Abstract
MTDH(metadherin), an important oncogene that is widely overexpressed in various cancers, is a potential biomarker of tumor malignancy. Variants in MTDH have been associated with susceptibility to breast cancer. However, no studies assessing MTDH gene polymorphisms and their potential relationship to ovarian cancer susceptibility have been reported. Thus, we investigated the association of MTDH (−470G>A) polymorphism with ovarian cancer development in 145 ovarian cancer patients and 254 matched control subjects, using sequence analysis. We found that the MTDH (−470G>A) polymorphism was statistically correlated with ovarian cancer risk (under the additive genetic model, GG vs. GA vs AA, P = 0.042). Compared with genotypes containing the G allele (GG and GA), the AA genotype may decrease the risk of ovarian cancer (P = 0.0198, OR = 0.33, 95% CI [0.12∼0.78]). Compared with the G allele, the A allele is protective against ovarian cancer risk (P = 0.01756, OR = 0.66, 95% CI [0.46∼0.93]). Furthermore, a statistically significant association between the GG and GA+AA genotypes and the clinical stage was observed (P = 0.038). These data suggest that MTDH (−470G>A) could be a useful molecular marker for assessing ovarian cancer risk and for predicting ovarian cancer patient prognosis.
Introduction
Ovarian cancer has the highest fatality rate of all female reproductive system malignancies, and in 2008 there were an estimated 225,500 new cases and 140,200 deaths worldwide [1]. As is the case for many malignancies, ovarian cancer is a multifactorial disease, and hormonal factors, wound healing and inflammation may all play a role in its development. Interactions between the environment and genetic factors also play significant roles [2]. Many studies have investigated the genetic basis of ovarian cancer susceptibility. For example, BRCA1, BRCA2, MLH1, MSH2, RAD51C, RAD51D, RB1, SMAD6, CASP8, and LIN28B have all been implicated in ovarian cancer [3], [4], [5], [6], [7], [8], [9], [10]. Recently, genome-wide association studies (GWAS) have found strong associations between ovarian cancer and several common susceptibility alleles in four loci [11], [12], [13]. Braem et al. reviewed 147 candidate genes, and the 3 GWAS studies published from 1990 to October 2010 identified approximately 1100 genetic variants in more than 200 candidate genes and 20 intergenic regions [8]. However, the relationships between known genetic variants and ovarian cancer are limited, and more studies need be performed to elucidate causal genetic variants and facilitate the identification of high risk subgroups within the general population [5].
MTDH, also known as astrocyte elevated gene-1 (AEG-1) and Lyric, was originally identified as an HIV-inducible gene in primary human fetal astrocytes [14]. MTDH is located at 8q22, consists of 12 exons and 11 introns and encodes a 582 amino acid protein with a calculated molecular mass of 64 kDa. MTDH over-expression has been detected in esophageal squamous cell carcinoma [15], gastric cancer [16], renal cancer [17], prostate cancer [18], non-small cell lung cancer [19], hepatocellular carcinoma [20], breast cancer [21]–[22], and neuroblastoma [23], compared to normal cells and the matched non-neoplastic regions [24]. The level of MTDH over-expression is significantly correlated with tumorigenesis, invasion, migration, progression, angiogenesis [25], EMT (epithelial mesenchymal transition), chemoresistance and radioresistance in various cancer types [17], [26], [27], [28], [29], [30], [31]. MTDH overe-xpression is correlated with peritoneal dissemination, lymph node metastasis, International Federation of Gynecology and Obstetrics stage, histological grade, presence of residual tumor and tumor recurrence in ovarian cancer [32], [33]. High MTDH expression was associated with the progression and prognosis of ovarian cancer [32], [33].
Our group has also previously found that variants in MTDH are significantly associated with breast cancer [34]. However, the association of MTDH variants with ovarian cancer susceptibility has not been investigated.
Results
The Relationship Between the MTDH (−470G>A) Polymorphism and Ovarian Cancer Risk
The participants in the case and control groups were all from mainland China. There were no significant clinical differences (i.e., body mass index [BMI], median age, menstrual history or other related parameters) between the 2 groups.
