Abstract
The 17,922 base pairs (bp) nucleotide sequence of the linear mitochondrial DNA (mtDNA) molecule of the freshwater jellyfish Craspedacusta sowerbyi (Hydrozoa,Trachylina, Limnomedusae) has been determined. This sequence exhibits surprisingly low A+T content (57.1%), containing genes for 13 energy pathway proteins, a small and a large subunit rRNAs, and methionine and tryptophan tRNAs. Mitochondrial ancestral medusozoan gene order (AMGO) was found in the C. sowerbyi, as those found in Cubaia aphrodite (Hydrozoa, Trachylina, Limnomedusae), discomedusan Scyphozoa and Staurozoa. The genes of C. sowerbyi mtDNA are arranged in two clusters with opposite transcriptional polarities, whereby transcription proceeds toward the ends of the DNA molecule. Identical inverted terminal repeats (ITRs) flank the ends of the mitochondrial DNA molecule, a characteristic typical of medusozoans. In addition, two open reading frames (ORFs) of 354 and 1611 bp in length were found downstream of the large subunit rRNA gene, similar to the two ORFs of ORF314 and polB discovered in the linear mtDNA of C. aphrodite, discomedusan Scyphozoa and Staurozoa. Phylogenetic analyses of C. sowerbyi and other cnidarians were carried out based on both nucleotide and inferred amino acid sequences of the 13 mitochondrial energy pathway genes. Our working hypothesis supports the monophyletic Medusozoa being a sister group to Octocorallia (Cnidaria, Anthozoa). Within Medusozoa, the phylogenetic analysis suggests that Staurozoa may be the earliest diverging class and the sister group of all other medusozoans. Cubozoa and coronate Scyphozoa form a clade that is the sister group of Hydrozoa plus discomedusan Scyphozoa. Hydrozoa is the sister group of discomedusan Scyphozoa. Semaeostomeae is a paraphyletic clade with Rhizostomeae, while Limnomedusae (Trachylina) is the sister group of hydroidolinans and may be the earliest diverging lineage among Hydrozoa.
Introduction
Characterization of mitochondrial DNA (mtDNA) genomes in cnidarians has become an increasing interest in evolutionary and phylogenetic studies. The phylum Cnidaria is divided into two large clades, Medusozoa and Anthozoa, the former of which contains four classes, including Hydrozoa, Cubozoa, Scyphozoa, and Staurozoa [1]. Species in the Medusozoa have a medusoid body form in their life cycle, as different from those in the Anthozoa, and a linear mtDNA molecule in contrast to a single circular molecule of 14 to 20 kilobases in metazoans [2], [3], [4], [5]. However, most available data on cnidarian mtDNA sequences are derived from species in the Anthozoa, with the availability of more than 40 complete mtDNA sequences in public databases, which shows a degree of similarity to metazoan mtDNA genomes in being a circular structure [6]. Meanwhile, it seems likely that there is an increasing amount of data on mtDNA genomes in medusozoans. So far, 26 linear mtDNA genomes, although not all fully complete, have been reported from species of all four classes in the Medusozoa [7], for example, Hydra oligactis [8], H. vulgaris, and H. magnipapillata [9] in Hydrozoa, Alatina moseri [10] in Cubozoa, Lucernaria janetae [7] in Staurozoa, and Aurelia aurita in Scyphozoa [11]. Interestingly, the linear mtDNA can be divided into two to eight molecules [7], [9], [10]. Kayal et al. (2012) firstly examined linear mtDNA in Medusozoa and reconstructed the evolutionary history of linear mtDNA in medusozoans by analyzing the mitochondrial genome structures [7]. The dramatic feature of two open reading frames (ORFs), polB and ORF314, was identified in mtDNA sequences of cubozoan, scyphozoan, staurozoan, and trachyline hydrozoan, which was thought to be remnants of a linear plasmid that invaded the mitochondrial genomes of the last common ancestor of Medusozoa and should be responsible for the linearity [7], [11]. However, hydroidolinan hydrozoans have lost the two ORFs and they instead have duplicated cox1 at each end of their mitochondrial chromosome(s) [7], [8], [9].
However, very few nucleotide sequences can be found from Craspedacusta in public databases, such as in GenBank. The nucleotide sequences referred to Craspedacusta mtDNA comprise only several cox1 and rnl sequences. Recently, a nearly complete mitochondrial genome sequence of Cubaia aphrodite (Hydrozoa, Trachylina, Limnomedusae) was reported [7]. C. aphrodite, living in marine water, belongs to the same order, Limnomedusae, as freshwater jellyfish Craspedacusta sowerbyi. Though with sequence of the identical inverted terminal repeats (ITRs) and partial polB being undetermined, its mitogenome was thought to possesses characteristically the mitochondrial ancestral medusozoan gene order (AMGO), as reported in scyphozoans and staurozoans [7], [11]. But, the mitochondrial genome organizations in non-trachyline hydrozoans contain one or several linear molecules, thus exhibiting a degree of variation, with probably the presence of other three gene orders [7], [8], [9]. Mitochondrial genome organizations in the Hydrozoa may be thus much more variable than in other medusozoans. The understanding of their mtDNA genomes may provide necessary information to reveal the diversity of mtDNA genome organization and their evolution. More mitogenomes from trachyline hydrozoans are certainly needed to test whether AMGO is typical of this group or whether other gene orders exist.
Freshwater jellyfish are frequently reported, with more than 20 species of freshwater jellyfish in the world [12]. Among them, Craspedacusta sowerbyi (Hydrozoa, Trachylina, Limnomedusae) is the most widely distributed species [13]. It is considered that this species is originated from the Yangtze River area in China [13], [14], but has been dispersed to many freshwater habitats in the world [15], [16], [17], [18], [19], [20]. The hydrozoan has two adult forms, the fixed polyp, and the free-living medusa, with the latter being the sexual form, occurring sporadically in different season of the year, although more commonly during summer in temperate regions [18]. It was thus considered possible to obtain samples of C. sowerbyi in China in order to examine its mtDNA genome [21]. In this report, we presented the complete sequence of the mitochondrial genome of C. sowerbyi, which represents the first complete mitochondrial genome for species in the Trachylina. Furthermore, we used this sequence together with other reported sequences of medusozoan mtDNA to analyze the phylogenetic relationship within the Cnidaria, and especially within the Medusozoa.
