Abstract
Cajal bodies (CB) are subnuclear domains that contain various proteins with diverse functions including the CB marker protein coilin. In this study, we investigate the proteolytic activity of calpain on coilin. Here we report a 28 kDa cleaved coilin fragment detected by two coilin antibodies that is cell cycle regulated, with levels that are consistently reduced during mitosis. We further show that an in vitro calpain assay with full length or C-terminal coilin recombinant protein releases the same size cleaved fragment. Furthermore, addition of exogenous RNA to purified coilin induces proteolysis by calpain. We also report that the relative levels of this cleaved coilin fragment are susceptible to changes induced by various cell stressors, and that coilin localization is affected by inhibition or knockdown of calpain both under normal and stressed conditions. Collectively, our data suggest that coilin is subjected to regulated specific proteolysis by calpain, and this processing may play a role in the regulation of coilin activity and CB formation.
Introduction
Cajal bodies (CBs) are subnuclear domains that are best characterized for their role in small nuclear ribonucleoprotein (snRNP) biogenesis, but also contain factors necessary for small nucleolar RNP (snoRNP) maturation, histone pre-mRNA processing and telomerase assembly (Spector et al. 1992; Gall 2000; Morris 2008; Matera et al. 2009) (Carmo-Fonseca 2002; Hebert 2010). Of note, CBs contain guide RNAs (small Cajal-body specific RNAs) that direct proper modification of the snRNA components of certain spliceosomal snRNPs (Darzacq et al. 2002; Jady et al. 2003). The marker protein for CBs is considered to be coilin, which is a phosphoprotein (Carmo-Fonseca et al. 1993) that also localizes to the nucleoplasm. Coilin phosphorylation increases during mitosis (Carmo-Fonseca et al. 1993), and this increased phosphorylation is correlated with altered protein interactions, reduced self-association and CB disassembly (Toyota et al. 2010) (Carmo-Fonseca et al. 1993; Hearst et al. 2009) (Hebert and Matera 2000). Mutation of coilin phosphorylation sites (known or predicted) has also been demonstrated to impact cellular proliferation and CB formation as well as coilin localization and stability (Lyon et al. 1997; Hebert and Matera 2000; Hearst et al. 2009; Gilder et al. 2011). Recently, bacterially expressed coilin has been shown to have RNase and DNA binding activity and be tightly associated with both RNA and DNA (Broome and Hebert 2012).
Another protein that can be found in the CB is SMN, the survival of motor neuron protein. Mutations in SMN lead to the disease spinal muscular atrophy (SMA), which is the leading genetic cause of infant mortality. SMN is also found in the cytoplasm, and while this cytoplasmic fraction is phosphorylated, nuclear SMN is relatively dephosphorylated (Grimmler et al. 2005), (Petri et al. 2007). A major function of SMN is the ordered assembly of core Sm proteins onto the snRNA of spliceosomal snRNPs during the cytoplasmic phase of snRNP biogenesis. SMN has been shown to be a target of calpains, proteases that regulate substrate activity by conducting limited cleavage of their targets (Walker et al. 2008), (Fuentes et al. 2010). Cleavage of SMN by calpain occurs in the cytoplasm (Fuentes et al. 2010), hinting at the possibility that aspects of the snRNP biogenesis role of cytoplasmic SMN could be impacted by this event and the resultant cleavage products.
In contrast to the abundance of data clarifying the role of SMN, the function of coilin is less well understood, but shown to be necessary for the proper formation and composition of CBs. In some cell lines and organisms (mouse and zebrafish), but not in a Drosophila model, coilin reduction or knockout decreases cellular proliferation and viability (Strzelecka et al. 2010; Liu et al. 2009; Tucker et al. 2001; Walker et al. 2009) (Lemm et al. 2006; Whittom et al. 2008). Although coilin and its interaction with other factors in the CB may be important for the formation of this nuclear structure, other studies have shown that additional components, as well as certain types of RNA, can nucleate a CB or impact its formation (Hebert 2010) (Tuma and Roth 1999; Kaiser et al. 2008) (Shevtsov and Dundr 2011) (Mahmoudi et al. 2010). Hence, while coilin plays a crucial role in the formation and composition of CBs, it is not the only factor needed to seed and grow a CB (Shevtsov and Dundr 2011).
In order to fully understand CB formation and function, it is crucial that all inhabitants of this nuclear structure are identified and modifications of these molecules fully characterized. Part of this process will require a thorough examination of the functional consequence of phosphorylation in the CB (Hebert 2010). Another aspect of this characterization is the elucidation of regulated specific proteolysis events, in contrast to proteolysis associated with total protein degradation, of a given CB protein. In 1993, Chan and colleagues described the generation of a rabbit polyclonal antibody (R288) against coilin (Andrade et al. 1993). The authors noted that this antibody, as well as some human anti-coilin serum, detected lower molecular weight fragments in addition to full-length coilin and stated “the identity of these lower molecular mass reactivities may be interesting and need to be determined.” It was suggested by these same authors that these lower molecular mass bands detected by R288 and other antibodies were degradation products or processed fragments of coilin. We have also observed these lower molecular weight fragments detected by coilin antibodies, and in this work describe the origin and regulation of the most prominent of these reactivities. Our findings demonstrate the existence of a “cleaved” fragment of coilin, most likely first identified in 1993 (Andrade et al. 1993), whose levels change during mitosis and during cell stress. In vitro studies demonstrate that calpain-1 processing of coilin can generate the cleaved fragment observed in cell lysate. We further show that the calpain inhibitor ALLN induces CB formation in primary and transformed cell lines and demonstrate that bound RNA on coilin increases the in vitro processing of coilin by calpain. We also observe that depletion of calpain levels attenuate coilin nucleolar localization. Given coilin’s central role in the formation and composition of CBs, the results presented here indicate that targeted processing of coilin is another variable to be considered in the context of CB assembly and activity. Additionally, or alternatively, the cleavage of coilin by calpain and the generated product, may impact the function of nucleoplasmic coilin, which is presently not known.
Materials and Methods
Cell culture, DNA constructs and transfections
Human cervical carcinoma (HeLa) and normal human fetal lung fibroblast (WI38) cells were obtained from the American Type Culture Collection (Manassas, VA). Human osteosarcoma cells (U2OS), from the American Type Culture Collection (Manassas, VA), were a gift from Dr. Luis Martinez (The University of Mississippi Medical Center, Jackson, MS). Cell lines were cultured as described (Sun et al. 2005). His-tagged coilin constructs, and GFP-coilin and Coilin-GFP constructs were previously described (Hebert and Matera 2000; Hearst et al. 2009; Toyota et al. 2010). The recombinant proteins were induced and purified as described previously (Gilder et al. 2011).
RNAi and drug treatment
Coilin siRNA (N004645.12.4) (Toyota et al. 2010) and calpain subunit 4 siRNA (N001749.12.1) were obtained from Integrated DNA Technology (Coralville, IA, USA). The non-targeting, control siRNA (Carrero et al. 2011) was obtained from Thermo Scientific (LaFayette, CO, USA). The siRNA transfections were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Mitotic cells were obtained from shake off or nocodazole (0.4 μg/ml) treatment for 16 hours. Drug treatments were as follows unless otherwise stated: actinomycin D (2.5 μg/ml) for 2 hours, cisplatin (3 μg/ml) for 16 hours or etoposide (20 μM) for 16 hours. The calpain-1 inhibitor, ALLN, was obtained from EMD Chemicals (Gibbstown, NJ, USA) and cells were treated at a final concentration of 10 uM ALLN or DMSO (vehicle) for 16 hrs.
Cell lysis and calpain assay
Cells were lysed using RIPA buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS, 1 mM EDTA) supplemented with protease inhibitor cocktail tablets (Roche, Indianapolis, IN, USA) and the lysates were prepared as described before (Hearst et al. 2009). Calpain assays were carried out either with cell lysate or bacterially expressed proteins with addition of 0.05–1.0 U of calpain-1 (EMD Chemicals, Gibbstown, NJ, USA) and 1 mM CaCl2 in each reaction. The mixture of cell lysate or bacterial expressed protein, CaCl2 and calpain was incubated at 30°C for 15 minutes. In addition to the reaction mixture, 0.5 mM ALLN or HeLa total RNA (300 ng) was added to each reaction wherever mentioned. After the incubation, the reactions were immediately stopped with addition of SDS sample loading buffer and boiled for 5 minutes at 95°C. The samples were then subjected to SDS-PAGE then either Coomassie stain or Western transfer.