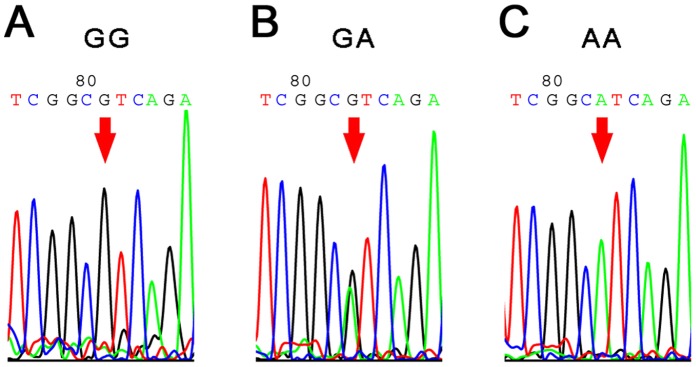
Hardy-Weinberg equilibrium showed that the chi-square values of the case group and control group were 0.1 and 3.94, respectively; both were p>0.05. As shown in Table 1, the MTDH (−470G>A) genotypes and allele distributions had a statistically significant difference between the case and control groups. We observed a statistically significant correlation with ovarian cancer risk (the additive genetic model, GG vs. GA vs AA, P = 0.042). Using the dominant genetic model (GG+GA vs AA), we observed a statistically significant difference in ovarian cancer risk (P = 0.0198, OR = 0.33, 95% CI [0.12 ∼0. 78]). These data showed that the homozygous AA genotype may be protective against ovarian cancer development and may decrease the risk of ovarian cancer. The A allele appears to be protective (P = 0. 01756, OR = 0.66, 95% CI [0.46∼0.93]) against ovarian cancer. Sequencing chromatograms from randomly chosen cases are used to illustrate the variants of MTDH (Fig. 1).
Table 1. The MTDH (−470G>A) genotype & Allele distribution in Ovarian Cancer and controls.
| Genotype | Ovarian Cancer n(%)† | Controls n(%)† | P-value | OR 95%CI |
| GG | 93(64.1) | 141(55.5) | 0.042 | 1.00 (reference) |
| GA | 47(32.4) | 88(34.6) | 0.81 (0.52 ∼1.26) | |
| AA | 5(3.45) | 25(9.84) | 0.30 (0.11 ∼0.82) | |
| GG+GA | 140(96.6 ) | 229(90.2 ) | 0.0198 | 1.00 (reference) |
| AA | 5(3.45 ) | 25(9.84 ) | 0.33 (0.12 ∼0.87) | |
| GG | 93(64.1 ) | 141(55.5 ) | 0.0924 | 1.00 (reference) |
| GA+AA | 52(35.9 ) | 113(44.5 ) | 0.70 (0.46 ∼1.06) | |
| G | 233(80.3 ) | 370(72.8 ) | 0.01756 | 1.00 (reference) |
| A | 57(19.7 ) | 138(27.2 ) | 0.66 (0.46 ∼0.93) |
The X2 for HWE of Ovarian Cancer group and control group is 0.1 and 3.94 respectively (both P>0.05).
Figure 1. Sequencing chromatograms of MTDH (−470G>A).
A–C, the sequencing chromatogram results of the genotype GG, GA and AA respectively. Samples were chosen randomly.
The Relationship between the MTDH (−470G>A) Polymorphism and Clinicopathological Variables
Table 2 shows the association of the GG and GA+AA genotypes with clinicopathological characteristics, including age at diagnosis, degree of tumor differentiation, clinical stage, lymph node metastasis, CA125 expression, tumor size and tumor histology. A statistically significant association with clinical stage was found (P = 0.038). No relationships between the polymorphism and age at diagnosis, degree of tumor differentiation, lymph node metastasis, CA125 expression, tumor size or tumor histology were found. Thus, the MTDH (−470G>A) polymorphism may be an indicator of clinical stage in ovarian cancer.
Table 2. Results of association analysis between rs16896059 and clinicopathological characteristics.