Results
Genome Structure, Organization, and Nucleotide Composition
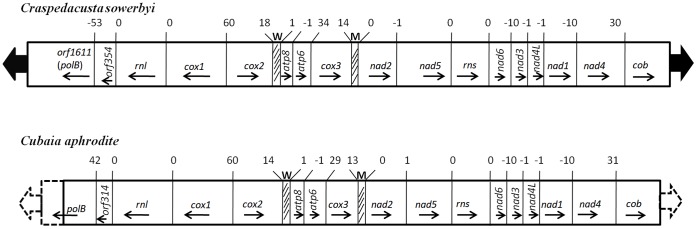
The sequence of a 17,922 bp segment was cloned from C. sowerbyi, which contains 13 known mitochondrial protein coding genes, two tRNAs, a large and a small subunit rRNAs, and two putative ORFs (Fig. 1). The genes in C. sowerbyi mitochondrial genome were arranged into two clusters of opposite transcriptional orientations. Transcription proceeded toward the ends of the DNA molecule, with 15 genes transcribed in one direction, and two known genes (cox1 and rnl) and two putative ORFs in the other. The change in the transcriptional polarity occurred between cox1 and cox2, where the longest non-coding intergenic region of 60 bp was located. The gene arrangement in C. sowerbyi mtDNA was consistent with AMGO, which was identical to that in trachyline C. aphrodite (Fig. 1), discomedusan Scyphozoa and Staurozoa, but different from those of all nontrachyline hydrozoan species [7].
Figure 1. Maps of the mitochondrial genomes of C. sowerbyi and C. aphrodite.
Protein and ribosomal genes (large open boxes) are abbreviated as presented in the text; tRNA genes (small hatched boxes) are identified by the one-letter code for their corresponding amino acid. Intergenic regions greater than 100 bp are shown by shaded boxes; ITR shown by two large arrows. Arrows within or under each box show the direction of transcription. Positive numbers at gene boundaries indicate the number of intergenic nucleotides; negative numbers indicate the number of overlapping nucleotides. The dash lines mean that the sequence of this part is not determined.
Interestingly, the nucleotide composition of C. sowerbyi mtDNA showed a small bias towards low A+T content (being 57.1%), when compared with those in the trachyline C. aphrodite (69%), other hydrozoans (>75%), and in general, metazoans (>70%) [7], [22]. The protein coding genes had the lowest A+T content (55.0%), while the rRNA genes showed the highest A+T content (62.7%) in the C. sowerbyi mtDNA (Table 1). The lowest value for A+T content was observed at the third codon positions (47.1%) in the protein coding genes, whilst in other cnidarians, A+T content at the third codon positions was generally the highest, compared with other two codon positions [9]. The discrepancy of A+T content between rRNA genes and third codon positions is contradictory to the deduction by Voigt et al., that A+T content in rRNA genes was generally correlated with the usage of A and T at third codon positions in all Cnidaria [9].
Table 1. Nucleotide composition data for different groups of genes, ORFs, and non-coding regions in C. sowerbyi mtDNA.
| Full sequence | Coding sequences | ORFs | rRNA -genes | tRNA -genes | Intergenic | ||
| Total | 3rd positions | ||||||
| %G | 25.33 | 20.7 | 20.6 | 24.2 | 19.5 | 21.1 | 16.6 |
| %A | 21.26 | 22.9 | 22.6 | 24.7 | 29.8 | 28.9 | 29.3 |
| %T | 31.82 | 32.1 | 24.5 | 34.5 | 32.9 | 30.3 | 32.5 |
| %C | 21.59 | 24.3 | 32.4 | 16.6 | 17.8 | 19.7 | 32.7 |
| %A+T | 57.14 | 55.0 | 47.1 | 59.2 | 62.7 | 59.2 | 61.8 |
| Total (bp) | 17922 | 12051 | 4017 | 1965 | 2812 | 142 | 157 |
Protein-coding Genes
Genes coding for proteins involved in respiration and oxidative phosphorylation (atp6, atp8, cob, cox1–3, nad1–6, and nad4L) in C. sowerbyi mtDNA were similar in size to their homologues in other medusozoans. atp8, nad2, and nad6 were the least conserved genes at the inferred amino acid level, showing only 21 to 64% sequence identity between C. sowerbyi and other representatives of Medusozoa; but, this percentage goes up to 92% in cox1, the most conserved gene (Table 2). Comparisons of thirteen energy pathway protein genes and 4 RNA genes in C. sowerbyi with their homologues in other medusozoans showed that these genes shared most nucleotide sequences with C. aphrodite (Table 2).
Table 2. Comparison of the mitochondrial protein coding genes and RNA genes of the freshwater jellyfish C. sowerbyi (C.s), the jellyfish A. aurita (A.a), the hydrozoan H. oligactis (H.o), cubozoan A. moseri (A.m), and the trachyline C. aphrodite (C.a).
