Abstract
The condensation state of chromosomes is a critical parameter in multiple processes within the cell. Failures in the maintenance of appropriate condensation states may lead to genomic instability, mis-expression of genes, and a number of disease states. During cell proliferation, replication of DNA represents an ongoing challenge for chromosome packaging as DNA must be unpackaged for replication and then faithfully repackaged. An integral member of the DNA replication machinery is the GINS complex which has been shown to stabilize the CMG complex which is required for processivity of the Mcm2–7 helicase complex during S phase. Through examination of the phenotypes associated with a null mutation in Psf2, a member of the evolutionarily conserved GINS complex, we find that Drosophila Psf2 likely has a role in establishing chromosome condensation and that the defects associated with this mis-condensation impact M phase progression, genomic stability, and transcriptional regulation.
Keywords/Phrases: GINS Complex, Psf2, DNA replication, Chromosome condensation
Introduction
Proper chromosome architecture and condensation are essential for the numerous processes associated with normal functioning organisms. The accurate partitioning of DNA during mitosis and meiosis depend the underlying condensation state of the chromosomes during metaphase. In addition defects in condensation may lead to genomic instability and the development of a multitude of disease states (Koshland and Strunnikov 1996). The maintenance of chromosome condensation is challenged every cell cycle by the existence of DNA replication where the entire nuclear genome must be unpackaged, accurately replicated, and then repackaged in preparation for M phase. There is a growing body of evidence which suggests that proteins initially identified as important for DNA replication also have roles in the establishment of chromosome condensation (Pflumm 2002). The GINS complex may be one of these players, initially characterized for its role in DNA replication, which may also have a role in chromosome biology.
The evolutionarily conserved GINS complex is a heterotetrameric complex that consists of: Sld5, Psf1, Psf2, and Psf3. Crystallographic studies of the GINS complex show a complex with a structure reminiscent of an elongated spindle with a central pore (Chang et al. 2007). This complex has been implicated along with Cdc45 and Mcm2–7 (the CMG complex) as making up the core of the replisome progression complex (RPC) (Aparicio et al. 2006; Moyer et al. 2006 ). The CMG complex is required for the initiation of DNA replication and the subsequent translocation of the replication fork (Aparicio et al. 2009). When the CMG complex is removed, during DNA replication, the replication fork stalls; the stalling of the replication fork is likely due to a decrease in the helicase activity of Mcm2–7 (Aparicio et al. 2006; Labib et al. 2007; Aparicio et al. 2009). The function of the GINS complex within the CMG complex has been suggested to be the stabilization of the interaction between Cdc45 and the Mcm2–7 hexamer (Aparicio et al. 2009).
The Psf2 subunit of the GINS complex was first identified in the budding yeast S. cerevisiae as a partner of Sld5, a gene shown to be involved via Dpb11 in the loading of DNA polymerase epsilon (Takayama et al. 2003; Araki et al. 1995). Subsequent studies have linked Psf2 to eye development in Xenopus, the segregation of chromosomes in S. pombe, and the development of cancer in humans (Obama et al. 2005; Huang et al. 2005; Walter et al. 2008). Here we present the first in vivo exploration of the function of Psf2 in Drosophila. Using a null mutation we find that Psf2 is an essential gene and that normal dosage of Psf2 is likely required for establishing proper chromosome condensation during S phase. Moreover we show that this mis-condensation of chromosomes causes problems in the packaging of metaphase chromosomes and potential problems with the maintenance and/or establishment of transcriptional regulation associated with heterochromatic and euchromatic chromatin states.