| Clinical data information | All(%) | Genotype | P-value | OR | |
| GG(%) | GA+AA(%) | ||||
| Age | |||||
| ≤50 | 43(31.9) | 31(23.0) | 12(8.89) | 0.608 | 1.00(reference) |
| >50 | 92(68.1) | 61(45.2) | 31(23.0) | 1.206 | |
| Degree of Differentiation | |||||
| Low | 89(84.0) | 58(54.7) | 31(29.2) | 0.617 | 1.00(reference) |
| Middle & High | 17(16.0) | 10(9.43) | 7(6.60) | 0.764 | |
| Clinical stage | |||||
| I & II | 36(28.8) | 18(14.4) | 18(14.4) | 0.038 | 1.00(reference) |
| III & IV | 89(71.2) | 62(49.6) | 27(21.6) | 2.296 | |
| Positive lymph node | |||||
| Negative | 37(66.1) | 20(35.7) | 17(30.4) | 0.515 | 1.00(reference) |
| Positive | 19(33.9) | 12(21.4) | 7(12.5) | 1.457 | |
| CA125 | |||||
| ≤65(U/ml) | 23(17.6) | 15(11.5) | 8(6.11) | 0.962 | 1.00(reference) |
| >65(U/ml) | 108(82.4) | 71(54.2) | 37(28.2) | 1.023 | |
| Size of tumor | |||||
| <10 cm | 80(59.3) | 54(40.0) | 26(19.3) | 0.496 | 1.00(reference) |
| ≥10 cm | 55(40.7) | 34(25.2) | 21(15.6) | 0.780 | |
| Tumor histology | |||||
| Serous | 89(65.4) | 58(42.6) | 31(22.8) | 0.877 | 1.00(reference) |
| Other | 47(34.6) | 30(22.1) | 17(12.5) | 0.943 | |
The Relationship Between the MTDH (−470G>A) Polymorphism and MTDH Protein Levels
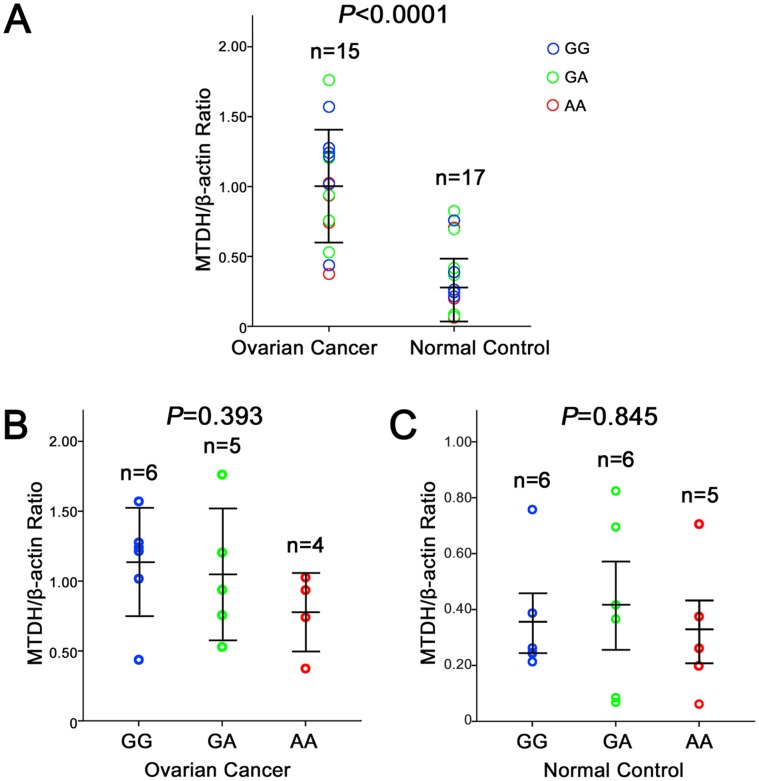
We further studied the possible relationship between the MTDH (−470G>A) polymorphism and the MTDH protein expression level in vivo. As shown in Fig. 2, the levels of MTDH protein in 15 ovarian cancer tissues were significantly higher than those in 17 normal tissues (P<0.01). As shown in Fig. 2B and 2C, the MTDH protein expression was higher in patients with the GG or GA genotypes than in those with the AA genotype, but these differences were not significant (P>0.05). These findings suggest that the SNP −470G>A may not significantly impact the expression of the MTDH gene.
Figure 2. Association of the −470G>A genotype and MTDH (−470G>A) protein expression.
A, Relative level of MTDH protein expression in ovarian cancer tissues compared to normal ovarian tissues. B, Relative level of MTDH protein expression in the ovarian cancer tissues of patients with different −470G>A genotypes. C, Relative level of MTDH protein expression in normal tissues of individuals with different −470G>A genotypes. One circle represents the mean of three independent measurements from one patient. The distribution of the three genotypes were random between the groups. N represents the samples number of respective group. Bars represent the standard deviation. Student’s t test was used to evaluate the differences in the expression levels of different constructs.
