pathogen |
IA3902 |
ADC28395.1 |
prevent-host-death family protein |
|
Campylobacter upsaliensis |
pathogen |
JV21 |
ZP_07894578.1* |
ChpA/MazF transcriptional modulator |
|
Shigella flexneri 2a str. |
pathogen |
301 |
NP_709188.1* |
PemK protein |
|
Shigella flexneri 5 str. |
|
8401 |
YP_690766.1 |
growth inhibitor, PemK-like, autoregulated |
|
Cronobacter sakazakii |
pathogen |
ATCC BAA-894 |
YP_001436394.1 |
bifunctional antitoxin/transcriptional repressor RelB |
|
Lactobacillus coleohominis |
probiotic |
101-4-CHN |
ZP_05553821.1* |
regulatory protein |
|
Bacillus megaterium |
probiotic |
QM B1551 |
YP_003560763.1* |
endoribonuclease EndoA |
| |
|
DSM 319 |
YP_003595503.1 |
|
|
Bacillus thuringiensis serovar israelensis |
probiotic |
ATCC 35646 |
ZP_00740711.1* |
MazF protein |
| |
KBAB4 |
YP_001643128.1 |
MazF protein |
|
|
Bacillus weihenstephanensis |
|
AH1134 |
ZP_03233130.1 |
PemK family |
|
Bacillus cereus |
|
|
|
|
|
Coprobacillus sp. |
probiotic |
29_1 |
ZP_08009764.1* |
PemK family transcriptional regulator |
|
Enterococcus faecalis |
probiotic |
V583 |
NP_814592.1 |
PemK family transcriptional regulator |
| |
|
TX0102 |
EFQ11602.1* |
|
| |
|
|
|
Toxin-antitoxin system, toxin compoment, MazF |
| |
|
TX2134 |
ZP_07557645.1* |
|
| |
|
|
|
Toxin-antitoxin system, toxin compoment, MazF |
| |
|
HH22 |
ZP_03985009.1 |
|
| |
|
|
|
PemK family transcriptional regulator |
|
Lactobacillus plantarum |
probiotic |
WCFS1 |
NP_786238.1* |
cell growth regulatory protein |
|
Lactobacillus rhamnosus |
probiotic |
LMS2-1 |
ZP_04441477.1* |
cell growth regulatory protein |
| |
|
Lc 705 |
YP_003175134.1 |
transcriptional modulator of MazE/toxin, MazF |
|
Lactobacillus johnsonii |
probiotic |
ATCC 33200 |
ZP_04007131.1* |
PemK family growth inhibitor |
|
Lactobacillus crispatus |
probiotics |
CTV-05 |
ZP_07789240.1* |
ppGpp-regulated growth inhibitor |
|
Lactobacillus gasseri |
probiotic |
ATCC 33323 |
YP_815460.1* |
toxin-antitoxin system, toxin component, MazF family |
| |
|
224-1 |
ZP_06262236.1 |
|
| |
|
MV-22 |
ZP_07711551.1 |
|
|
Lactobacillus casei |
probiotic |
BL23 |
YP_001986080.1 |
Cell growth regulatory protein |
|
Lactobacillus amylovorus |
probiotic |
GRL1118 |
YP_005854914.1* |
transcriptional modulator of MazE/toxin MazF |
|
Vibrio cholerae |
probiotic |
1587 |
ZP_01950611.1* |
transcriptional modulator of MazE/toxin, MazF |
| |
|
NCTC 8457 |
ZP_01969676.1 |
|
|
Pediococcus pentosaceus |
probiotic |
ATCC 25745 |
YP_805020.1* |
toxin-antitoxin addiction module toxin component MazF (an endoRNAse) |
|
Bacillus amyloliquefaciens |
probiotic |
Y2 |
YP_006327269.1* |
Endoribonuclease |
|
Bacillus pumilus |
probiotic |
SAFR-032 |
YP_001485694.1* |
PemK family growth inhibitor endoribonuclease EndoA |
| |
|
ATCC 7061 |
ZP_03054587.1 |
|
|
Lactobacillus sakei subsp. sakai |
probiotic |
Sakei 23K |
YP_396224.1 |
DNA-binding protein PemK family |
|
Leuconostoc mesenteroides subsp. mesenteroides |
probiotic |
ATCC 8293 |
YP_819271.1* |
toxin-antitoxin addiction module toxin component MazF |
| |
|
ATCC 19254 |
ZP_03913232.1 |
PemK family growth inhibitor |
|
Leuconostoc mesenteroides subsp. cremoris |
|
|
|
|
|
Enterococcus faecalis |
probiotic |
TX0102 |
EFQ11602.1* |
toxin-antitoxin system, toxin component, MazF family |
| |
|
TX0031 |
EFT96737.1 |
|
|
Bifidobacterium animalis subsp. lactis |
probiotic |
HN019 |
ZP_02964075.1* |
transcriptional modulator of MazE/toxin, MazF |
|
Bifidobacterium bifidum |
probiotic |
NCIMB 41171 |
ZP_03645723.1 |
RelB antitoxin |
|
Bifidobacterium longum subsp. longum |