Electro elution and dialysis of coilin
GST-tagged coilin protein was purified as previously described (Broome and Hebert 2012), with minor changes: prior to separation by SDS-PAGE, protein was incubated at 37°C for 30 min with DNase I and RNase A/T1 cocktail (Invitrogen, Carlsbad, CA, USA). The concentrated electro-eluted coilin protein was incubated on ice for 30 min in a final concentration of 21 mM α-cyclodextrin, 259 mM NaCl and 1.34 μM coilin. Following incubation, reaction was centrifuged for 5 m at 16000 g, and the supernatant dialyzed against 1X PBS high salt buffer containing 10 mM Na2HPO4, 1.8 mM NaH2PO4 and 250 mM NaCl. Dialysis was performed using a 10000 MWCO Pierce Slide-A-Lyzer Dialysis Cassette from Thermo Scientific (LaFayette, CO, USA) following the manufacturer’s suggested protocol. Dialyzed coilin was incubated with or without HeLa total RNA prior to incubation with calpain.
Cell lysate fractionation
HeLa cells were incubated for 16 h in a final concentration of 0.4μg/mL nocodazole to achieve mitotic arrest. Nuclear isolation and cell fractionation was performed as previously described (Citterio et al. 2004; Remboutsika et al. 1999) on both nocodazole and control treated cells, with modifications. Resulting pellets from nuclei isolation protocol were digested with 4 units DNaseI (Invitrogen, Carlsbad, CA, USA) per mL in buffer B (10 mM PIPES pH 6.8, 100 mM KCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA) for 45 min at 37°C followed by centrifugation at 1,500 × g for 10 min at 4°C. Supernatant at this point corresponds to DNase fraction. The pellet was extracted with 250 mM ammonium sulfate in buffer B and centrifuged at 4,000 × g for 5 min at 4°C, releasing the ammonium sulfate (AS) fraction. Next, the pellet was solubilized with 2 M NaCl in buffer B, followed by centrifugation at 350,000 × g for 20 min. The final pellet was solubilized in 4X SDS sample loading buffer (200 mM Tris-HCl pH 6.8, 8% SDS, 6% DTT), sonicated and boiled prior to loading onto SDS PAGE gel. All other fractions were diluted to 1X in sample loading buffer (50 mM Tris-HCl pH 6.8, 2% SDS, 1.5% DTT) and boiled prior to loading onto SDS-PAGE gel.
Western blotting, immunofluorescence and antibodies
Western blotting, immunofluorescence and image acquisition were performed as previously described (Sun et al. 2005). The following antibodies were used: Rabbit polyclonal anti-coilin H300 (1:500 Western, 1:200 IF) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), mouse monoclonal anti-coilin pδ (1:500) (Abcam, Cambridge, MA, USA), mouse monoclonal anti-β-tubulin (1:1000) (Sigma-Aldrich, St. Louis, MO, USA), mouse monoclonal anti-SMN (1:200) (BD Transduction Laboratories, San Jose, CA, USA), mouse monoclonal anti-fibrillarin (1:50), rabbit polyclonal anti-acetylated histone H3 (H3K14) (1:1000) (Upstate, Lake placid, NY, USA), mouse monoclonal anti-GFP (1:500) (Roche, Mannheim, Germany) and mouse monoclonal anti-calpain small subunit (calpain 4) (1:1000 western, 1:200 IF) (Millipore, Temecula, CA, USA).
Results
Identification of a cell cycle regulated coilin fragment
To begin our investigation into the putative processed fragments detected with coilin antibodies, we examined the ratio of the “cleaved” signal to full length coilin in interphase and mitotic lysates from both HeLa and U2OS cell lines (Figure 1A). A polyclonal antibody raised against the C-terminal 300 amino acids (aa 276–576) of human coilin was used to detect both full length coilin as well as any lower molecular weight reactivities. This antibody (H300 from Santa Cruz) should therefore react with similar epitopes as found for R288, which was generated against a 14 kDa. C-terminal coilin fragment and used initially to describe these lower molecular weight reactivities (Andrade et al. 1993). In both cell lines, a distinct signal can be detected from interphase lysates approximately 28 kDa. in size. Relative to full length coilin, the amount of this “cleaved” signal is reduced in both lines during mitosis. To demonstrate that this cleaved signal is derived from coilin and not from antibody cross reactivity to another protein, we knocked down coilin message with coilin siRNA for 24, 48 and 72 hr and observed a decrease in both full length coilin and the cleaved product when compared to control siRNA (Figure 1B and Supp. Figure 1A). The ratio of full-length coilin and the coilin cleaved product to tubulin in the presence of coilin siRNA was then analyzed and normalized to the ratios found with control siRNA (Supp. Figure 1B). These findings indicate that the coilin cleavage fragment has a longer half-life than full-length coilin. We also used a coilin monoclonal antibody (pδ, (Almeida et al. 1998)) and could detect the same size cleaved fragment with this antibody as found for the coilin polyclonal antibody H300 (Figure 1C). Epitope mapping has demonstrated that pδ reacts with epitopes located between aa 363–481 of coilin (Almeida et al. 1998), so this region of coilin, or at least a part of it, must be present in the cleaved fragment detected by Western blotting.
Figure 1.
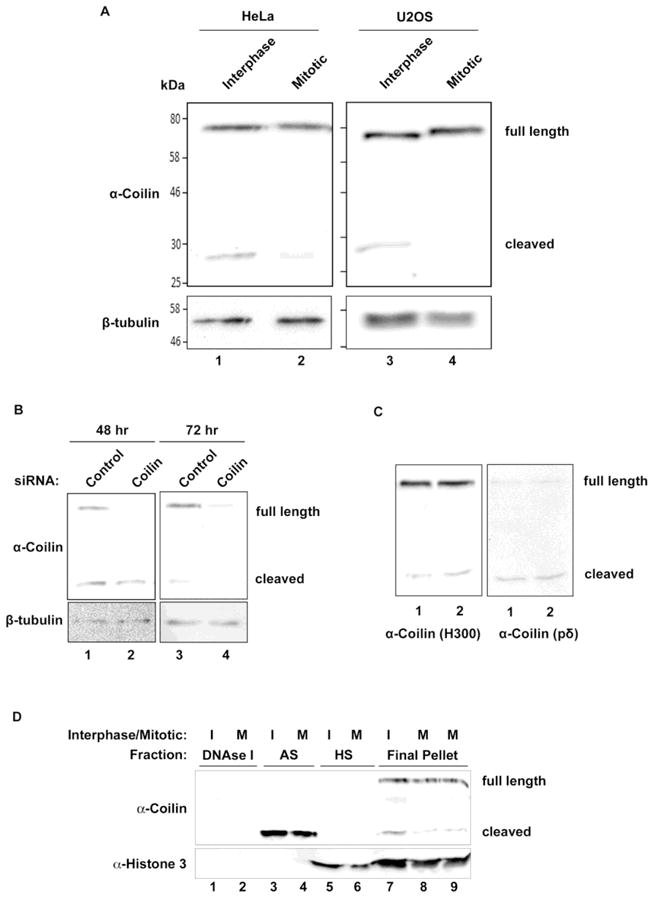
Full length coilin cleaved to 28 kDa. fragment in cells. (A) Cleaved coilin fragment levels differ between interphase and mitotic lysates of HeLa and U2OS cells. HeLa and U2OS cells were untreated or treated with nocodazole to achieve mitotic arrest. Interphase and mitotic cell lysates were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin and anti-β-tubulin antibodies. (B) Coilin siRNA treatment reduces cleaved coilin fragment levels. HeLa cells were harvested 48 and 72 hr post-transfection with control or coilin-targeted siRNA, lysates were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin and anti-β-tubulin antibodies. (C) Cleaved fragment recognized by both poly- and mono-clonal coilin antibodies. U2OS lysates were subjected to SDS-PAGE and Western transfer. The blot was probed with anti-coilin H300 antibodies, followed by stripping and reprobing of the blot with anti-coilin pδ antibodies. (D) Cleaved coilin fragment solubility is different from full length coilin. HeLa cells were untreated or nocodazole treated to achieve interphase (I) and mitotic cells (M). Cells were fractionated as described in methods, and equal volumes of each fraction (DNase I, ammonium sulfate (AS), high salt (HS) and final pellet) were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin antibodies (top panel) and anti-acetyalated histone 3 (H3K14) antibodies (bottom panel).