Results
Psf2 mutant allele
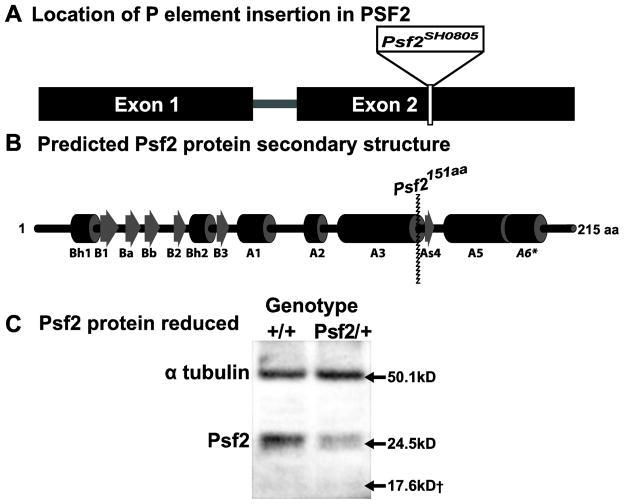
The Psf2 gene is found on the left arm of chromosome 2 in the Drosophila genome. Transcript models for the gene indicate that it is made up of 2 exons with an single 58bp intron (FBgn0261976)(Tweedie et al. 2009)(Figure 1A). One, recessive lethal P element insertion line; Psf2SH0805, has been identified for the Psf2 coding region through a large scale screen aimed at identifying novel essential genes in Drosophila (Figure 1A)(Oh et al. 2003). Amplification and sequencing the DNA flanking the P element insertion reveals that the P element is inserted at position 4,388,852 in the second chromosome. Insertion at this position disrupts the coding sequence of Psf2 by removing 64 amino acids from the C terminal. The orientation of the P element results in the addition of a single alanine residue followed by a stop codon. The 64 amino acid residues that are removed from the C terminal of the native Psf2 protein by the P element insertion in the Psf2SH0805 allele represent a portion of Psf2 that is critical for interaction with Sld5 and the stability of the GINS complex (Figure 1B)(Chang et al. 2007). Of additional interest is that sequence alignments of Drosophila Psf2 with Human Psf2, combined with the published crystal structure of human Psf2 (Chang et al. 2007), reveals that the Drosophila Psf2 protein may contain an additional C terminal alpha helical domain of unknown function (A6, Figure 1C).
Figure 1.
Nature of the Psf2 mutation. (A) Schematic of Psf2 gene region on chromosome 2L with P-element insertion site indicated for the Psf2SH0805 mutant allele used in this study. (B) Predicted secondary protein structure for Psf2 gene region with impact of P-element insertion site indicated at amino acid 151. (C) Western blot representing maternal loading in 0–2 hour embryos laid by heterozygous females showing reduction in the full length protein normalized to α-tubulin as a loading control. No band migrates at 17.6kD corresponding to the predicted size of the truncated Psf2 protein generated by the Psf2 mutant allele
To determine whether or not a truncated version of the protein was translated from the mutant allele we performed Western blots using anti-Psf2 polyclonal antibody (Moyer et al. 2006). We probed extracts from 0–2 hour old embryos deposited by fertilized heterozygous females, thereby allowing us to detect maternal loading of the Psf2 protein. Our results indicate that the truncated protein is not present, as no band was observed to migrate at the predicted 17.6kD (Figure 1C). Moreover, the levels of full length protein are lowered in the embryos derived from heterozygous females versus wild-type control females. The results of repeated western blots normalized for loading with alpha tubulin, show that Psf2 protein levels in Psf2SH0805 /+ animals are at 49.8% ±4.1% of wild-type protein levels. When taken all together our results suggest that the Psf2SH0805 allele likely represents a null allele of Psf2.
Psf2 mutant viability
Individuals homozygous for the Psf2SH0805 allele arrested at the late embryo/first instar larval stages. The lethality of the Psf2SH0805 allele was not rescued by crosses to deficiencies that span the Psf2 containing genomic region but was rescued by the introduction of a Psf2 transgene driven by a native promoter (see materials and methods) indicating that the lethality observed for the Psf2SH0805 is due to the absence of Psf2. In order to investigate the nature of the homozygous arrest, the mutant allele was placed over a GFP expressing balancer to allow for early identification of homozygous embryos and larvae. Both homozygous and wild-type control embryos were stained with DAPI to visualize chromosome dynamics. Embryos homozygous for the Psf2 mutation displayed aberrant distribution of nuclei within the embryonic syncytium as well as cell cycle asynchrony as compared to wild-type (Figure 2A–D). The nuclear drop out is indicative of cell cycle failure in populations of nuclei within the syncytium of the homozygous embryos (Figure 2B). Closer examination of the nuclei found in homozygous embryos demonstrated deviation from normally synchronous nuclear divisions as well as the presence of mitotic defects in the form of anaphase bridges (Figure 2D, arrows).
Figure 2.