probiotic |
JDM301 |
YP_003660624.1* |
RelB antitoxin |
| |
|
DSM 20213 |
ZP_06596513.1 |
toxin-antitoxin system protein |
|
Bifidobacterium breve |
|
|
|
|
|
Bifidobacterium bifidum |
probiotic |
S17 |
YP_003938101.1 |
hypothetical protein with RelB antitoxin domain |
|
Lactobacillus casei |
Probiotic |
ATCC 334 |
YP_807723.1 |
toxin-antitoxin addiction module toxin component MazF (an endoRNAse) |
|
Lactobacillus casei |
Probiotic |
BL23 |
YP_001988635.1 |
Growth inhibitor |
|
Lactobacillus paracasei subsp. paracasei |
Probiotic |
ATCC 25302 |
ZP_03963082.1* |
PemK family transcriptional regulator |
|
Lactobacillus casei str. |
probiotic |
Zhang |
YP_003789562.1 |
toxin-antitoxin addiction module toxin component MazF |
|
Lachnospiraceae bacterium |
probiotic |
3_1_57FAA_CT1 |
ZP_08609193.1* |
hypothetical protein HMPREF0994_05199 |
|
Leuconostoc citreum |
Probiotic |
KM20 |
YP_001727503.1* |
growth inhibitor |
|
Lactobacillus reuteri |
probiotic |
DSM 20016 |
YP_001270863.1* |
transcriptional modulator of MazE/toxin, MazF |
| |
|
JCM 1112 |
YP_001841242.1 |
hypothetical protein LAR_0246 |
| |
|
MM2-3 |
ZP_03847756.1 |
PemK family growth inhibitor |
|
Streptococcus sp. oral taxon 071 str. |
probiotic |
73H25AP |
ZP_07458552.1* |
ChpA/MazF transcriptional modulator |
|
Bacillus selenitireducens |
probiotic |
MLS10 |
YP_003700699.1* |
transcriptional modulator of MazE/toxin, MazF |
|
Bacillus halodurans |
probiotic |
C-125 |
NP_244588.1* |
ppGpp-regulated growth inhibitor (ChpA/MazF) |
|
Enterococcus faecium |
probiotic |
DO |
ZP_05714797.1* |
PemK family protein |
| |
|
TX0133a04 |
ZP_07845579.1 |
toxin component, MazF family |
| TX0133C | ZP_07850444.1 | toxin component, MazF family |
*: data are presented in Figure 1.
Phylogenetic analyses and cluster analysis of MazF/antitoxin protein, a growth inhibitor
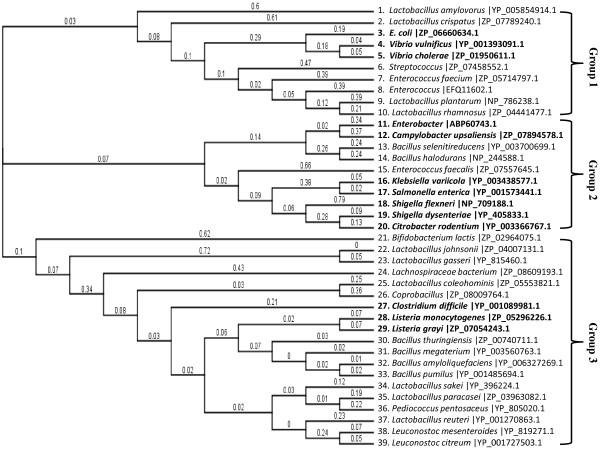
The phylogenetic tree in Figure 1 displays the phylogentic relationships of many well-recognized genera within Enterobacteriaceae, including several important foodborne pathogens such as pathogenic E. coli, Salmonella, Listeria, and Campylobacter, as well as some major probiotic strains. To build a phylogenetic tree from the data in Table 2, the amino acid sequences of all the MazF or growth inhibitor proteins were analyzed using the Geneious software package v5.5.7 with Neighbor-joining (NJ) method by applying ClustalW for sequence alignment (Figure 1). Three main clusters (viz., groups 1, 2, and 3) are given in Figure 1. At least two representatives of potential probiotic strains are listed for each group, depending on the complexity of groups. In group 1, the potential probiotic, non-pathogenic strains, such as Lactobacillus amylovorus, Lactobacillus crispatus, Streptococcus, Enterococcus faecium, Enterococcus, Lactobacillus plantarum, and Lactobacillus rhamnosus, are grouped with the foodborne pathogens E. coli, Vibrio vulnificus, and Vibrio cholerae. In group 2, the foodborne pathogens Enterobacter, Campylobacter upsaliensis, Klebsiella variicola, Salmonella enterica, Shigella flexneri, Shigella dysenteriae, and Citrobacter rodentium were shown to be genetically closer to Bacillus selenitireducens, Bacillus halodurans, and Enterococcus faecalis In group 3, some probiotic strains, such as the Bacillus, Lactobacillus, Leuconostoc, and the Bifidobacterium genera are categorized along with other major foodborne pathogens, such as Clostridium difficile, Listeria monocytogenes, Listeria grayi, and an emerging foodborne pathogen Pediococcus pentosaceus.