In the work that first described lower mobility signals reactive to coilin antibodies, the solubility of coilin during the cell cycle was examined using 150 mM NaCl, 10 mM Tris-HCl, pH 7.2, 0.5% Nonidet P-40 to lyse cells and obtain soluble and pellet fractions (Andrade et al. 1993). It was shown in this work that the majority of coilin was in the pellet fraction during G1 and S phase, but predominantly found in the soluble fraction during mitosis (Andrade et al. 1993). Interestingly, the most prominent lower mobility band identified by Andrade et al., which corresponds to the size of the cleaved fragment we report here, is exclusively found in the soluble fraction, regardless of cell cycle (Andrade et al. 1993). To extend these studies, we conducted sequential extraction of HeLa interphase nuclei and mitotic cells using progressively more stringent conditions in order to ascertain if full length coilin possesses different solubility characteristics as compared to the cleaved fragment. Indeed, we found that the majority of the cleaved fragment is found in the ammonium sulfate fractions whereas the bulk of full length coilin remains in the insoluble pellet (Figure 1D). Taken together, these data demonstrate that a putative cleaved fragment of coilin can be detected by different coilin antibodies, reduced by coilin siRNA and cell cycle regulated. These observations were similar in both cell lines, one of which (U2OS) has a functional p53 response to DNA damage (Liu et al. 2008), while the other (HeLa) lacks this capability. We also show that the cleaved coilin fragment is more soluble than full length coilin.
Coilin is an in vitro target of calpain-1 cleavage
Recent work has demonstrated that SMN is subject to Ca2+-dependent calpain cleavage (Walker et al. 2008), (Fuentes et al. 2010). In contrast to proteases that act to fully degrade a substrate protein, calpains are calcium dependent cysteine proteases that perform limited digestion of the target protein, thereby regulating the activity of that protein. Calpains are involved in many different cellular processes, and calpain-1 (μ-calpain) has been shown to cleave SMN (Walker et al. 2008) (Fuentes et al. 2010). To determine if calpains also play a role in the generation of the coilin lower molecular weight fragment, we incubated bacterially expressed recombinant His-tagged coilin with calpain-1. As shown in Figure 2A, a cleaved product approximately the same size as that observed in lysate is detected upon incubation of bacterially purified His-coilin with CaCl2 and calpain-1. An additional coilin fragment (arrow) is also observed at around 50 kDa. Since this fragment is detectable in the reactions lacking calpain, it most likely represents a coilin degradation fragment isolated along with full length coilin during protein isolation. We next examined the effect of adding a large amount of exogenous calpain-1 to lysate and observed that excess protease can completely degrade both coilin and the cleaved product (Figure 2B, lane 3). The addition of N-Acetyl-Leu-Leu-Norleu-al (ALLN), an inhibitor of calpain-1 with secondary targets of calpain-II and other neutral cysteine proteases, completely abolished the processing activity of calpain-1 (lane 4). In order to map the region of coilin from which the cleaved product detected by anti-coilin antibodies is derived, we incubated bacterially purified His-tagged full length coilin or fragments thereof with CaCl2 in the presence or absence of calpain-1 (Figure 3). The cleaved product could be observed when full length coilin or coilin 94–576 was incubated with calpain (Figure 3B, lane 2; Figure 3C, lane 2 and 3). However, a similar sized cleaved product was not detected when the coilin N-terminal 362 aa (N362), C-terminal 214 aa (C214), coilin 94–450 or coilin 94–475 fragments were exposed to calpain-1 (Figure 3B, lanes 4 and 6; Figure 3C, lane 5, 6, 8 and 9). Thus these coilin fragments are not sufficient to liberate the cleaved product seen upon full length coilin and coilin 94–576 processing by calpain-1. It should be noted that N362 is a better substrate for calpain relative to C214 when considering the extent of degradation compared to the respective no calpain control. It should also be noted that higher amounts of calpain lead to increased amounts of degradation products that are smaller than the 28 kDa. cleaved product. Also, at these higher amounts of calpain, the level of the 28 kDa. cleaved fragment is reduced compared to reactions with less calpain, presumably because the 28 kDa. fragment itself is subject to calpain (in Figure 3C, compare the intensity of the 28 kDa. cleaved fragment and smaller fragments in lane 2 to that in lane 3).
Figure 2.
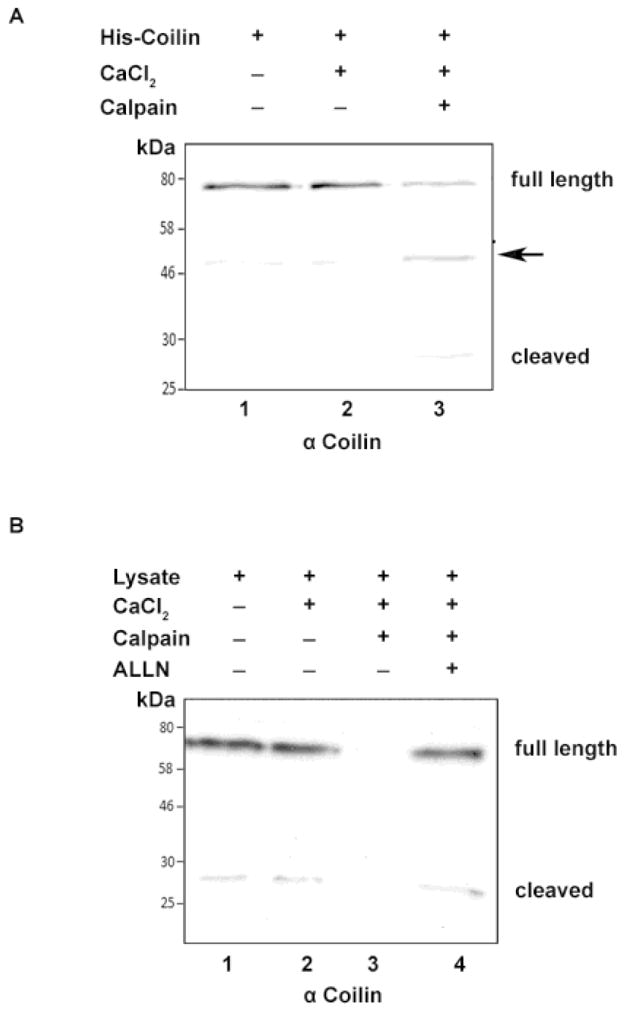
Coilin is a substrate for calpain-1. (A) Calpain digests bacterial expressed His-T7-tagged coilin. His-coilin was incubated alone, with 1mM CaCl2 or with both 1mM CaCl2 and calpain-1 (0.5 U) at 30°C for 15 minutes. Following incubation, reactions were subjected to SDS-PAGE, Western transfer, and blots probed with polyclonal H300 anti-coilin antibodies. An arrow marks an additional coilin fragment of around 50 kDa. (B) Calpain digests endogenous coilin. U2OS cell lysates were incubated alone, or with a combination of 1mM CaCl2, calpain (1U), and the calpain inhibitor ALLN (0.5 mM) at 30°C for 15 minutes. Following incubation, reactions were subjected to SDS-PAGE, Western transfer, and blots probed with polyclonal anti-coilin H300 antibodies.
Figure 3.