Discussion
MTDH plays a critical role in tumor biology, and it is involved in a variety of tumor biological behaviors. Our group was the first to discover a significant association between MTDH and tumor susceptibility: that is, that 2 MTDH variants are associated with breast cancer [34]. In this study, we further analyzed various SNPs in MTDH and found another SNP (rs16896059, −470G>A) with statistically significant differences between ovarian cancer case and control groups. Furthermore, rs16896059 was statistically significantly associated with the clinical stage of the tumor. To the best of our knowledge, this is the first variant association study of MTDH SNPs and ovarian cancer risk. We expect that MTDH will be a useful molecular marker for assessing ovarian cancer risk and for predicting ovarian cancer patient prognosis. However, this finding should be verified in a larger sample.
The SNP rs16896059 is located in the promoter of MTDH and thus polymorphisms do not alter the sequence or structure of the protein. We investigated the protein expression level of MTDH by western blotting. As previously reported, the expression levels of MTDH in ovarian cancer tissues were significantly higher than the levels in adjacent normal tissues (P<0.0001) [32], [33]. However, we did not find any statistically significant effect of the −470G>A SNP on the protein expression of the MTDH gene in ovarian cancer tissues or the normal tissues. Thus, no impact of the SNP on MTDH expression was evident. Because of the −470G>A SNP was located in the promoter region, and then it could also affect promoter activeity. Therefore, the association of the MTDH (−470G>A) polymorphism with MTDH promoter activeity and its effect on ovarian cancer development should be studied in vitro to further investigate the molecular mechanisms involved.
As indicated above, most patients who participated in our study were living in Shandong Province, China. Due to the general genetic homogeneity of this ethnic population, we speculate that these findings will be consistent in larger sample sizes across China. However, the relationship between MTDH polymorphism and ovarian cancer risk requires further investigation in different ethnic populations [34].
In conclusion, the A allele of the MTDH SNP rs16896059 (−470G>A) is protective against ovarian cancer, and the homozygous AA genotype may be a protective genotype. The polymorphism is statistically significantly associated with clinical stage.
Materials and Methods
Patients and Samples
The study was approved by the Ethical Committee of Shandong University. All participants gave written informed consent to participate in this study. 145 patients (mean age of 51.8±13.1 years) participated in the study, diagnosed with ovarian cancer in Qilu Hospital of Shandong University between September 2008 and July 2011. Clinical data information, including age at diagnosis, degree of differentiation, clinical stage, positive lymph node, CA125, size of tumor and tumor histology were obtained from patients’ medical records. 254 age-matched healthy women (mean age of 49.2±12.8 years) were recruited as control. Most participants were Han Chinese residing in Shandong Province, China. DNA from peripheral blood cells s was extracted with TIANamp Genomic DNA Kit (Tiangen, Beijing, China), by instructions. The DNA purity and concentration were measured by ultraviolet spectrophotometer (GE Healthcare, USA). DNA samples were conventionally stored at −80°C as previously described [34], [35].
Genotyping Analysis of the MTDH (−470G>A)
Genotyping of the SNP rs16896059 (−470G>A) polymorphism was determined by PCR and sequencing method. The sequence of MTDH gene was obtained from NCBI (Gene ID: 92140, Nucleotide: AC_000140.1, GI: 157734173). Primers were designed with Primer Premier 5 according to the sequence of rs16896059 as follows: forward primer 5′- CTGGCAACTGGTAGGCACGC -3′ and reverse primer 5′- GAGGGACTCGCAGGATGACG -3′. The PCR productions size was 893 bp. PCR amplification was performed in 50 µl reaction systems containing 1 µl genomic DNA (100 ng/µl), 4 µl of 2.5 mM dNTPs, 10 µl buffer, 2 µl of each primer(0.4 µM) and 0.5 U PrimeSTAR HS DNA Polymerase (TAKARA, DALIAN, China). The PCR amplification conditions were: 94°C for 5 min, 35 cycles at 98°C for 10 seconds, 55°C for 15 seconds, and 72°C for 2 min, and a final extension step of 72°C for 5 mins. All PCR productions were sequenced by BioSune Biotechnology Co., Ltd. (Shanghai, China), and the sequence data were analyzed with Chromas 2.31 and MegAlign 7.0 software.