Figure 1.
Phylogenetic analyses of Enterobacteriaceae based on MazF (toxin), a growth inhibitor. The NCBI accession numbers are shown after each taxon name.
Phylogenetic identification of bacterial toxin MazF protein motifs and the relationship between gene structure and phylogenetic classification
In Figure 2, it is shown that the number of candidate motifs are slightly different in each group, but with a high degree of amino acid sequence variability within conserved “hot spots” between groups 1, 2, and 3. To determine which motifs are the best candidates for the engineered MazF construction (discussed in next section) in each individual group, recent studies [43] suggest that the N-terminal (the first 80 amino acids in Figure 2) of the MazF protein are the most suitable motifs. The other motifs in the C-terminal might be referred to as ‘incorrect’ motifs. There are three criteria for evaluating the remaining motifs as “incorrect and without biological significance”, and this is related to: the mean hydrophobicity, identity, and the gene structure. In Figure 2, in the analysis for the phylogenetic identification of bacterial Toxin MazF protein motifs, the mean hydrophobicity sequence and identity were computed and compared for each sequence; it is interesting to find that all the compared MazF proteins have a higher degree of mean hydrophobicity, identity, and more conserved gene structure among several conserved amino acid regions, particular in the N-terminal. The selection of potential functional MazF motifs is discussed in the next section. The functional importance of the mean hydrophobicity has also been discussed to involve mRNA and protein degradation and slower translation of mRNA for disordered proteins (http://employees.csbsju.edu/hjakubowski/classes/ch331/protstructure/olprotfold.html).
Figure 2.
ClustalW output (default settings) for the aligned amino acid sequences of MazF. The pdf images of the alignments were generated using Geneious v5.5.7. Conserved amino acids are highlighted in colors; the numbers (1 to 39) in this Figure correspond to the taxon names in Figure 1 .
Bacterial probiotic cocktail strains
It is important to note that the benefits of probiotics are strain-specific [64,65]. A commercial product, VSL#3, developed by Sigma-Tau Pharmaceuticals, Inc., provides a mixture of probiotic bacteria (B. breve, B. infantis, B. longum, L. acidophilus, L. bulgaricus, L. casei, L. plantarum, and Streptococcus thermophilus) to help protect GI tract integrity [66]. In this study, the bacterial probiotic cocktail strains we propose would be comprised of all representatives of groups 1, 2, and 3 shown in Figure 1. The principal basis behind the composition of these probiotic cocktail strains is the assumption that a combination of organisms might be more effective than the application of a single strain, which potentially could suppress many foodborne pathogens, such as the E. coli and vibros in Group 1, several foodborne pathogens, such as Enterobacter, Klebsiella variicola, Salmonella enterica, Shigella flexneri, Shigella dysenteriae, Citrobacter rodentium, and Campylobacter upsaliensis, (one of the most common Campylobacter strain found in people with diarrhea in Group 2), Clostridium difficile, Listeria monocytogenes, and Listeria grayi, in Group 3 of Figure 1. Table 1 shows an astonishing diversity in the genomic GC-content and genome size among Lactobacillus species and their diverse distribution in all groups within Figure 1, which indicates a potential to further identify other closely-related Lactobacillus species (not listed in Table 1) into the three previously-described groups. The hypothesis underlying our approach is that probiotic strains found within the same group with foodborne pathogens will have a reasonable degree of genetic and molecular phylogenetic compatibility and could bridge a relationship similar to a “symbiosis” of entities, including exchanging toxin/antitoxin molecules among the probiotic and pathogenic strains. Lactobacillus species are known to produce antimicrobial substances, including bacteriocins, lactic acid, and hydrogen peroxide. The MazEF toxin component may provide a basis for bacterial growth inhibition within the same group (Figure 1 and 2). Therefore, this toxin-antitoxin module may have great potential to inhibit the growth of potentially-pathogenic bacteria through a possible competitive exclusion due to selective inhibition [46]. Figures 1 and 2 list all possible cocktails of these probiotic strains. In reality, a foodborne outbreak is more likely to be associated with one particular foodborne pathogen in particular foods. For example, there is a low incidence of Campylobacter in ground beef and pork, while Campylobacter is the major foodborne pathogens associated with poultry, therefore, a single food-borne pathogen with the application of mixed probiotics will be considered initially.