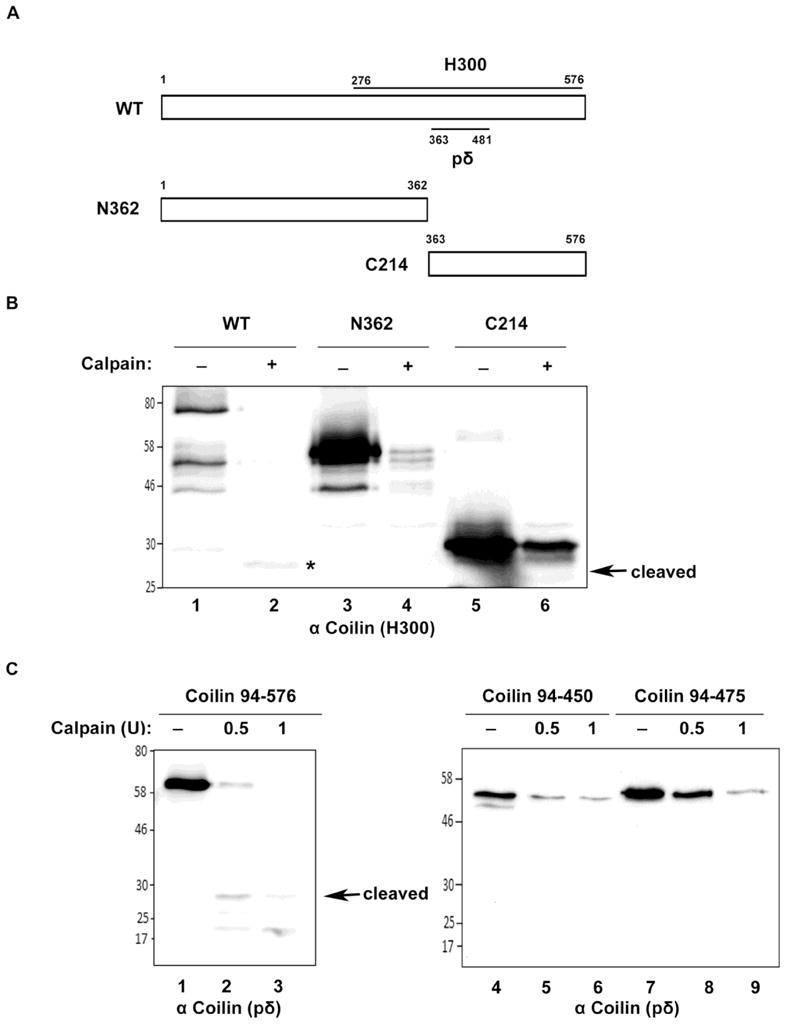
Mapping of coilin digestion by calpain. (A) Schematic representation of full length coilin (WT) and N-terminal (N362) and C-terminal (C214) coilin fragments. The location of the epitopes to which the anti-coilin antibodies H300 and pδ react are shown. (B) Full length coilin, but not N362 or C214, releases cleaved coilin fragment. His-tagged WT, N362 and C214 were incubated with or without calpain in presence of 1mM CaCl2 at 30°C for 15 minutes. The reactions were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin H300 antibodies. Asterisk (*) indicates the cleaved coilin fragment. (C) Coilin 94–576 releases the 28 kDa. cleaved coilin fragment. His-tagged coilin constructs containing amino acids 94–576, 94–450 or 94–475 were incubated with 0.5 or 1 U calpain in presence of 1mM CaCl2 at 30°C for 15 min. Reactions were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin pδ antibodies.
The region of coilin detected by the coilin monoclonal antibody pδ, and the location of the aa 276–576 fragment used to generate the coilin H300 polyclonal antibody are shown in Figure 3A. Since these antibodies react with the C-terminal half of coilin, it is possible that the cleaved product generated by calpain is the result of processing events in this region of the protein (from aa 276–576). In order to provide additional evidence that the cleaved fragment arises from an internal region of coilin, we utilized GFP-coilin and Coilin-GFP expressed in HeLa cells. Following a 24 h transfection, the cells were harvested and lysates subjected to calpain digestion, SDS-PAGE, Western transfer, and probed with anti-GFP and anti-coilin antibodies. As shown in Supp. Figure 2, the 28 kDa coilin fragment is present in the blot probed with anti-coilin but is not visible with anti-GFP. These results further suggest that the cleaved coilin fragment is processed from internal sites of full length coilin, and does not contain the extreme N- or C-terminus. The preferential detection of coilin-GFP compared to GFP-coilin by GFP antibodies may indicate differing GFP epitope accessibility on the membrane. Reprobing of the same blot with anti-coilin antibodies clearly shows that GFP-coilin and coilin-GFP are equally present in the lysate. Online calpain cleavage site predictors, such as the Calpain for Modulatory Proteolysis Database (CaMPDB), indicate multiple locations within coilin susceptible to calpain cleavage. The highest scoring of these is centered around aa 136. Consequently, it possible that calpain cleaves in this location and in the C-terminus between residues 475 and 576 in order to generate the 28 kDa. cleaved product detected here.
In vitro RNA addition aids calpain cleavage of coilin
We have found that recombinant coilin isolated from bacteria co-purifies with nucleic acids (Broome and Hebert 2012), and therefore hypothesized that presence of RNA might influence the cleavage of coilin by calpain. To investigate if RNA impacts the ability of calpain to cleave coilin, bacterially expressed coilin was purified in the absence of nucleic acids by treatment with DNase I and RNase A/T1 before isolation by SDS-PAGE, electroelution and dialysis (see Methods). After purifying nucleic acid-free coilin, incubations were set up with CaCl2, increasing concentrations of calpain (0.05 or 0.2 U) and exogenous RNA (Figure 4A). The amount of full length coilin is most reduced in the reactions containing exogenous RNA, evident by comparison of lanes 2 and 4 to lanes 3 and 5 of a Coomassie stained SDS-polyacrylamide gel (Figure 4A). Since the coilin degradation fragments are not easily observed upon Coomassie staining, a portion of the same reactions shown in Figure 4A were also subjected to Western transfer after SDS-PAGE and probed with both polyclonal and monoclonal coilin antibodies (Figure 4B). In addition to the 28 kDa. cleaved coilin fragment, additional coilin fragments are released upon calpain digestion (Figure 4B). These fragments (labeled a, b, and c) are differentially detected by both coilin antibodies, and the smallest of these fragments (c) is at its most abundant with 0.2 U calpain and RNA. As observed previously in Figure 3C, higher levels of calpain generate reductions in the amount of the 28 kDa. cleaved fragment and corresponding increases in the amount of small degradation products, such as those marked by (c) in Figure 4B. These results indicate that coilin may be sequentially degraded and the 28 kDa cleaved product may be more stable than the other degradation fragments, and hence is detectable in lysate. These results also indicate that RNA renders coilin more susceptible to calpain cleavage in vitro.
Figure 4.
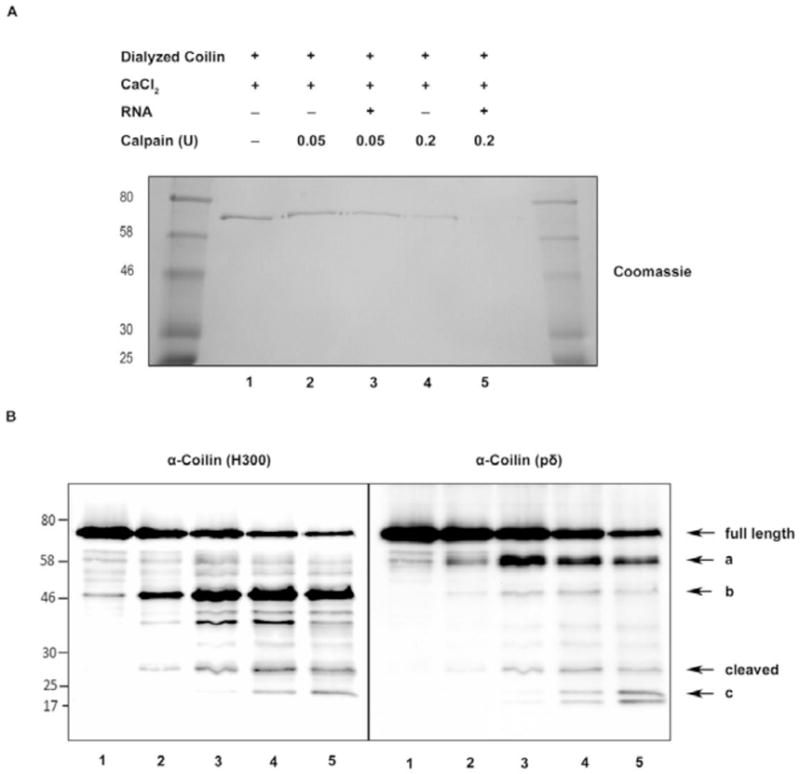
RNA addition makes coilin more vulnerable to calpain digestion. (A) Coomassie blue stained SDS-polyacrylamide gel shows untreated and calpain digested coilin levels. DNA and RNA free bacterial expressed dialyzed coilin was digested with increasing concentrations of calpain in the presence of 1 mM CaCl2 at 30°C for 15 min, following pre-incubation (5 min) of dialyzed coilin with or without HeLa total RNA. A portion of the reactions was subjected to SDS-PAGE and stained with Coomassie. (B) The remainder of the reactions was subjected to SDS-PAGE then Western transfer, and blots probed with anti-coilin H300 antibodies (left panel) and anti-coilin pδ antibodies (right panel). Note the lane assignments in (B) are the same as in (A). The letters a, b and c indicate other fragments released from full length coilin upon calpain digestion in addition to the cleaved fragment.