| Genes | Number of encoded nucleotidesa | Percent sequence identity | |||||||
| C.s | A.a | H.o | A.m | C.a | C.s/A.a | C.s/H.o | C.s/A.m | C.s/C.a | |
| Atp6 | 705 | 705 | 702 | 711 | 702 | 69% | 46% | 53% | 80% |
| Atp8 | 207 | 204 | 207 | 198 | 204 | 30% | 21% | 33% | 56% |
| Cob | 1185 | 1140 | 1140 | 1146 | 1143 | 68% | 67% | 71% | 83% |
| Cox1 | 1566 | 1581 | 1575 | 1566 | 1563 | 82% | 78% | 82% | 92% |
| Cox2 | 738 | 726 | 756 | 732 | 738 | 65% | 57% | 68% | 84% |
| Cox3 | 786 | 786 | 786 | 783 | 783 | 76% | 58% | 78% | 87% |
| Nad1 | 999 | 972 | 990 | 960 | 993 | 65% | 62% | 62% | 80% |
| Nad2 | 1350 | 1320 | 1305 | 1338 | 1350 | 34% | 30% | 32% | 50% |
| Nad3 | 357 | 360 | 348 | 339 | 354 | 67% | 59% | 54% | 72% |
| Nad4 | 1461 | 1443 | 1458 | 1443 | 1458 | 55% | 55% | 46% | 73% |
| Nad4L | 309 | 303 | 300 | 282 | 297 | 57% | 56% | 57% | 80% |
| Nad5 | 1833 | 1818 | 1833 | 1821 | 1830 | 51% | 52% | 49% | 70% |
| Nad6 | 564 | 573 | 558 | 543 | 561 | 38% | 37% | 34% | 64% |
| rns | 995 | 960 | 977 | 893 | 994 | 63% | 68% | 57% | 76% |
| rnl | 1817 | 1817 | 1745 | 1679 | 1761 | 36% | 39% | 36% | 72% |
| Trn-M | 71 | 70 | 71 | 70 | 71 | 45% | 68% | 64% | 93% |
| Trn-W | 71 | 71 | 70 | nd | 71 | 45% | 75% | nd | 88% |
Two overlapping ORFs of 354 and 1611 nucleotides have been found downstream of rnl, close to the end of the C. sowerbyi linear mtDNA molecule. Similar ORFs were found at one end of the mtDNA molecule in all species of Staurozoa, discomedusan Scyphozoa, and the trachyline hydrozoan C. aphrodite, and also found on a separate mitochondrial chromosome in Cubozoa [7], [10]. The deduced amino acid sequence of ORF354 shared the most sequence elements, but with a low level of identity (only 32%) with the poorly conserved analogue ORF314 from C. aphrodite, which may act as terminal protein (TP), as suggested by Kayal et al. [7]. The deduced amino acid sequence of ORF1611 displayed extensive sequence similarity with the family B DNA polymerases, and this ORF has been identified as polB. The deduced POLB containing domains involved in the 3′-5′exonuclease (Exo I to III) and polymerase (Pol I to Pol V) activities was the longest inferred POLB sequence reported for cnidarians. As discovered in the linear mtDNA of some other medusozoans, the deduced POLB shared extremely low similarity with any other species, even with C. aphrodite [7], [11].
Analyses of the codon usage among the 13 energy pathway protein genes and, separately, in the two ORFs were shown in Table 3. This table was compiled based on a minimally modified genetic code, with TGA = tryptophan being the only deviation deduced for mitochondrial protein synthesis in C. sowerbyi. Among the 13 energy pathway protein genes, only one codon, CGG, was never used, and all protein-coding genes were assumed to have complete termination codons (either TAA or TAG). All protein-coding genes in C. sowerbyi mtDNA started with a conventional ATG initiation codon. The proportion of GC-rich amino acids (the “GARP” amino acids: glycine, alanine, arginine, and proline) was 19.4%. Considering the dramatic high GC content (45%) of mitochondrial protein genes in C. sowerbyi mtDNA, this result seems not to be consistent with the previous study that a strong correlation was found between nucleotide composition and the proportion of the “GARP” amino acids [7]. The reason may be that in C. sowerbyi mtDNA the high GC content of the coding sequence appears largely at the third codon positions, whereas in other reported cnidarians G or C preferred to appear at the first or second codon positions that always determine the kind of amino acid to be coded for.
Table 3. Codon usage among the 13 energy pathway protein genes and Two ORFs.
| C.s * | ORFs# | C.s * | ORFs# | C.s * | ORFs# | C.s * | ORFs# | ||||||||
| Phe | TTT | 109 | 10 | Ser | TCT | 93 | 9 | Tyr | TAT | 75 | 12 | Cys | TGT | 30 | 3 |
| TTC | 235 | 30 | TCC | 60 | 13 | TAC | 84 | 19 | TGC | 12 | 3 | ||||
| Leu | TTA | 98 | 19 | TCA | 26 | 11 | TER | TAA | 8 | 1 | Trp | TGA | 80 | 4 | |
| TTG | 148 | 9 | TCG | 48 | 8 | TAG | 5 | 0 | TGG | 24 | 0 | ||||
| Leu | CTT | 77 | 9 | Pro | CCT | 53 | 6 | His | CAT | 46 | 14 | Arg | CGT | 11 | 3 |
| CTC | 114 | 16 | CCC | 60 | 19 | CAC | 59 | 15 | CGC | 5 | 1 | ||||
| CTA | 82 | 12 | CCA | 16 | 2 | Gln | CAA | 57 | 14 | CGA | 22 | 5 | |||
| CTG | 79 | 10 | CCG | 18 | 3 | CAG | 28 | 7 | CGG | 0 | 1 | ||||
| Ile | ATT | 72 | 12 | Thr | ACT | 75 | 9 | Asn | AAT | 41 | 12 | Ser | AGT | 37 | 5 |
| ATC | 172 | 26 | ACC | 84 | 13 | AAC | 76 | 22 | AGC | 50 | 7 | ||||
| ATA | 98 | 18 | ACA | 17 | 3 | Lys | AAA | 52 | 45 | Arg | AGA | 49 | 23 | ||
| Met | ATG | 141 | 14 | ACG | 51 | 4 | AAG | 45 | 16 | AGG | 13 | 4 | |||
| Val | GTT | 65 | 5 | Ala | GCT | 95 | 2 | Asp | GAT | 45 | 5 | Gly | GGT | 57 | 5 |
| GTC | 138 | 8 | GCC | 104 | 7 | GAC | 49 | 16 | GGC | 24 | 2 | ||||
| GTA | 50 | 8 | GCA | 44 | 8 | Glu | GAA | 71 | 28 | GGA | 130 | 7 | |||
| GTG | 76 | 7 | GCG | 34 | 4 | GAG | 44 | 8 | GGG | 56 | 13 |
Codon usage among the 13 energy pathway protein genes of C. sowerbyi;
Codon usage of the genes ORF1611 and ORF354 of C. sowerbyi.