Molecular recombination techniques: construction of genetically engineered synthetic probiotic strains
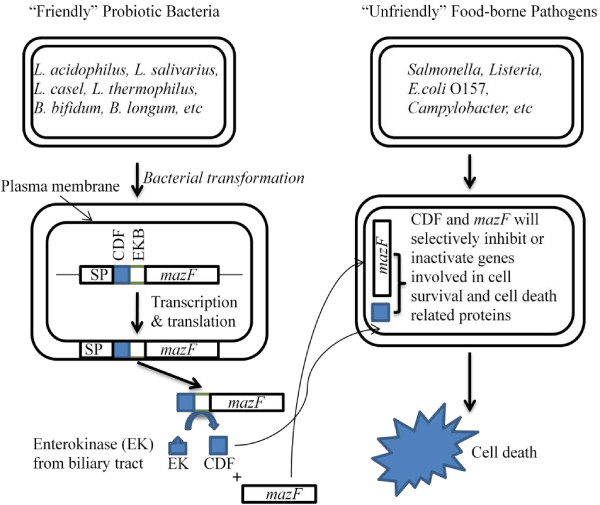
Figure 3 details an engineered probiotic strain bearing a recombinant plasmid containing a stress-induced promoter, followed by an in-frame gene fusion accomplished by fusing an appropriate signal peptide, a functional cell death peptide/factor (CDF), an enterokinase (EK) binding site, and a genetically-modified engineered MazF gene, which will be constructed and transformed into the probiotic bacteria. In this recombinant probiotic strain, environmental stress, relevant to food environments will trigger gene expression of this fusion protein under an environmental stress-inducible promoter. The signal peptide directs the encoded fusion protein to the extracellular space of the engineered probiotic strains. The signal peptide depleted fusion proteins will be cleaved by a biliary tract enterokinase directly into the digestive system to release the functionally-active CDF and MazF. This event will occur based on the ability of probiotic strains to form stable, non-infectious biofilm-type aggregates, which may attach with other bacteria, including foodborne pathogens, in the urogenital and intestinal tracts [22]. These active species-specific CDF and MazF proteins will bind and pass through “unfriendly” bacterial cell surface barriers into the cytoplasm. These processed CDFs and MazF proteins will selectively inactivate and/or inhibit proteins involved in cell survival and induce the synthesis of more cell death-related proteins with activity against foodborne pathogens, eventually controlling, inhibiting, or inactivating “unfriendly” cells. Although antitoxin MazE could reverse the bacteriocidal effect of the overexpressed MazF, MazE cannot impede the downstream cascade already initiated by MazF at early stages of the MazF-mediated cascade [42].
Figure 3.
A Schematic representation of the engineered, secreted CDF and MazF-mediated cell death during food processing and storage. SP: promoter, CDF: cell death factor/peptide, EKB: enterokinase binding site.
The overall hypothesis for this experiment is graphically presented in Figure 3. The engineered MazF gene sequence will be designed based on the genomic sequences of all publically-available foodborne pathogenic strains by using reasoned random gene biosynthesis and/or genuine gene reshuffling to rapidly combine functions and properties of parental genes for the development of improved gene specificity and generality, molecular modeling (transcription factor binding site identification, etc.), and systems biology technologies/tools.