Cellular stress influences the level of the coilin cleavage product
Previous work has shown that coilin localization changes in response to cell stress, such as that induced by treatment with the transcription inhibitor actinomycin D (ActD) (Carmo-Fonseca et al. 1992) or the anti-cancer agents cisplatin and etoposide (Gilder et al. 2011). Specifically, the amount of coilin signal around and within the nucleolus increases upon exposure to these compounds. To examine if this change in coilin localization correlates with alterations in the amount of the coilin cleaved product, we treated both HeLa and U2OS cells with these three compounds and monitored the level of the cleaved product. As shown in Figure 5, compared to the untreated ratio, the relative amount of the cleaved product to full length coilin in HeLa cells is increased upon ActD treatment but decreased when exposed to cisplatin or etoposide (Figure 5A and B). Cisplatin- and etoposide-mediated reductions in the level of the cleaved product to full length coilin was also observed in U2OS cells (Figure 5C and D). However, in contrast to HeLa, the relative amount of cleaved product did not increase in ActD treated U2OS cells. We have observed that U2OS cells have a greater percentage of variability in regards to coilin localization compared to HeLa cells. Whereas the majority of HeLa cells contain coilin in typically 4 to 6 CBs, and lack cells with nucleolar coilin signal, U2OS cells contain a more broad distribution of coilin in which approximately 25% of cells lack CBs or contain nucleolar coilin. It is possible that this wide range of coilin signal is indicative of the overall level of the cleaved fragment considering that the relative amount of the cleaved product is three times greater in U2OS compared to HeLa cells (Figure 5E). It should be pointed out that the conditions used to generate the lysate in these and the previous experiments (RIPA + sonication) are expected to fully extract both full length coilin and the cleaved fragment and indeed, centrifugation of the lysate after sonication for 10 minutes at 16000 g does not result in a detectable pellet. Therefore, observed alterations in the level of the cleaved fragment are not likely caused by differential extraction.
Figure 5.
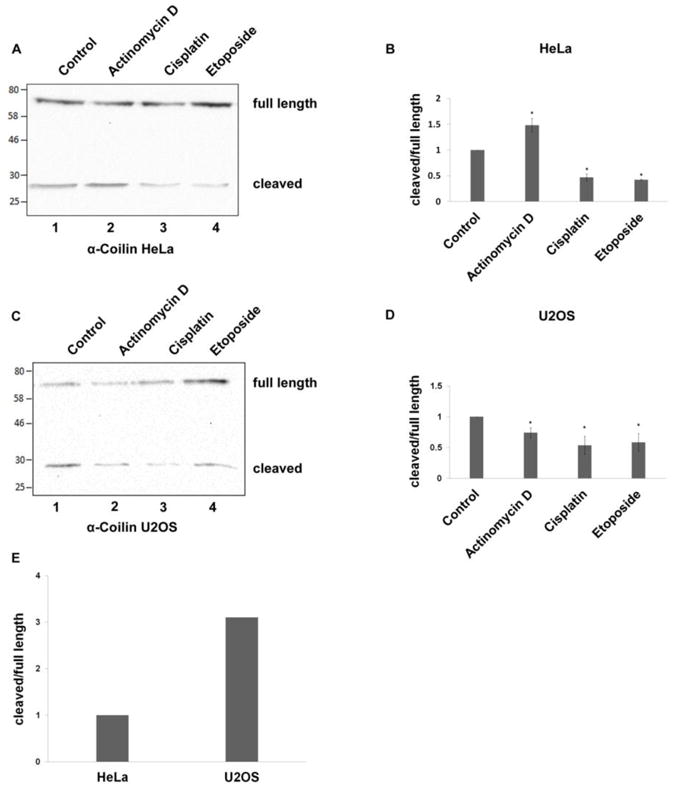
Cellular stress influences cleaved coilin fragment levels. Cleaved coilin fragment levels in HeLa (A and B) and U2OS (C and D) cells untreated or treated with actinomycin D, cisplatin, or etoposide. The treated and untreated cell lysates were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin antibodies. The ratio of cleaved coilin fragment to full length coilin was quantified by densitometric analysis (average of three independent experiments (n=3), error bars represent mean±SD). (E) Typical ratio of cleaved coilin fragment to full length coilin in untreated HeLa and U2OS cells. HeLa and U2OS cell lysates were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin H300 antibodies. The ratio of cleaved coilin fragment to full length coilin was quantified with densitometric analysis. Each ratio represents the average of three independent experiments (n=3).
The calpain inhibitor ALLN increases CB number in U2OS and WI-38 cells
We have found that coilin is a target for calpain-1 and the level of the cleaved product relative to full-length coilin is cell cycle regulated and can vary upon specific treatments. Additionally, the relative amount of the cleaved product is greater in U2OS cells, which have a more broadly distributed coilin localization compared to HeLa, in which the majority of cells contain coilin in CBs. These findings led us to test if calpain inhibition influences CB number and coilin localization. For these studies, we utilized the U2OS line as well as the primary fibroblast WI-38 cell line, which contain CBs only in a small percentage of cells. Cells were exposed to DMSO (vehicle) or ALLN, followed by immunofluorescence using coilin and SMN antibodies (Figure 6). In both cell lines the percentage of cells with CBs or coilin foci (which lack SMN colocalization) increased after ALLN treatment (Figure 6A and B). To test if this increase correlated with a change in the relative amount of the cleaved product, cells were unexposed or treated with DMSO or ALLN, followed by lysate generation and Western blotting for coilin and tubulin. As shown in Figure 6C, the level of cleaved product did not appear to decrease appreciably in the U2OS cell line upon ALLN exposure, but a visible decrease was seen in ALLN treated WI-38 cells. Cisplatin treatment was also shown to reduce the level of the 28 kDa. cleavage fragment in WI-38 cells (Figure 6C, Supp. Figure 3A and B). In contrast, SMN, a known substrate for calpains that accumulates in the CB (Walker, 2008), does not show any obvious changes in levels upon ALLN treatment (Supp. Figure 3C).
Figure 6.
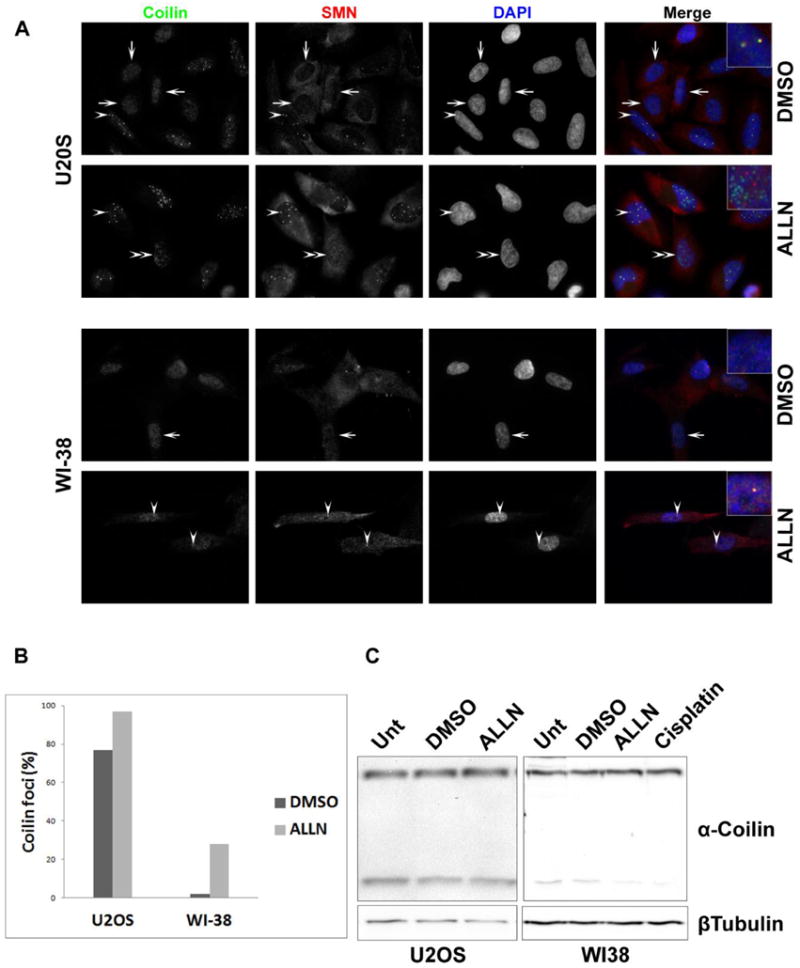
Calpain inhibition induces coilin foci and alters cleaved coilin fragment levels. (A and B) ALLN treatment induces coilin foci in U2OS and WI-38 cells. U2OS and WI-38 cells were treated with ALLN or DMSO (vehicle) for 16 hr. The treated cells were fixed and immunostained for coilin (green), SMN (red) and nuclei (DAPI) (blue). Coilin, SMN and DAPI images were overlaid in merge panel. In U2OS and WI-38 cells, arrows indicate cells with predominantly nucleoplasmic coilin, arrowheads indicate coilin foci with SMN (Cajal body) and double arrowheads mark coilin foci without SMN (A). More than 100 cells were counted for each treatment and cell type and scored for the presence or absence of coilin foci. These percentages are presented in (B). (C) ALLN treatment decreases cleaved coilin fragment levels in WI-38 cells. U2OS and WI-38 cells were treated with DMSO or ALLN for 16 hr. Also shown is 16 hr cisplatin treatment of WI-38 cells. Cell lysates were subjected to SDS-PAGE, Western transfer, and blots probed with anti-coilin antibodies H300 (upper panel) and anti-β-tubulin antibodies (lower panel). Results are representative of three independent experiments.