RNA Genes
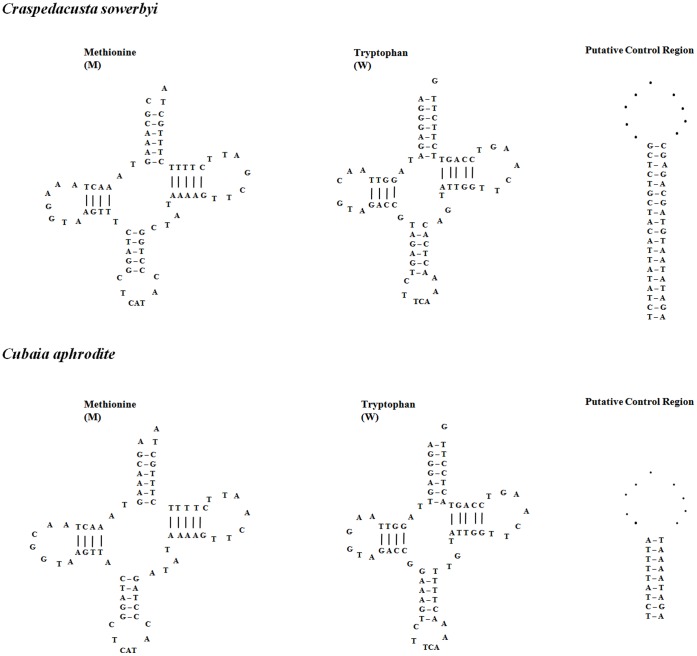
Like many other cnidarians studied to date, C. sowerbyi mtDNA possessed only two tRNA genes for methionine (trnM; CAU) and tryptophan (trnW; UCA), and two rRNA genes coding for a large and a small subunit rRNA (rnl and rns). The nucleotide composition of RNA genes was slightly AT-richer than that of the protein coding regions. The sequences of C. sowerbyi trnM (cau) and trnW (uca) were both 71 bp in size as same as in C. aphrodite, which were well conserved with high similarity in cnidarians (Table 2) [7]. Inferred secondary structures of tRNAs in C. sowerbyi and C. aphrodite mtDNAs were shown in Fig. 2. The C. sowerbyi small and large subunit ribosomal RNA, rns and rnl, were 995 and 1,817 bp in size, respectively, similar to their counterparts in other cnidarians. These two genes were located more than 7 kilobases apart in C. sowerbyi mtDNA and had opposite transcriptional polarities.
Figure 2. Secondary structure models predicted for tRNA and intergenic putative control region of C. sowerbyi and C. aphrodite.
Intergenic Regions and Terminal Sequences
Intergenic regions constituted less than 1%, with 157 bp in total, in the C. sowerbyi mtDNA genome; the largest of these (60 bp) existed between cox1 and cox2, which appeared coincidentally with the change in transcriptional polarity of C. sowerby mtDNA genes. As shown in Fig. 2, this AT-rich (68.3%) region can be folded into a conserved stem-loop motif with 19 bp in the stem; while in C. aphrodite mtDNA, a conserved stem-loop motif with only 9 bp in the stem was also discovered. Similar AT-rich regions were detected in other medusozoans [7]. This region was named as putative control region, and may be involved in the control of mtDNA transcription and/or replication [7].The second largest non-coding region was located upstream of cox3, with only 34 bp in size. The scarcity of non-coding nucleotides in the mitochondrial genome of C. sowerbyi was similar to the mtDNA of other reported medusozoans.
In addition to non-coding intergenic regions, non-coding flanking regions were present in the mitochondrial genome of C. sowerbyi. The identical inverted terminal repeats (ITRs), in which the sequence at one end of a determined segment is repeated in an inverted orientation at the other end, occurred on the C. sowerbyi linear mtDNA ends, as in other medusozoans [7], [8], [9], [10], [11]. The ITR region contained 435 bp, including a part of 3′ end of cob (59 bp). In the mtDNA of A. aurita, only 20 bp of the 3′ end of cob was included in the ITR region. The ITRs of C. sowerbyi lacked any obvious potential secondary structure and did not show any significant similarity to known sequences. In addition to C. sowerbyi, ITRs have determined for other four medusozoans: H. oligactis, H. magnipapillata, A. aurita, and A. moseri. They possessed feature of organelle telomeres and contained subtelomeric regions of fragmented and sometimes duplicate genes [10]. Though poorly understood, ITRs, together with the two ORFs of polB and ORF314, were suggested as remnants of a linear plasmid which invaded the mitochondrial genomes of the last common ancestor of Medusozoa and was involved in the mechanism of replication and maintenance for linear chromosomes similar to the type III linear mtDNA as found in yeasts, linear plasmids, and adenoviral DNA [7], [11].