Conclusions
Survival of foodborne pathogens in cultures or in animal GI tracts may be very genus- or species-specific. Data presented in this paper can be explored to develop effective intervention strategies applied directly during food processing and preparation, as well as in the animal feed supply, which may lead to an overall reduction in use of antibiotic growth promoters (AGP) throughout the world. Recent research in molecular biology and genomics has provided potential applications of probiotic strains as dietary supplements, which could replace AGP in animal diets or as biotherapeutic agents in cases of antibiotic-associated diarrhea in travelers and in childhood diarrhea and other bacterial gastrointestinal illnesses. Experiments relating to this potential probiotic application may reveal a further greater range of potential benefits. For many of these potential benefits, current research is limited, and only preliminary results are available. All effects can only be attributed to the individual strain(s) tested. Testing of a specific supplement may not be extrapolated to benefits from any other strain of the same species, and testing results do not imply that comparable benefits will be imparted from other LAB (or other probiotics). In this study, we have computationally explored several potential intervention strategies to control foodborne pathogens, either by using a cocktail of probiotic strains or an engineered probiotic strain.
The inhibition of pathogens by probiotic strains is mainly due to the production of antibacterial peptides [67], the release of short-chain fatty acids, or reduction of the pH within the lumen [68,69] by the production of organic acids or by decreasing pathogen adherence to intestinal epithelial cells [70]. Therefore, the benefits of probiotics could be very strain- or species-specific, and probiotic strains may rely on different mechanisms to suppress growth, attachment, or other metabolic processes, inherent to pathogenic bacteria. Moreover, the optimal effects of probiotic strains may involve the simultaneous use of more than one strain. Our contention in this paper is not experimental proof but, rather, a clear scientifically-backed hypothesis in the form of a detailed accompanying method that multi-probiotic strain composites with diverse genetic backgrounds may complement [71] one another as vectors of competitive exclusion and, therefore, could maximize the potential to inhibit an array of common foodborne pathogens [72] in the gastrointestinal tract of humans or livestock, as well as in foods and animal feed.
The use of probiotic bacteria with the ability to produce CDFs and engineered MazF to selectively inactivate pathogens is a novel approach to controlling pathogens in foods, and possibly treating human infections. A number of studies suggest that this project could have practical significance and be a potentially new approach for the development of novel and cost-effective food safety intervention technologies for the control of foodborne pathogens and improving public health [73-75].
The approaches described above represent a first attempt to describe a systematic approach or method to test the hypothesis that “friendly” bacteria can be used to inactivate or inhibit pathogens in food based on expression of MazF. There are many specific cell death factors that may be associated with bacterial programmed cell death and multi-cellular behavior mechanisms in foodborne pathogens. Through computational modeling, remodeling, genetic recombination, or further gene reshuffling, and exploring experimental approaches, it may be possible to evaluate and elucidate more effective CDFs and MazF to be used for controlling foodborne pathogens, which will ultimately result in a reduction in the use of antimicrobial compounds in humans and animals, as well as during food processing and storage.
Materials and methods
Genomic sequences
All of the genome and gene sequences examined in this study (Table 1 &2) are available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/GenbankOverview.html).
Identification and analysis of bacterial toxin-antitoxin modules, MazE/MazF
The prediction accuracy of the best chromosomal-encoded MazF orthologs among relatively distinct genome strains is critical for the performance of molecular phylogenetic analysis. We used a sequential BLAST workflow based on pairwise comparison by applying either an E. coli programmed cell death toxin MazF (Genbank accession: ZP_06660634.1), an endoribonuclease MazF, a MazF protein from Vibrio cholerae (ZP_01950611.1), or a PemK family transcriptional regulator protein (ZP_05296226.1) from Listeria monocytogenes to perform a BLASTP homology search of these combined protein sequences with all 75 publically available foodborne pathogen and probiotic strain genomic sequences (presented in Table 1) publically available databases such as NCBI and Uniprot database (http://www.uniprot.org/). BLAST search results were used to list MazF or MazF-like proteins from 75 different strains being represented as 39 different species (Table 2).
Phylogenetic analysis and computational identification of phylogenetic motifs of the MazF protein
In order to identify potential functional motifs of MazF proteins, phylogentic analyses of MazF proteins were conducted by applying the embedded multiple sequence alignment ClustalW program in the Geneious software package v 5.5.7 [76,77] with the Neighbor Joining method. ClustalW output for the aligned amino acid sequences and the pdf images of the alignments were generated using the Geneious software package v 5.5.7.
Competing interests
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Authors’ contributions
XY conceived the study. XY and JH performed the bioinformatics study and XY wrote the paper. All authors read and approved the manuscript.
Contributor Information
Xianghe Yan, Email: xianghe.yan@ars.usda.gov.
Joshua B Gurtler, Email: joshua.gurtler@ars.usda.gov.
Pina M Fratamico, Email: pina.fratamico@ars.usda.gov.
Jing Hu, Email: jing.hu@fandm.edu.