Calpain 4 reduction rescues coilin from cisplatin-induced nucleolar localization
Since we observed that the calpain inhibitor ALLN induces coilin foci in U2OS and WI-38 cells (Figure 6), we tested more directly the impact of calpain reduction on coilin localization by targeting the calpain common small subunit calpain 4 with siRNA. As stated above, unlike HeLa, U2OS cells have a substantial percent of cells displaying nucleolar coilin localization. Upon reduction of calpain 4 by RNAi, the percent of cells with nucleolar coilin signal was reduced compared to control siRNA treatment (Figure 7, upper panels). As we reported previously, cisplatin treatment induces nucleolar localization of coilin in various cell lines (Gilder et al. 2011). To monitor if the reduction of calpain activity would have any bearing on coilin localization in the presence of cisplatin, U2OS cells were transfected with control or calpain 4 siRNA, followed by treatment with cisplatin (Figure 7, lower panels). The nucleolar localization of coilin in the presence of cisplatin was blunted upon calpain 4 reduction, suggesting that the processing of coilin by calpain regulates coilin localization in response to cisplatin (Supp. Figure 4A). A similar result was observed in HeLa cells (Figure 8 and Supp. Figure 4B). The efficient knockdown of calpain 4 in cisplatin treated or untreated U2OS and HeLa cell lines is shown in Figure 9.
Figure 7.
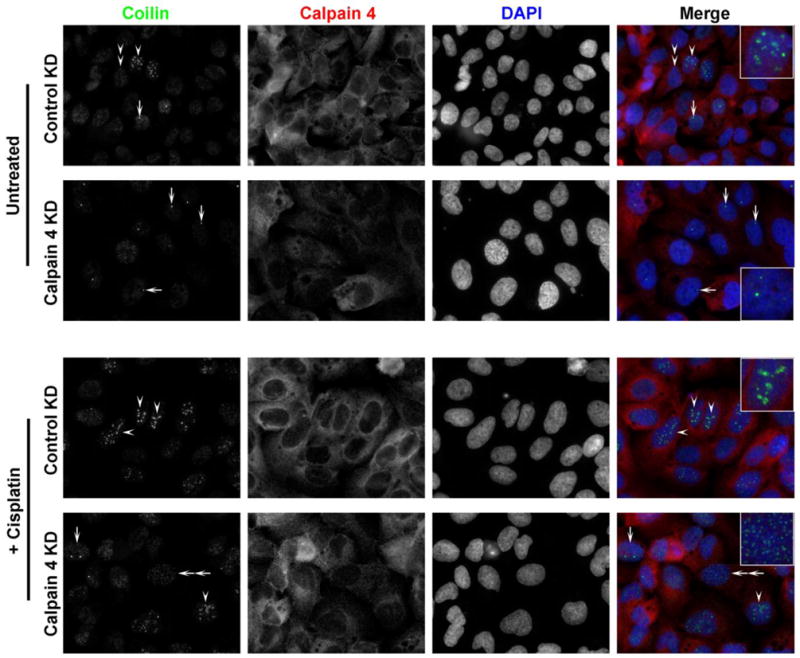
Calpain 4 knockdown abrogates cisplatin-induced nucleolar accumulation of coilin. U2OS cells were transfected with control or calpain-targeted siRNA. 32 hr post-transfection, cells were treated with cisplatin or left untreated. 48 hr post-transfection, cells were fixed and immunostained for coilin (green), calpain 4 (red) and nuclei were stained with DAPI (blue). The arrow indicates cells with normal CB coilin localization, arrowhead indicates cell with predominantly nucleolar coilin, double arrowhead indicates cells with predominantly nucleoplasmic coilin and double arrow indicates cells with small coilin foci.
Figure 8.
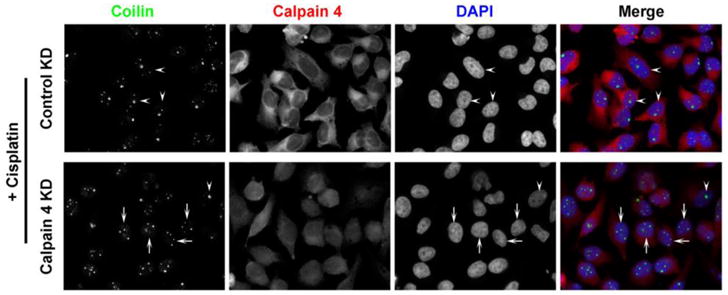
Calpain 4 knockdown reduces cisplatin-induced nucleolar localization of coilin in HeLa cells. Cells were transfected with control or calpain-targeted siRNA. 32 hr post-transfection, cells were treated with cisplatin. 48hr post-transfection, cells were fixed and immunostained for coilin (green), calpain 4 (red) and nuclei were stained with DAPI (blue). Arrows indicate cells with normal Cajal body localization of coilin and arrowheads indicate cells containing significant nucleolar coilin.
Figure 9.
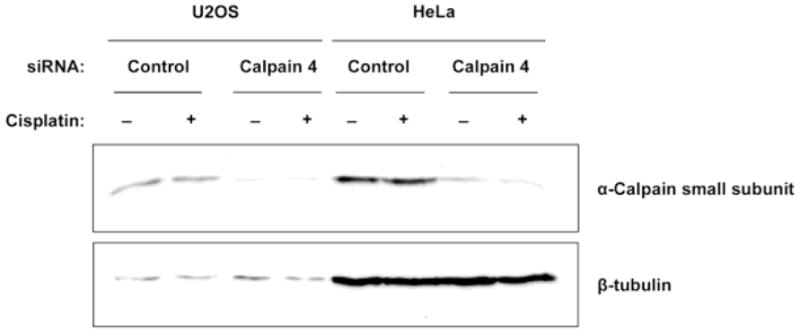
Calpain 4 siRNA transfection effectively knocks down calpain small subunit levels. U2OS and HeLa cells were transfected with control or calpain 4-targeted siRNA for 48 hr. 32 hr post-transfection, the cells were treated with cisplatin (+) or left untreated (−). Cells were harvested 48 hr post-transfection, and lysates subjected to SDS-PAGE, Western transfer, and probed with anti-calpain small subunit (calpain 4, upper panel) and anti-β-tubulin antibodies (lower panel).
Discussion
The data provided here demonstrate that a lower mobility signal first detected by coilin antibodies in 1993 (Andrade et al. 1993) may be the result of specific processing of coilin by calpain. The amount of this cleaved coilin fragment, which is more soluble than full-length coilin, changes during the cell cycle and upon exposure to ActD, cisplatin and etoposide. The functional consequence of this cleaved fragment, and the mechanisms by which its levels are regulated, are unknown. Given the difference in solubility between coilin and its cleaved fragment, it is possible that this fragment has activities distinct from coilin. Unfortunately, the role of coilin is poorly understood and most well characterized in the context of its essential role in the formation and composition of CBs. Hence, without a clear understanding of the role of coilin, it is difficult to postulate about the function of the cleaved fragment. Our recent work (Broome and Hebert 2012) demonstrates that coilin has RNase and nucleic acid binding activities. Consequently, it is possible, though far from proven, that coilin may participate more directly in RNP biogenesis than previously believed. Studies into the regulation of these coilin functions by calpain mediated targeted proteolysis should be very informative.
Regarding the function of the cleavage product, insight into what this fragment may be doing is gained when considering that the antibodies that detect it recognize the C-terminus of coilin, which contains an atypical tudor-like domain (Shanbhag et al. 2010) and interacts with SMN and Sm proteins (Hebert et al. 2001) (Xu et al. 2005) (Toyota et al. 2010). Thus, the coilin cleaved product may compete with full length coilin for SMN and Sm protein binding and therefore impact the dynamics of protein traffic through the CB. Alternatively, the cleaved product may impact the function of nucleoplasmic coilin, which is presently not known, or influence full length coilin RNase and nucleic binding activities. The finding that the level of this fragment decreases relative to full length coilin during mitosis (Figure 1A) suggests it possess anti-mitotic properties or its presence is in some way detrimental during this phase of the cell cycle. Similarly, reduced levels of the coilin cleaved fragment upon exposure to cisplatin and etoposide (Figure 5) hint that this fragment plays an unknown role in the response to DNA damage. Current and future studies will seek to clearly delineate the region of coilin present in the cleaved fragment and identify the signals that result in its generation. Studies are also being conducted to determine the turnover rate of this product. Previous work has shown that full length coilin is stable up to 5h after exposure to the translation inhibitor cycloheximide (Barcaroli et al. 2006), and its levels do not vary during the cell cycle (Andrade et al. 1993). Since we know from the data presented here that the cleaved fragment is reduced during mitosis, it is possible that it is turned over more quickly than full length coilin.