Phylogenetic Analysis
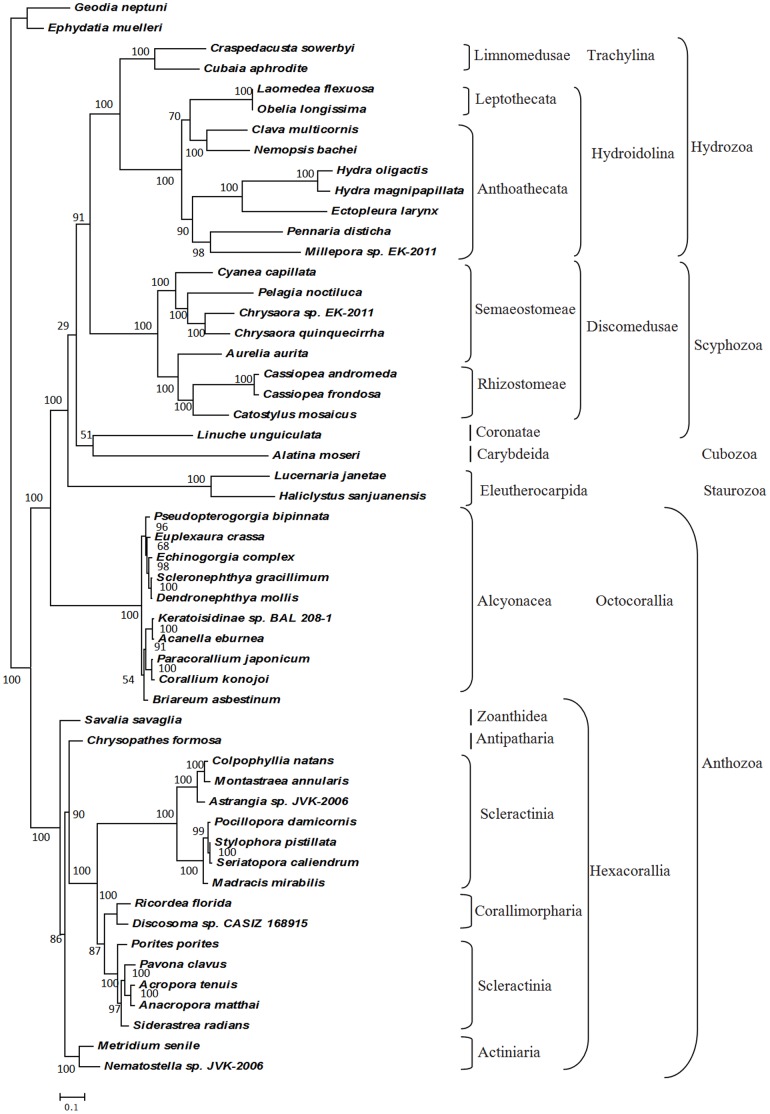
The acquisition of a complete sequence of C. sowerbyi mtDNA in this study, as well as additional cnidarian mtDNA from GenBank, allowed us to investigate the phylogenetic relationship at the level of orders within the phylum Cnidaria. Maximum likelihood (ML) analyses of nucleotide and inferred amino acids data were conducted based on 13 energy pathway protein genes in 51 cnidarian species, with two sequences from species of the Demospongia as the outgroup. The tree topologies derived from phylogenetic trees based on nucleotide (Fig. S1) and inferred amino acids (Fig. 3) data were substantially congruent with just a few variations. As shown in Fig. 3, the Cnidaria appeared to be monophyletic, but was divided into two primary groups, with one group containing Medusozoa and octocorallian Anthozoa, and the other the hexacorallian Anthozoa.
Figure 3. Phylogenetic analyses by ML methods based on mitochondrial inferred amino acid data.
ML tree obtained from the analysis of inferred amino acids of 13 energy pathway protein genes under the MTMAM model. The branch support values for each node are shown as ML bootstrap percentage. Scale bars indicate number of changes per site.
In the subclass Hexacorallia, scleractinian corals were reconstructed as a paraphyletic group that included naked corals Ricordia and Discosoma. This result was consistent with the phylogenetic analysis (based on only mitochondrial amino acids data) by Medina et al., who deduced skeleton loss in naked corals [23]. The phylogenetic relationships found for octocorals in the present study are identical to those reported previously except that the scleraxonian Briareum asbestinum was placed at the base of Octocorallia in Park’s analyses [6]. Our analyses placed B. asbestinum as sister group of the other two scleraxonians plus two calcaxonians with weak support values, and the sampled octocorals formed two obvious clades.
All medusozoans here formed a clade containing species from all the four classes, Hydrozoa, Cubozoa, Scyphozoa, and Staurozoa. Staurozoa was the sister group of all other medusozoans (based on inferred amino acids data; Fig. 3). Interestingly, the only one cubozoan Alatina moseri and the only one coronate scyphozoan Linuche unguiculata fell into a clade, with weak support, which was then the sister group of other scyphozoans and hydrozoans, also with a low bootstrap support based on amino acids data (Fig. 3). Based on nucleotide data, cubozoan, staurozoans, and coronate scyphozoan formed a complex clade, and it was hard to determine the phylogenetic relationships exactly. Furthermore, discomedusan scyphozoans fell into monophyletic group, being a sister group with all hydrozoans. It appeared that Scyphozoa was paraphyletic. Semaeostomeae was reconstructed as paraphyletic, with the jellyfish A. aurita being more closely related to the Rhizostomeae than to the Semaeostomeae.
In the class Hydrozoa, the trachyline C. sowerbyi and C. aphrodite formed a clade, which was in turn a sister group to the monophylectic Hydroidolina (Fig. 3 and Fig. S1). Species in the Hydroidolina formed two clades, one clade containing species from Aplanulata and Capitata, while the other having species from Filifera III, Filifera IV and Leptothecata. In the phylogentic trees, Anthoathecata, consisting of Aplanulata, Capitata and Filifera III-IV, is paraphyletic with respect to leptothecates. Aplanulata containing Hydra and Ectopleura larynx was the sister group of non-Aplanulata Capitata.
Discussion
In this study, we present the complete mtDNA sequence of the trachyline C. sowerbyi. Its mtDNA possessed the typical characteristics as reported for some other medusozoans: (a) a linear mtDNA molecule [3], [4]; (b) AMGO as identical to the gene order found in A. aurita [7], [11]; (c) a relatively compact genomic organization lacking introns [7]–[11]; (d) the presence of only two tRNA genes, trnM and trnW [24]; (e) flanking ITRs at the ends of the linear mtDNA [7], [8], [9], [10], [11]; (f) two extra ORFs of polB and ORF314 at one end [7], [10], [11].