Western Blot
Western blot analysis was performed as previously described [31], [36], Tissue was dissolved in RIPA buffer (1×PBS, 0.1% sodium dodecyl sulfate, 1% NP40, 5 mM EDTA, 1 mM sodium orthovanadate, 0.5% sodium deoxycholate and protease inhibitors). 40 µg of proteins were electrophoresed on 10% SDS-PAGE and transferred onto PVDF (Millipore) using a Mini Trans-Blot Cell (Bio-Rad Laboratories, Hercules, CA, USA). Immunoblotting was carried out using a primary antibody MTDH (ab45338, 1∶500, Abcam, Cambridge, MA, USA) and β-actin (1∶5,000, Sigma-Aldrich, St. Louis, MO, USA). The secondary antibodies (1∶5,000, KPL, Gaithersburg, MD, USA) were labeled by HRP (horseradish peroxidase). Membranes are visualized using an ECL kit (Merck, Darmstadt, Germany). The loading control was β-actin.
Statistical Analysis
The statistical data were analyzed as previously described [34]. Hardy-Weinberg equilibrium test was performed d using a web-based statistical tool OEGE (http://www.oege.org/software/hwe-mr-calc.shtml). The genotype and allele distributions in the ovarian cancer groups and control groups were analyzed using chi-squared tests, and Fisher’s exact test was used when one cell count was <5. The risk of ovarian cancer development was estimated as an odds ratio (OR) with a 95% confidence interval (CI) using an unconditional logistic regression analysis. All statistical tests were two-sided with a significance level of p≤0.05. The statistical data were analyzed with SPSS 17.0 (SPSS, Chicago, Illinois, USA).
Acknowledgments
We appreciate Ning Yang, Xiao Guan for collecting data for this study.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81001166, 30872738), Natural Science Foundation of Shandong Province (ZR2010HQ050), Foundation of Shandong Public Health Department (2009Q2012), China Postdoctoral Science Foundation (201104636, 20100471551), Independent Innovation Foundation of Shandong University (2012TS142). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62: 10–29. [DOI] [PubMed] [Google Scholar]
- 2. Risch HA (1998) Hormonal etiology of epithelial ovarian cancer, with a hypothesis concerning the role of androgens and progesterone. J Natl Cancer Inst 90: 1774–1786. [DOI] [PubMed] [Google Scholar]
- 3. Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, et al. (2010) Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42: 410–414. [DOI] [PubMed] [Google Scholar]
- 4. Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, et al. (2011) Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet 43: 879–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin J, Lu K, Lin J, Wu L, Hildebrandt MA, et al. (2011) Genetic variants in TGF-beta pathway are associated with ovarian cancer risk. PLoS One 6: e25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma X, Zhang J, Liu S, Huang Y, Chen B, et al. (2011) Polymorphisms in the CASP8 gene and the risk of epithelial ovarian cancer. Gynecol Oncol 122: 554–559. [DOI] [PubMed] [Google Scholar]
- 7. Permuth-Wey J, Kim D, Tsai YY, Lin HY, Chen YA, et al. (2011) LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res 71: 3896–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braem MG, Schouten LJ, Peeters PH, den Brandt PA, Onland-Moret NC (2011) Genetic susceptibility to sporadic ovarian cancer: A systematic review. Biochim Biophys Acta 1816: 132–146. [DOI] [PubMed] [Google Scholar]
- 9. Pelttari LM, Heikkinen T, Thompson D, Kallioniemi A, Schleutker J, et al. (2011) RAD51C is a susceptibility gene for ovarian cancer. Hum Mol Genet 20: 3278–3288. [DOI] [PubMed] [Google Scholar]
- 10.Ramus SJ, Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, et al.. (2012) Ovarian cancer susceptibility alleles and risk of ovarian cancer in BRCA1 and BRCA2 mutation carriers. Hum Mutat. [DOI] [PMC free article] [PubMed]
- 11. Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, et al. (2010) Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet 42: 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, et al. (2010) A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet 42: 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, et al. (2009) A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet 41: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ, et al. (2005) Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 353: 8–15. [DOI] [PubMed] [Google Scholar]