Vijay K Juneja, Email: vijay.juneja@ars.usda.gov.
Acknowledgements
We express our appreciation to Dr. James Smith for his critical review of the manuscript. The mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
References
- World Health Organization (WHO), Food and Agriculture Organization of the United Nations, and the World Organization for Animal Health. Expert workshop on non-human antimicrobial usage and antimicrobial resistance. Geneva; 2003. http://www.who.int/foodsafety/publications/micro/en/amr.pdf. [Google Scholar]
- CDC. Are antibacterial-containing products (soaps, household cleaners, etc.) better for preventing the spread of infection? Does their use add to the problem of resistance? Antibiotic Resistance Questions & Answers, Centers for Disease Control and Prevention, Atlanta; 2009. [Google Scholar]
- Ferber D. Antibiotic resistance. Livestock feed ban preserves drugs’ power. Science. 2002;295:27–28. doi: 10.1126/science.295.5552.27a. [DOI] [PubMed] [Google Scholar]
- Goossens H, Ferech M, Vander Stichele R, Elseviers M. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- Mathew AG, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis. 2007;4:115–133. doi: 10.1089/fpd.2006.0066. [DOI] [PubMed] [Google Scholar]
- WHO. Use of antimicrobials outside human medicine and resultant antimicrobial resistance in humans [R] World Health Organization; 2002. http://www.who.int/mediacentre/factsheets/fs268/en/index.html. [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S, Grootaert C, Vermeiren J, Gross G, Marzorati M, Verstraete W, Van de Wiele T. The intestinal environment in health and disease - recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15:2051–2065. doi: 10.2174/138161209788489159. [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, Konkel ME, Angly F, Dinsdale EA, Edwards RA, Nelson KE, White BA. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS One. 2008;3:e2945. doi: 10.1371/journal.pone.0002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok VA, Ophel-Keller K, Loo M, Hughes RJ. Application of methods for identifying broiler chicken gut bacterial species linked with increased energy metabolism. Appl Environ Microbiol. 2008;74:783–791. doi: 10.1128/AEM.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, Dore J, Leclerc M. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Cabana MD, Shane AL, Chao C, Oliva-Hemker M. Probiotics in primary care pediatrics. Clin Pediatr (Phila) 2006;45:405–410. doi: 10.1177/0009922806289614. [DOI] [PubMed] [Google Scholar]
- Cadieux P, Wind A, Sommer P, Schaefer L, Crowley K, Britton RA, Reid G. Evaluation of reuterin production in urogenital probiotic Lactobacillus reuteri RC-14. Appl Environ Microbiol. 2008;74:4645–4649. doi: 10.1128/AEM.00139-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux PA, Burton J, Devillard E, Reid G. Lactobacillus by-products inhibit the growth and virulence of uropathogenic Escherichia coli. J Physiol Pharmacol. 2009;60(Suppl 6):13–18. [PubMed] [Google Scholar]
- Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infect Immun. 2008;76:3360–3373. doi: 10.1128/IAI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillor O, Etzion A, Riley MA. The dual role of bacteriocins as anti- and probiotics. Appl Microbiol Biotechnol. 2008;81:591–606. doi: 10.1007/s00253-008-1726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbeck A, Edlund C, Gustafsson JA, Kager L, Nord CE. Impact of Lactobacillus acidophilus on the normal intestinal microflora after administration of two antimicrobial agents. Infection. 1988;16:329–336. doi: 10.1007/BF01644541. [DOI] [PubMed] [Google Scholar]
- Ng SC, Hart AL, Kamm MA, Stagg AJ, Knight SC. Mechanisms of action of probiotics: recent advances. Inflamm Bowel Dis. 2009;15:300–310. doi: 10.1002/ibd.20602. [DOI] [PubMed] [Google Scholar]
- Reid G, Bruce AW. Urogenital infections in women: can probiotics help? Postgrad Med J. 2003;79:428–432. doi: 10.1136/pmj.79.934.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Jass J, Sebulsky MT, McCormick JK. Potential uses of probiotics in clinical practice. Clin Microbiol Rev. 2003;16:658–672. doi: 10.1128/CMR.16.4.658-672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Sanders ME, Gaskins HR, Gibson GR, Mercenier A, Rastall R, Roberfroid M, Rowland I, Cherbut C, Klaenhammer TR. New scientific paradigms for probiotics and prebiotics. J Clin Gastroenterol. 2003;37:105–118. doi: 10.1097/00004836-200308000-00004. [DOI] [PubMed] [Google Scholar]