Thus far, only two mitochondrial genomes were determined in the order of Limnomedusae, with one from the marine species C. aphrodite and another from the freshwater jellyfish C. sowerbyi. The mtDNA sequences of these two species show high similarity with each other, especially in the presence of same gene order and the closest relationship in the phylogenetic tree. The AMGO and the two ORFs similar to polB and ORF314 were found in mitogenomes of limnomedusans, as reported in staurozoans and discomedusan scyphozoans [7], [11]. However, the mitochondrial genomes of hydroidolinans have at least three gene orders similar to AMGO and lack the two ORFs [7], [8], [9]. Like other hydrozoans, Limnomedusae has a biphysic life history. It consists of a planula stage developing into a benthic polyp and a medusa stage developing via the entocodon. It appears to be easily characterized by two symplesiomorphies, i.e., ecto-endodermal statocysts and a life cycle that includes a polyp stage, shared with other members of Trachylina and Hydroidolina, respectively [2], [25]. Limnomedusae was once classified into a group named Hydroidomedusa (including Anthomedusae, Laingiomedusae, Leptomedusae, Limnomedusae, and Siphonophorae), which was characterized with a polyp stage budding medusae through a medusary nodule [26]. In the cladistic analysis based on 87 morphological and life history characters of medusozoans by Marques and Collins [1], Limnomedusae was identified as either the earliest diverging hydrozoan lineage or as the basal group of either Trachylina or Hydroidolina. However, it was eventually placed at the base of the subclass Trachylina rather than among the hydroidolinan groups according to the molecular data of mitochondrial 16 S and nuclear small and large subunit ribosomal genes [2], [5], [27], [28]. Our ML analyses based on the 13 energy pathway protein genes from mtDNA placed the limnomedusans at the base of Hydrozoa. Considering limnomedusans possess the gene order of AMGO and the shared ancestral characters of Trachylina and Hydroidolina, together with phylogenetic analyses in previous studies, we suggest that Limnomedusae be the earliest diverging lineage among Hydrozoa. However, this suggestion needs to be further examined by analyzing more mitogenomes from other hydrozoans, especially from other trachyline hydrozoans (Narcomedusae, Trachymedusae and Actinulida).
The complete mtDNA sequence of C. sowerbyi and other recently determined mtDNA of medusozoans allow us to analyze the phylogenetic relationship and mitochondrial evolution in Cnidaria, especially in Medusozoa. Generally, our findings in the ML trees were largely congruent with the previous reports based on mitochondrial and/or nuclear data [2], [5], [6], [7], [8], [29], [30]. As indicated in previous studies also employing mitogenomic data, the paraphyly of Anthozoa is further confirmed in the present analyses with octocorallians being closer to the medusozoans than to the hexacorallians [6], [8]. The evolutionary positions of the four medusozoan classes showing in our ML trees, were largely consistent with the speculation based on mitogenome organizations [7] and the phylogenetic analyses based on nuclear and mitochondrial ribosomal data [2], [5] in general. The monophyletic clade of hydrozoans was clustered as a sister group to the monophyletic discomedusan Scyphozoa. Hydrozoa appears to be composed of two well supported groups, Hydroidolina and Trachylina (Limnomedusae). In the group of Hydroidolina, our ML analyses provided strong support for a clade of capitates to the exclusion of aplanulate taxa, which supported the previous studies that divided the Capitata into clades of Aplanulata and non-Aplanulata Capitata [30]. As the newly recognized class in Medusozoa, Staurozoa was removed from Scyphozoa by Marques and Collins [1] for its discrete history and features correspondingly distinct from other cnidarian classes. In the present study we recognize that Staurozoa should be the sister group of all other medusozoans as shown in the ML tree based on amino acids with 100 bootstrap percentage. Previous studies that employed other molecular data [2], [5], [7] and morphological evidence [31] also generated similar hypothesis. Cubozoans were removed from Scyphozoa and given ‘class’ status by Werner [32]. However, the position of Cubozoa within Medusozoa has been rather contentious, with some favoring a hydrozoan affinity [33], [34] and others a closer relationship to scyphozoans [1], [35], [36], [37]. Molecular analyses based on large and small ribosomal genes placed Cubozoa and Scyphozoa at a clade that was the sister group of Hydrozoa [2]. When referred to the mitogenome, the only cubozoan representative with eight linear mitochondrial chromosomes fell into a clade with the only representative of coronate scyphozoans in the phylogenetic analysis based on amino acids. Our analyses had Cubozoa and coronate Scyphozoa forming a clade that was the sister group of the rest scyphozoans and hydrozoans, and it seems that Coronatae is not as close to the rest scyphozoans as we supposed. However, the very limited species from these interesting groups make the hypothesis not compelling enough and further studies including more species need to be carried out.
Materials and Methods
Ethics Statement
No specific permits were required for collection the samples of medusae of Craspedacusta for scientific intention in China. The sampling location was not privately-owned or protected in any way, and no specific permissions were required for the location. All the field studies did not involve endangered or protected species.
Collection of Medusae and Isolation of DNA
Medusae of Craspedacusta were collected from a lake (30° N 29′ 37.21′′, 114°E 43′ 40′′) in Wuhan, Hubei province, China. Specimens were identified as C. sowerbyi according to the description by Jankowski [12] and the sequences alignments of internal transcribed spacers and cytochrome oxidase subunit I (COI). Individual medusae were washed in double-distilled water before being stored in 90% ethanol. Specimens were then soaked for 1 day in TE buffer (pH 8.0) to remove ethanol before the isolation of DNA. Genomic DNA was extracted using a standard sodium dodecylsulfate-proteinase K procedure described by Sambrook et al. [38].
Polymerase Chain Reaction (PCR) and Genome Sequencing
Primers were designed to match highly conserved regions of animal mtDNA genes and were used to amplify short fragments from the conserved genes of cox1, rnl, nad4 [11], cox3 and cob [39]. Then, specific primers (Table S1) were designed and used with Takara® LA PCR kits to amplify mtDNA between rnl and cob in several PCR reactions. The peripheral regions of the molecule were amplified using the hiTAIL-PCR method [40] with specific primers and anchor primers (Table S1). PCR products were cloned into pGEM-T vector (Promega, USA) and sequenced. C. sowerbyi mtDNA sequences are available via Genbank accession number JN593332.