- 15. Yu C, Chen K, Zheng H, Guo X, Jia W, et al. (2009) Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis 30: 894–901. [DOI] [PubMed] [Google Scholar]
- 16. Jian-bo X, Hui W, Yu-long H, Chang-hua Z, Long-juan Z, et al. (2011) Astrocyte-elevated gene-1 overexpression is associated with poor prognosis in gastric cancer. Med Oncol 28: 455–462. [DOI] [PubMed] [Google Scholar]
- 17. Chen W, Ke Z, Shi H, Yang S, Wang L (2010) Overexpression of AEG-1 in renal cell carcinoma and its correlation with tumor nuclear grade and progression. Neoplasma 57: 522–529. [DOI] [PubMed] [Google Scholar]
- 18. Thirkettle HJ, Girling J, Warren AY, Mills IG, Sahadevan K, et al. (2009) LYRIC/AEG-1 is targeted to different subcellular compartments by ubiquitinylation and intrinsic nuclear localization signals. Clin Cancer Res 15: 3003–3013. [DOI] [PubMed] [Google Scholar]
- 19. Song L, Li W, Zhang H, Liao W, Dai T, et al. (2009) Over-expression of AEG-1 significantly associates with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. J Pathol 219: 317–326. [DOI] [PubMed] [Google Scholar]
- 20. Yoo BK, Emdad L, Su ZZ, Villanueva A, Chiang DY, et al. (2009) Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest 119: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown DM, Ruoslahti E (2004) Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 5: 365–374. [DOI] [PubMed] [Google Scholar]
- 22. Li J, Zhang N, Song LB, Liao WT, Jiang LL, et al. (2008) Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res 14: 3319–3326. [DOI] [PubMed] [Google Scholar]
- 23. Lee SG, Jeon HY, Su ZZ, Richards JE, Vozhilla N, et al. (2009) Astrocyte elevated gene-1 contributes to the pathogenesis of neuroblastoma. Oncogene 28: 2476–2484. [DOI] [PubMed] [Google Scholar]
- 24. Gnosa S, Shen YM, Wang CJ, Zhang H, Stratmann J, et al. (2012) Expression of AEG-1 mRNA and protein in colorectal cancer patients and colon cancer cell lines. J Transl Med 10: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche H, et al. (2009) Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci U S A 106: 21300–21305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao Y, Moran MS, Yang Q, Liu Q, Yuan C, et al. (2012) Metadherin regulates radioresistance in cervical cancer cells. Oncol Rep 27: 1520–1526. [DOI] [PubMed] [Google Scholar]
- 27. Zhang B, Liu XX, He JR, Zhou CX, Guo M, et al. (2011) Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 32: 2–9. [DOI] [PubMed] [Google Scholar]
- 28. Hu G, Wei Y, Kang Y (2009) The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res 15: 5615–5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srivastava J, Siddiq A, Emdad L, Santhekadur PK, Chen D, et al.. (2012) Astrocyte elevated gene-1 (AEG-1) promotes hepatocarcinogenesis: Novel insights from a mouse model. Hepatology. [DOI] [PMC free article] [PubMed]
- 30. Li C, Li Y, Wang X, Wang Z, Cai J, et al. (2012) Elevated expression of astrocyte elevated gene-1 (AEG-1) is correlated with cisplatin-based chemoresistance and shortened outcome in patients with stages III-IV serous ovarian carcinoma. Histopathology 60: 953–963. [DOI] [PubMed] [Google Scholar]
- 31. Zhao Y, Kong X, Li X, Yan S, Yuan C, et al. (2011) Metadherin mediates lipopolysaccharide-induced migration and invasion of breast cancer cells. PLoS One 6: e29363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li C, Liu J, Lu R, Yu G, Wang X, et al. (2011) AEG -1 overexpression: a novel indicator for peritoneal dissemination and lymph node metastasis in epithelial ovarian cancers. Int J Gynecol Cancer 21: 602–608. [DOI] [PubMed] [Google Scholar]
- 33. Meng F, Luo C, Ma L, Hu Y, Lou G (2011) Clinical significance of astrocyte elevated gene-1 expression in human epithelial ovarian carcinoma. Int J Gynecol Pathol 30: 145–150. [DOI] [PubMed] [Google Scholar]
- 34. Liu X, Zhang N, Li X, Moran MS, Yuan C, et al. (2011) Identification of novel variants of metadherin in breast cancer. PLoS One 6: e17582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang N, Li X, Tao K, Jiang L, Ma T, et al. (2011) BCL-2 (-938C>A) polymorphism is associated with breast cancer susceptibility. BMC Med Genet 12: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan C, Jiao L, Yang L, Ying W, Hu Z, et al. (2008) The up-regulation of 14–3-3 proteins in Smad4 deficient epidermis and hair follicles at catagen. Proteomics 8: 2230–2243. [DOI] [PubMed] [Google Scholar]