- Grady R, Hayes F. Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Ohtsubo E. Mapping and disruption of the chpB locus in Escherichia coli. J Bacteriol. 1994;176:5861–5863. doi: 10.1128/jb.176.18.5861-5863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Verdiger R, Shlosberg-Fedida A, Engelberg-Kulka H. A differential effect of E. coli toxin-antitoxin systems on cell death in liquid media and biofilm formation. PLoS One. 2009;4:e6785. doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittenhuber G. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J Mol Microbiol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- Sat B, Hazan R, Fisher T, Khaner H, Glaser G, Engelberg-Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EP, Strom AR, Helinski DR. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Sierra S, Pardo-Abarrio C, Giraldo R, Diaz-Orejas R. Genetic identification of two functional regions in the antitoxin of the parD killer system of plasmid R1. FEMS Microbiol Lett. 2002;206:115–119. doi: 10.1111/j.1574-6968.2002.tb10995.x. [DOI] [PubMed] [Google Scholar]
- Sobecky PA, Easter CL, Bear PD, Helinski DR. Characterization of the stable maintenance properties of the par region of broad-host-range plasmid RK2. J Bacteriol. 1996;178:2086–2093. doi: 10.1128/jb.178.7.2086-2093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu L, Zhang J, Inouye M. Characterization of ChpBK, an mRNA interferase from Escherichia coli. J Biol Chem. 2005;280:26080–26088. doi: 10.1074/jbc.M502050200. [DOI] [PubMed] [Google Scholar]
- Bech FW, Jorgensen ST, Diderichsen B, Karlstrom OH. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Gunn JS, Miller SI. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J Bacteriol. 1996;178:6857–6864. doi: 10.1128/jb.178.23.6857-6864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippa AM, Goulian M. Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS Genet. 2009;5:e1000788. doi: 10.1371/journal.pgen.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. Comparison of the PhoPQ regulon in Escherichia coli and Salmonella typhimurium. J Mol Evol. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci U S A. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, Engelberg-Kulka H. Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins”. PLoS Genet. 2009;5(3):e1000390. doi: 10.1371/journal.pgen.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Yassin Y, Engelberg-Kulka H. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol. 2004;186:8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Yamaguchi Y, Inouye M. Intramolecular regulation of the sequence-specific mRNA interferase activity of MazF fused to a MazE fragment with a linker cleavable by specific proteases. Appl Environ Microbiol. 2012;78(11):3794–3799. doi: 10.1128/AEM.00364-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky M, Avshalom H, Erental A, Yelin I, Kumar S, London N, Sperber M, Schueler-Furman O, Engelberg-Kulka H. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2012;41(6):625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Sat B, Reches M, Amitai S, Hazan R. Bacterial programmed cell death systems as targets for antibiotics. Trends Microbiol. 2004;12(2):66–71. doi: 10.1016/j.tim.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Rothenbacher FP, Suzuki M, Hurley JM, Montville TJ, Kirn TJ, Ouyang M, Woychik NA. Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J Bacteriol. 2012;194(13):3464–3474. doi: 10.1128/JB.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja VK, Dwivedi HP, Yan X. Novel natural food antimicrobials. Annu Rev Food Sci Technol. 2012;3:381–403. doi: 10.1146/annurev-food-022811-101241. [DOI] [PubMed] [Google Scholar]
- de Vuyst L, Vandamme E. In: Bacteriocins of Lactic Acid Bacteria. Microbiology, Genetics and Applications. Vuyst L, Vandamme EJ, editor. Blackie Acad Prof, London; 1994. Antimicrobial potential of lactic acid bacteria; pp. 91–142. [Google Scholar]
- Shornikova AV, Casas IA, Isolauri E, Mykkanen H, Vesikari T. Lactobacillus reuteri as a therapeutic agent in acute diarrhea in young children. J Pediatr Gastroenterol Nutr. 1997;24:399–404. doi: 10.1097/00005176-199704000-00008. [DOI] [PubMed] [Google Scholar]
- Williams MD, Ha CY, Ciorba MA. Probiotics as therapy in gastroenterology: a study of physician opinions and recommendations. J Clin Gastroenterol. 2010;44:631–636. doi: 10.1097/MCG.0b013e3181d47f5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin BR, Gorbach SL, Saxelin M, Barakat S, Gualtieri L, Salminen S. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci. 1992;37:121–128. doi: 10.1007/BF01308354. [DOI] [PubMed] [Google Scholar]