Another major component of the CB, SMN, has also been shown to be subject to specific processing events by calpain (Walker et al. 2008), (Fuentes et al. 2010). In addition to its nuclear accumulations, SMN also localizes to the cytoplasm, and it is this fraction that is subject to calpain cleavage (Fuentes et al. 2010). Since SMN is phosphorylated in the cytoplasm and dephosphorylated in the nucleus (Grimmler et al. 2005), (Petri et al. 2007), it is possible that this modification influences calpain cleavage. Coilin is also a phosphoprotein, and this phosphorylation increases during mitosis (Carmo-Fonseca et al. 1993). Hence it is possible that, like SMN, calpain cleavage of coilin is regulated by phosphorylation and additional studies are currently underway to address this possibility. Since we have shown here that RNA can impact the digestion of coilin by calpain (Figure 4), future work will also explore the interplay between coilin nucleic acid binding and phosphorylation in regulating coilin processing events by calpain.
The antibodies we use here to detect the cleavage fragment map to the C-terminal region of coilin, thus it possible that an N-terminal fragment is also present but not detected by the antibodies used in this study. Unfortunately, antibodies raised to the N-terminus of coilin do not work well and this precludes us from the identification of any putative N-terminal fragments. Using bacterially expressed coilin proteins, we found that both full length coilin and coilin 94–576, but not fragments 1–362 or 362–576 (N362 and C214), release the 28 kDa cleaved coilin fragment upon incubation with calpain (Figure 3). Additionally, cleaved coilin was not released from fragments 94–450 or 94–475. Taken together, these results suggest that two cleavage sites, located in the N-terminus after residue 94 and in the C-terminus perhaps between residues 475 and 576, result in generation of the cleaved coilin fragment. It has been shown that the C-terminus of coilin influences CB number in that fusion of GFP to the C-terminus of coilin generates numerous CBs (Hebert and Matera 2000; Shpargel et al. 2003). One possibility for this result is that GFP fusion to the C-terminus of coilin disrupts coilin structure such that calpain cleavage is reduced. Although the majority of coilin is predicted to be intrinsically disordered, some structure is predicted in its N- and C-terminus (Broome and Hebert 2012). Interestingly, two hallmarks of intrinsically disordered proteins are their susceptibility to protein degradation and aberrant mobility on SDS-PAGE (Receveur-Brechot et al. 2006). With a mobility on SDS-PAGE closer to 80 kDa than the expected 62 kDa, and the presence of lower molecular weight fragments, coilin has both of these hallmarks.
We have previously shown that bacterially expressed coilin remains bound to RNA throughout a stringent purification protocol (Broome and Hebert 2012), suggesting a high affinity between the two molecules. Considering our observation that RNA appears to increase the activity of calpain on coilin (Figure 4), we hypothesize that in the presence of RNA, coilin may undergo specific protein folding that leads to additional sites for proteolysis. The hypothesis is supported by the fact that calpains are not amino acid or sequence specific, but they hydrolyze bonds between protein domains in a limited manner to modulate the protein properties (Saido et al. 1994; Sorimachi et al. 1994).
Since we found that the levels of the cleaved coilin fragment change during the cell cycle (Figure 1), and it has been previously reported that localization of coilin changes when cells are exposed to exogenous stressors (Gilder et al. 2011), (Cioce et al. 2006), we hypothesized that there may be a dynamic flux between levels of full length and cleaved coilin which can be influenced by cell stressors. In fact, we found that ActD treatment differentially affects relative cleaved coilin levels in HeLa and U2OS cells, while cisplatin and etoposide cause a reduction in relative cleaved coilin levels in both cell lines. Due to this correlation between the known localization changes of coilin induced by cell stress and the changes in relative cleaved coilin levels we observed with the same stressors, we next wanted to explore how calpain inhibition would affect coilin localization. First, we found that U2OS cells contain a relatively higher amount of cleaved product compared to that found in HeLa cells (Figure 5E). Second, we noted that the U2OS cells contain a larger percent of cells that lack CBs or have nucleolar accumulation of coilin compared to HeLa. We therefore suspected that reduction of the cleaved fragment would increase the percent of cells with CBs in U2OS. We also tested the primary line WI-38, which normally has a low percentage of cells with CBs. As predicted, ALLN treatment induced CB number in both cell lines (Figure 6A, B). The relative amount of cleaved fragment did not change in U2OS cells, but the cleaved fragment levels were reduced in WI-38 cells upon ALLN treatment (Figure 6C). Previous work has shown that ALLN inhibits cell cycle progression (Sherwood et al. 1993). It is possible, therefore, that the induction of CBs by ALLN is not due to alterations in the amount of coilin processing, but rather is reflective of a more general response to alterations in the cell cycle. CBs disassemble during mitosis and reform early- to mid-G1, reaching their largest size in S and G2 (Andrade et al. 1993). Hence it is possible that cell cycle arrest in response to ALLN treatment allows for more visible CBs, and the level of the cleaved coilin fragment is unrelated to their detection.
Finally, we wanted to explore what affect the lack of calpain activity would have on localization of coilin specifically under stressed cellular conditions. Since the calpain inhibitor ALLN treatment induces coilin foci, we speculated that calpain might have a regulatory role in coilin localization. As we reported previously, cisplatin treatment or γ-irradiation induces nucleolar localization of coilin which suppresses RNA polymerase I activity (Gilder et al. 2011). We observe that calpain 4 KD reduces the cisplatin-induced nucleolar localization of coilin (Figures 7 and 8). This suggests that either coilin cannot be processed to localize to the nucleolus in the calpain 4 KD background or the mechanisms that keep coilin in the nucleolus in the presence of cisplatin might be abolished. Further studies are required to elucidate the mechanistic role of the coilin cleavage fragment in conditions of stress, such as that induced by cisplatin treatment, and the role calpain plays in coilin localization.
In conclusion, the results shown here demonstrate that coilin is subjected to regulated specific proteolysis by calpains. This is not a rare event considering that the cleaved fragment is easily detectable by coilin antibodies in cell lysate. In vitro, calpain-1 was shown to generate the same sized cleavage fragment as observed in cell lysate, suggesting that this protease is involved in coilin processing. How this processing impacts coilin function, with particular emphasis on its role in CB formation, composition and activity, is unknown, but clarification of these events will provide another insight into the inner-workings of the nucleus.
Supplementary Material
Coilin siRNA treatment reduces full length and cleaved coilin fragment levels. (A) U2OS cells were transfected with control or coilin siRNA. 24hr post transfection, the cells were harvested and the lysates subjected to SDS-PAGE, Western transfer, then probed with anti-coilin and anti-β-tubulin antibodies. (B) Quantification of knockdown from (A) and Figure 1B (48 and 72 h timepoints). The full-length coilin and cleaved coilin signals were divided by the tubulin signal. The values for coilin siRNA were then normalized to the results for the control siRNA.
The cleaved coilin fragment does not arise from the extreme N- or C-terminus of full length coilin. HeLa cells were transfected with GFP-coilin or Coilin-GFP. 24hr post transfection, the cells were harvested and the lysates were incubated with calpain as described in Materials and Methods. The reactions were subjected to SDS-PAGE, Western transfer, and then probed with anti-GFP and anti-coilin antibodies. The letter ‘a’ indicates full length GFP-coilin or Coilin-GFP, ‘b’ indicates endogenous full length coilin and ‘c’ indicates cleaved coilin fragment.
Protein bands detected by anti-coilin H300 antibody from Figure 6C were quantified by densitometric analysis. Cleaved coilin fragment to full length coilin ratios were calculated for U2OS (A) and WI38 (B) cells. We have observed similar results in three experiments. (C) ALLN treatment does not influence SMN levels. HeLa cells were treated with DMSO or ALLN for 16 hr. The harvested cell lysates subjected to SDS-PAGE, Western transfer, then probed with anti-SMN and anti-β-tubulin antibodies.