Gene Annotation and Analysis
Transfer RNA (tRNA) genes in the mtDNA were identified by the tRNAscan-SE program [41]; other genes were identified by similarity searches on the NCBI web site using the BLAST program. The secondary structures of tRNA-Met and tRNA-Trp were deduced by comparison with their homologues in A. aurita and H. oligactis.
For analysis of sequence similarities, individual genes were aligned with ClustalW 1.82 [42] using default parameters; alignments were manually adjusted based on either amino acid alignments (for protein coding genes) or inferred rRNA or tRNA secondary structures (for RNA-coding genes). Sequence identities were calculated using local scripts based on BioPerl modules [43]. Codon usage was calculated with the CUSP program in the EMBOSS package [44].
Phylogenetic Analyses
Phylogenetic analyses were conducted on both amino acid and nucleotide sequences of the coding regions of 13 genes related to respiration and oxidative phosphorylation. The inferred sequences were derived from the GenBank files listed in Table S2.
Fifty-one mtDNA coding sequences of the species in Cnidaria and two sequences of the species in Demospongia were used for phylogenetic analysis based on nucleotides and inferred amino acids data, respectively. Mitochondrial coding sequences were aligned based on amino acid alignments using CodonAlign [45] and recoded using the “neutral transition model” coding strategy [46]. Amino-acid sequences of individual proteins were aligned three times with ClustalW 1.82 [41] using different combinations of opening/extension gap penalties: 10/0.2 (default), 12/4 and 5/1. The three alignments were compared using SOAP [47], and only positions that were identical among them were included in phylogenetic analyses. A maximum likelihood (ML) search was performed for the best tree using RAxML 2.2.3 [48]. The program ProtTest version 1.3 suggested mtMAM model of sequence evolution [49], [50] for the ML analyses of the amino acid data, while the program of ModelTest suggested that a GTR model with gamma [51] was the best for ML analyses of the nucleotide data. One hundred replicates for rapid bootstrap analyses [52] were also performed using RAxML, and a 50% majority rule consensus was calculated to determine the support values for each node.
Supporting Information
Phylogenetic analyses by ML methods based on mitochondrial nucleotide data. ML tree obtained from the analysis of aligned and recoded nucleotides of 13 energy pathway protein genes under the GTR+GAMMA model. The branch support values for each node are shown as ML bootstrap percentage. Scale bars indicate number of changes per site.
(TIF)
Primers for amplification of C. sowerbyi .
(DOC)
List of GenBank accession numbers for mitochondrial genomes.
(DOC)
Acknowledgments
The authors thank Dr. Qian Gao for his assistance in the collection and identification of the specimens of the freshwater jellyfish. Thanks also go Mr. Ming Zou and Bei Huang for their help in phylogenetic analysis.
Funding Statement
This research was supported by the National Technology System for Conventional Freshwater Fish Industries (CARS-46-08) and the National Program on Key Basic Research Project (Grant no. 2009CB118705). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Marques AC, Collins AG (2004) Cladistic analysis of Medusozoa and cnidarian evolution. Invert. Biol. 123: 23–42. [Google Scholar]
- 2. Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, et al. (2006) Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol 55: 97–115. [DOI] [PubMed] [Google Scholar]
- 3. Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW (1992) Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci USA 89: 8750–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bridge D, Cunningham CW, DeSalle R, Buss LW (1995) Class-level relationships in the phylum Cnidaria: molecular and morphological evidence. Mol Biol Evol 12: 679–689. [DOI] [PubMed] [Google Scholar]
- 5. Collins AG (2002) Phylogeny of Medusozoa and the evolution of cnidarian life cycles. J Evol Biol 15: 418–432. [Google Scholar]
- 6. Park E, Hwang DS, Lee JS, Song JI, Seo TK, et al. (2012) Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol 62(1): 329–45. [DOI] [PubMed] [Google Scholar]
- 7. Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, et al. (2012) Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol 4(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kayal E, Lavrov DV (2008) The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene 410: 177–186. [DOI] [PubMed] [Google Scholar]
- 9. Voigt O, Erpenbeck D, Wörheide G (2008) A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata . BMC Genomics 9: 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith DR, Kayal E, Yanagihara AA, Collins AG, Pirro S, et al. (2012) First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol 4(1): 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shao Z, Graf S, Chaga OY, Lavrov DV (2006) Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 381: 92–101. [DOI] [PubMed] [Google Scholar]
- 12. Jankowski T (2001) The freshwater medusae of the world: A taxonomic and systematic literature study with some remarks on other inland water jellyfish. Hydrobiologia 462: 91–113. [Google Scholar]
- 13. Kramp PL (1950) Freshwater medusae in China. Proc Zool Soc Lond 120: 165–184. [Google Scholar]
- 14. Kramp PL (1961) Synopsis of the Medusae of the world. Order Limnomedusae. J Mar Biol Ass UK 40: 213–236. [Google Scholar]
- 15. Acker TS, Muscat AM (1976) The ecology of Craspedacusta sowerbii Lankester, a freshwater hydrozoan. Am Midl Nat 95: 323–336. [Google Scholar]
- 16. Boothroyd IKG, Etheredge MK, Green JD (2002) Spatial distribution, size structure, and prey of Craspedacusta sowerbyi Lankester in a shallow New Zealand lake. Hydrobiologia 468: 23–32. [Google Scholar]
- 17. Pennak RW (1956) The fresh-water jellyfish Craspedacusta in Colorado with some remarks on its ecology and morphological degenerataion. T Am Microsc Soc 75: 324–331. [Google Scholar]
- 18. Silva WM, Roche KF (2007) Occurrence of the freshwater jellyfish Craspedacusta sowerbii (Lankester, 1880) (Hydrozoa, Limnomedusae) in a calcareous lake in Mato Frosse do Sul, Brazil Biota Neotropica. 7: 227–229. [Google Scholar]
- 19. Jankowski T, Collins AG, Campbell R (2008) Global diversity of inland water cnidarians. Hydrobiologia 595: 35–40. [Google Scholar]
- 20. Fritz GB, Pfannkuchen M, Reuner A, Schill RO, Brümmer F (2009) Craspedacusta sowerbii, Lankester 1880– population dispersal analysis using COI and ITS sequences. J Limnol 68: 46–52. [Google Scholar]
- 21. Zhang LQ, Wang GT, Yao WJ, Li WX, Gao Q (2009) Molecular systematics of medusae in the genus Craspedacusta (Cnidaria: Hydrozoa: Limnomedusae) in China with the reference to the identity of species. J Plankton Res 31: 563–570. [Google Scholar]
- 22. Lavrov DV, Forget L, Kelly M, Lang BF (2005) Mitochondrial genomes of two demosponges provide insights into an early stage of animal evolution. Mol Biol Evol 22: 1231–1239. [DOI] [PubMed] [Google Scholar]
- 23. Medina M, Collins AG, Takaoka TL, Kuehl JV, Boore JL (2006) Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci U. S. A. . 103: 9096–9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Oppen MJ, Catmull J, McDonald BJ, Hislop NR, Hagerman PJ, et al. (2002) The mitochondrial genome of Acropora tenuis (Cnidaria; Scleractinia) contains a large group I intron and a candidate control region. J Mol Evol 55: 1–13. [DOI] [PubMed] [Google Scholar]
- 25.Petersen KW (1979) Development of coloniality in Hydrozoa. In: Larviood G and Rosen BR. Biology and systematics of colonial organisms. New York: Academic Press. 105–139.