- Plummer S, Weaver MA, Harris JC, Dee P, Hunter J. Clostridium difficile pilot study: effects of probiotic supplementation on the incidence of C. difficile diarrhoea. Int Microbiol. 2004;7:59–62. [PubMed] [Google Scholar]
- Vanderhoof JA, Whitney DB, Antonson DL, Hanner TL, Lupo JV, Young RJ. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J Pediatr. 1999;135:564–568. doi: 10.1016/S0022-3476(99)70053-3. [DOI] [PubMed] [Google Scholar]
- Vanderhoof JA, Young RJ. Current and potential uses of probiotics. Ann Allergy Asthma Immunol. 2004;93:S33–S37. doi: 10.1016/S1081-1206(10)61730-9. [DOI] [PubMed] [Google Scholar]
- Arber W. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- Saito H, Kashida S, Inoue T, Shiba K. The role of peptide motifs in the evolution of a protein network. Nucleic Acids Res. 2007;35:6357–6366. doi: 10.1093/nar/gkm692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn B, Steiner N, Sumida C, Peters TJ. Intestinal enterokinase. Mechanisms of tts “secretion” into the lumen of the small intestine. Lancet. 1971;1:165–166. doi: 10.1016/s0140-6736(71)91936-2. [DOI] [PubMed] [Google Scholar]
- Kitamoto Y, Yuan X, Wu Q, McCourt DW, Sadler JE. Enterokinase, the initiator of intestinal digestion, is a mosaic protease composed of a distinctive assortment of domains. Proc Natl Acad Sci U S A. 1994;91:7588–7592. doi: 10.1073/pnas.91.16.7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HW, Choi SI, Seong BL. Engineered recombinant enteropeptidase catalytic subunit: effect of N-terminal modification. Arch Biochem Biophys. 2002;400:1–6. doi: 10.1006/abbi.2001.2737. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Vankemmelbeke MN, Holland LE, Walker DC, James R, Penfold CN. Investigating early events in receptor binding and translocation of colicin E9 using synchronized cell killing and proteolytic cleavage. J Bacteriol. 2008;190:4342–4350. doi: 10.1128/JB.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. Evidence of Selection upon Genomic GC-Content in Bacteria. PLoS Genet. 2010;6(9):e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canani RB, Cirillo P, Terrin G, Cesarano L, Spagnuolo MI, De Vincenzo A, Albano F, Passariello A, De Marco G, Manguso F, Guarino A. Probiotics for treatment of acute diarrhoea in children: randomised clinical trial of five different preparations. BMJ. 2007;335:340. doi: 10.1136/bmj.39272.581736.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weizman Z, Asli G, Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics. 2005;115:5–9. doi: 10.1542/peds.2004-1815. [DOI] [PubMed] [Google Scholar]
- Quigley EMM, Sanders ME. Probiotic foods for gastrointestinal health. Gastroenterology & Endoscopy News Special Edition. 2009;7:27–33. [Google Scholar]
- Corr SC, Gahan CG, Hill C. Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol Med Microbiol. 2007;50:380–388. doi: 10.1111/j.1574-695X.2007.00264.x. [DOI] [PubMed] [Google Scholar]
- Marteau P, Seksik P. Tolerance of probiotics and prebiotics. J Clin Gastroenterol. 2004;38:S67–S69. doi: 10.1097/01.mcg.0000128929.37156.a7. [DOI] [PubMed] [Google Scholar]
- Marteau P, Seksik P, Lepage P, Dore J. Cellular and physiological effects of probiotics and prebiotics. Mini Rev Med Chem. 2004;4:889–896. doi: 10.2174/1389557043403369. [DOI] [PubMed] [Google Scholar]
- Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol. 1999;276:G941–G950. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- Klaenhammer TR, Kullen MJ. Selection and design of probiotics. Int J Food Microbiol. 1999;50:45–57. doi: 10.1016/S0168-1605(99)00076-8. [DOI] [PubMed] [Google Scholar]
- Cursino L, Smajs D, Smarda J, Nardi RM, Nicoli JR, Chartone-Souza E, Nascimento AM. Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. J Appl Microbiol. 2006;100:821–829. doi: 10.1111/j.1365-2672.2006.02834.x. [DOI] [PubMed] [Google Scholar]
- Iacono A, Raso GM, Canani RB, Calignano A, Meli R. Probiotics as an emerging therapeutic strategy to treat NAFLD: focus on molecular and biochemical mechanisms. J Nutr Biochem. 2010;22:699–711. doi: 10.1016/j.jnutbio.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Natl Acad Sci U S A. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG, Norris JS. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother. 2003;47:1301–1307. doi: 10.1128/AAC.47.4.1301-1307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Buxton S. Geneious v5.4. 2011. http://www.geneious.com.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]