Calpain4 KD decreases nucleolar accumulation of coilin: quantification of Figures 7 and 8. U2OS (A) and HeLa (B) cells were counted for each treatment and the percentages of cells with nucleolar coilin were presented; more than fifty cells were counted for each treatment.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM081448. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J Cell Biol. 1998;142 (4):899–912. doi: 10.1083/jcb.142.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D, Dinsdale D, Neale MH, Bongiorno-Borbone L, Ranalli M, Munarriz E, Sayan AE, McWilliam JM, Smith TM, Fava E, Knight RA, Melino G, De Laurenzi V. FLASH is an essential component of Cajal bodies. Proc Natl Acad Sci U S A. 2006;103(40):14802–14807. doi: 10.1073/pnas.0604225103. 0604225103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome HJ, Hebert MD. In Vitro RNase and Nucleic Acid Binding Activities Implicate Coilin in U snRNA Processing. PLoS One. 2012;7(4):e36300. doi: 10.1371/journal.pone.0036300. PONE-D-11-24062 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M. New clues to the function of the Cajal body. EMBO Rep. 2002;3 (8):726–727. doi: 10.1093/embo-reports/kvf154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing Coiled Bodies Is Regulated in Interphase and Mitosis - Evidence that the Coiled Body Is a Kinetic Nuclear Structure. J Cell Biol. 1993;120 (4):841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero ZI, Velma V, Douglas HE, Hebert MD. Coilin phosphomutants disrupt cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS One. 2011;6(10):e25743. doi: 10.1371/journal.pone.0025743. PONE-D-11-12853 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol. 2006;175(3):401–413. doi: 10.1083/jcb.200604099. jcb.200604099 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Papait R, Nicassio F, Vecchi M, Gomiero P, Mantovani R, Di Fiore PP, Bonapace IM. Np95 is a histone-binding protein endowed with ubiquitin ligase activity. Mol Cell Biol. 2004;24 (6):2526–2535. doi: 10.1128/MCB.24.6.2526-2535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. Embo J. 2002;21 (11):2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes JL, Strayer MS, Matera AG. Molecular determinants of survival motor neuron (SMN) protein cleavage by the calcium-activated protease, calpain. PLoS One. 2010;5(12):e15769. doi: 10.1371/journal.pone.0015769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gilder AS, Do PM, Carrero ZI, Cosman AM, Broome HJ, Velma V, Martinez LA, Hebert MD. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol Biol Cell. 2011;22(7):1070–1079. doi: 10.1091/mbc.E10-08-0731. mbc.E10-08-0731 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Bauer L, Nousiainen M, Korner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6 (1):70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122(Pt 11):1872–1881. doi: 10.1242/jcs.044040. jcs.044040 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys. 2010;496(2):69–76. doi: 10.1016/j.abb.2010.02.012. S0003-9861(10)00075-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11 (12):4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;15 (20):2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J. 2003;22 (8):1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322(5908):1713–1717. doi: 10.1126/science.1165216. 1165216 [pii] [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17(7):3221–3231. doi: 10.1091/mbc.E06-03-0247. E06-03-0247 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20(6):1661–1670. doi: 10.1091/mbc.E08-05-0525. E08-05-0525 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Parry JA, Chin A, Duensing S, Duensing A. Soluble histone H2AX is induced by DNA replication stress and sensitizes cells to undergo apoptosis. Mol Cancer. 2008;7:61. doi: 10.1186/1476-4598-7-61. 1476-4598-7-61 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010;8(11):e1000521. doi: 10.1371/journal.pbio.1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17(5):639–647. doi: 10.1016/j.devcel.2009.10.017. S1534-5807(09)00439-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783(11):2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. S0167-4889(08)00267-X [pii] [DOI] [PubMed] [Google Scholar]
- Petri S, Grimmler M, Over S, Fischer U, Gruss OJ. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J Cell Biol. 2007;179 (3):451–465. doi: 10.1083/jcb.200704163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62(1):24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Remboutsika E, Lutz Y, Gansmuller A, Vonesch JL, Losson R, Chambon P. The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J Cell Sci. 1999;112 (Pt 11):1671–1683. doi: 10.1242/jcs.112.11.1671. [DOI] [PubMed] [Google Scholar]
- Saido TC, Sorimachi H, Suzuki K. Calpain: new perspectives in molecular diversity and physiological-pathological involvement. Faseb J. 1994;8 (11):814–822. [PubMed] [Google Scholar]
- Shanbhag R, Kurabi A, Kwan JJ, Donaldson LW. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett. 2010;584(20):4351–4356. doi: 10.1016/j.febslet.2010.09.034. S0014-5793(10)00771-4 [pii] [DOI] [PubMed] [Google Scholar]
- Sherwood SW, Kung AL, Roitelman J, Simoni RD, Schimke RT. In vivo inhibition of cyclin B degradation and induction of cell-cycle arrest in mammalian cells by the neutral cysteine protease inhibitor N-acetylleucylleucylnorleucinal. Proc Natl Acad Sci U S A. 1993;90 (8):3353–3357. doi: 10.1073/pnas.90.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13(2):167–173. doi: 10.1038/ncb2157. ncb2157 [pii] [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci. 2003;116 (Pt 2):303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- Sorimachi H, Saido TC, Suzuki K. New era of calpain research. Discovery of tissue-specific calpains. FEBS Lett. 1994;343(1):1–5. doi: 10.1016/0014-5793(94)80595-4. 0014-5793(94)80595-4 [pii] [DOI] [PubMed] [Google Scholar]
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell. 1992;3:555–569. doi: 10.1091/mbc.3.5.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Oates A, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embyogenesis. Nucleus. 2010 doi: 10.4161/nucl.1.1.10680. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu H, Subramony SH, Hebert MD. Interactions between Coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci. 2005;118 (Pt 21):4995–5003. doi: 10.1242/jcs.02613. [DOI] [PubMed] [Google Scholar]
- Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB’. Chromosoma. 2010;119(2):205–215. doi: 10.1007/s00412-009-0249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154 (2):293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma RS, Roth MB. Induction of coiled body-like structures in Xenopus oocytes by U7 snRNA. Chromosoma. 1999;108(6):337–344. doi: 10.1007/s004120050385. 91080337.412 [pii] [DOI] [PubMed] [Google Scholar]
- Walker MP, Rajendra TK, Saieva L, Fuentes JL, Pellizzoni L, Matera AG. SMN complex localizes to the sarcomeric Z-disc and is a proteolytic target of calpain. Hum Mol Genet. 2008;17(21):3399–3410. doi: 10.1093/hmg/ddn234. ddn234 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4(7):e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittom AA, Xu H, Hebert MD. Coilin levels and modifications influence artificial reporter splicing. Cell Mol Life Sci. 2008;65 (7–8):1256–1271. doi: 10.1007/s00018-008-7587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;114 (3):155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Coilin siRNA treatment reduces full length and cleaved coilin fragment levels. (A) U2OS cells were transfected with control or coilin siRNA. 24hr post transfection, the cells were harvested and the lysates subjected to SDS-PAGE, Western transfer, then probed with anti-coilin and anti-β-tubulin antibodies. (B) Quantification of knockdown from (A) and Figure 1B (48 and 72 h timepoints). The full-length coilin and cleaved coilin signals were divided by the tubulin signal. The values for coilin siRNA were then normalized to the results for the control siRNA.
The cleaved coilin fragment does not arise from the extreme N- or C-terminus of full length coilin. HeLa cells were transfected with GFP-coilin or Coilin-GFP. 24hr post transfection, the cells were harvested and the lysates were incubated with calpain as described in Materials and Methods. The reactions were subjected to SDS-PAGE, Western transfer, and then probed with anti-GFP and anti-coilin antibodies. The letter ‘a’ indicates full length GFP-coilin or Coilin-GFP, ‘b’ indicates endogenous full length coilin and ‘c’ indicates cleaved coilin fragment.
Protein bands detected by anti-coilin H300 antibody from Figure 6C were quantified by densitometric analysis. Cleaved coilin fragment to full length coilin ratios were calculated for U2OS (A) and WI38 (B) cells. We have observed similar results in three experiments. (C) ALLN treatment does not influence SMN levels. HeLa cells were treated with DMSO or ALLN for 16 hr. The harvested cell lysates subjected to SDS-PAGE, Western transfer, then probed with anti-SMN and anti-β-tubulin antibodies.
Calpain4 KD decreases nucleolar accumulation of coilin: quantification of Figures 7 and 8. U2OS (A) and HeLa (B) cells were counted for each treatment and the percentages of cells with nucleolar coilin were presented; more than fifty cells were counted for each treatment.