- 26. Bouillon J, Boero F (2000) The Hydrozoa: a new classification in the light of old knowledge. Thalass Salent 24: 3–5. [Google Scholar]
- 27. Collins AG (2000) Towards understanding the phylogenetic history of Hydrozoa: hypothesis testing with 18 S gene sequence data. Scient Mar 64: 5–22. [Google Scholar]
- 28. Collins AG, Bentlage B, Lindner A, Lindsay D, Haddock SHD, et al. (2008) Phylogenetics of Trachylina (Cnidaria: Hydrozoa) with new insights on the evolution of some problematical taxa. J Mar Biol Assoc 88: 1673–1685. [Google Scholar]
- 29. Kim J, Kim W, Cunningham CW (1999) A new perspective on lower metazoan relationships from 18 S rDNA sequences. Mol Biol Evol 16: 423–427. [DOI] [PubMed] [Google Scholar]
- 30. Cartwright P, Evans NM, Dunn CW, Marques AC, Miglietta MP, et al. (2008) Phylogenetics of Hydroidolina (Hydrozoa: Cnidaria). J Mar Biol Assoc 88: 1663–1672. [Google Scholar]
- 31. Van Iten H, De Moraes LJ, Simoes MG, Marques AC, Collins AG (2006) Reassessment of the phylogenetic position of conulariids (?Ediacaran-Triassic) within the subphylum medusozoa (Phylum Cnidaria). J Syst Palaeontol 4: 109–118. [Google Scholar]
- 32. Werner B (1973) New investigations on systematics and evolution of the class Scyphozoa and the phylum Cnidaria. Pub Seto Mar Biol Lab 20: 35–61. [Google Scholar]
- 33. Petersen KW (1990) Evolution and taxonomy in capitate hydroids and medusae (Cnidaria: Hydrozoa). Zool J Linn Soc 100: 101–231. [Google Scholar]
- 34. Leonard J L (1980) Cubomedusae belong to the class Cubozoa, not Scyphozoa. Nature 284: 377. [Google Scholar]
- 35. Salvini-Plawen (1978) On the origin and evolution of the lower Metazoa. Z Zool Syst Evolutions 16: 40–88. [Google Scholar]
- 36. Satterlie RA, Spencer AN (1980) Reply to Leonard’s paper Cubomedusae belong to the class Cubozoa, not Scyphozoa. Nature 284: 377. [Google Scholar]
- 37. Schuchert P (1993) Phylogenetic analysis of the Cnidaria. J ZOOL SYST EVOL RES 31: 161–173. [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T (eds) (1989) Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press.
- 39. Boore JL, Brown WM (2000) Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol 17: 87–106. [DOI] [PubMed] [Google Scholar]
- 40. Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–656. [DOI] [PubMed] [Google Scholar]
- 41. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stajich JE, Block D, Boulez K (2002) The bioperl toolkit: perl modules for the life sciences. Genome Res 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 45.Hall BG (2004) Phylogenetic Trees Made Easy: A How-to Manual, 2nd ed. Sunderland, Massachusetts: Sinauer Associates.
- 46. Hassanin A, Leger N, Deutsch J (2005) Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst Biol 54: 277–298. [DOI] [PubMed] [Google Scholar]
- 47. Löytynoja A, Milinkovitch MC (2001) SOAP, cleaning multiple alignments from unstable blocks. Bioinformatics 17: 573–574. [DOI] [PubMed] [Google Scholar]
- 48. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 49. Cao Y, Janke A, Waddell PJ, Westerman M, Takenaka O, et al. (1998) Conflict amongst individual mitochondrial proteins in resolving the phylogeny of eutherian orders. J Mol Evol 47: 307–322. [DOI] [PubMed] [Google Scholar]
- 50. Yang Z, Nielsen R, Hasegawa M (1998) Models of amino acid substitution and applications to mitochondrial protein evolution. Mol Biol Evol 15: 1600–1611. [DOI] [PubMed] [Google Scholar]
- 51. Posada D, Crandall KA (1998) Modeltest: Testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 52. Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57: 758–771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analyses by ML methods based on mitochondrial nucleotide data. ML tree obtained from the analysis of aligned and recoded nucleotides of 13 energy pathway protein genes under the GTR+GAMMA model. The branch support values for each node are shown as ML bootstrap percentage. Scale bars indicate number of changes per site.
(TIF)
Primers for amplification of C. sowerbyi .
(DOC)
List of GenBank accession numbers for mitochondrial genomes.
(DOC)