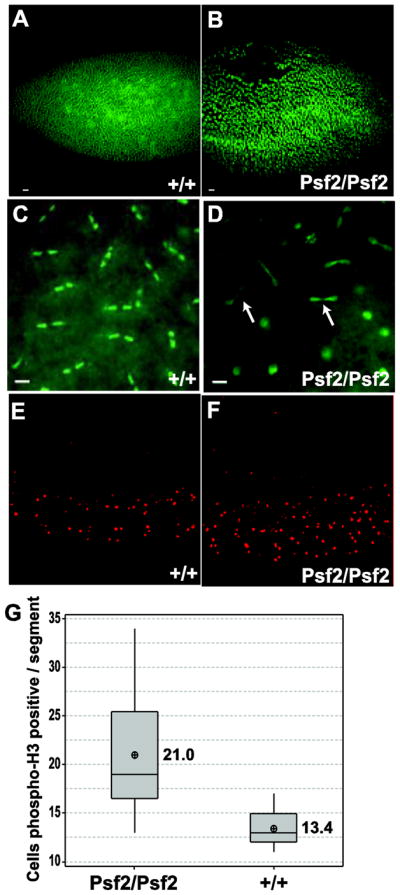
Embryonic phenotypes associated with Psf2 mutation. Early embryo defects in homozygous Psf2 individuals. (B, D) compared to wild-type (A,C) where Psf2/Psf2 embryos show nuclear fallout (B) and cell cycle asynchrony (D) and anaphase bridges (D arrows). (E, F) Representative micrographs of embryos stained for phospho H3, a marker for cells in mitosis. Psf2/Psf2 embryos show a significant (p=0.001) increase in mitotic cells per segment as compared to wild-type (G). Scale bar = 10μm
Despite the defects observed in early embryos homozygous for the Psf2 mutant allele, some embryos progress through cellularization and segmentation. Of these, less than 10% hatch to become small and nearly immobile 1st instar larvae that readily die. Morphologically, the segmented embryos appear normal. However, given the mitotic defects observed in the majority of embryos we stained with antibodies against phospho-H3 to determine if there was a skew in proportion of cells in mitosis in the homozygous embryos compared to wild-type. Visualization and quantitation suggested that there is a significant increase in the proportion of cells in mitosis in the homozygous embryos relative to wild-type (p=0.001)(Figure 2E–G).
Our previous, and ongoing, work with other DNA replication mutants suggests that dosage of these proteins are important for viability as many of these mutant alleles display dominant semi-lethality (Apger et al. 2010; Gouge and Christensen 2010b)(our unpublished data). Consistent with our other findings the Psf2SH0805 allele is also shows dominant semi-lethality. In self-crosses of Psf2SH0805/+b, only 66% (531/810) of the heterozygous flies were recovered relative to expected, indicating a 34% mortality rate for individuals heterozygous for the Psf2 mutation.
Mitotic defects
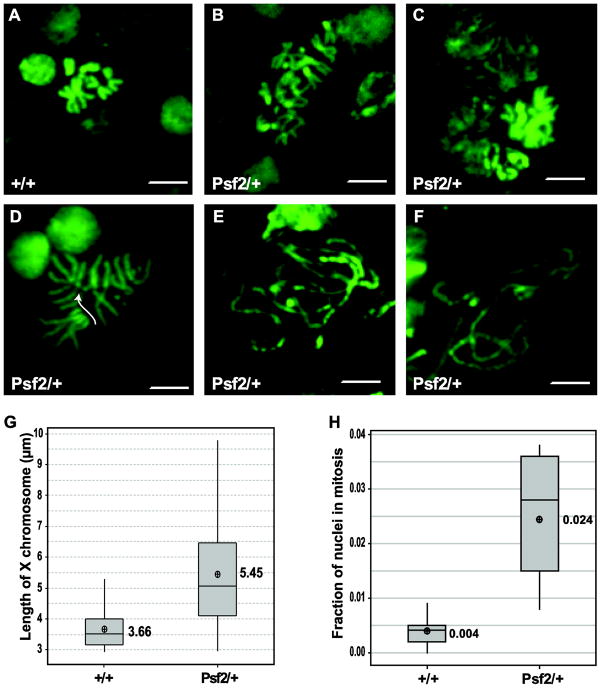
Given the chromosome defects observed in early embryos, the dominant semi-lethalitynature of the Psf2 mutation, and previous work that has shown that reduced dosage of Sld5 results in mitotic defects, we sought to determine if there were mitotic defects in the brains of 3rd instar wandering larva heterozygous for Psf2SH0805 (Gouge and Christensen 2010b). A suite of mitotic defects were observed in brain squash preparations from Psf2SH0805 heterozygotes as compared to wild-type controls (Figure 3A–F). Wild-type chromosome spreads are characterized by well packaged mitotic figures, well connected sister chromatids, and a 2n number of chromosomes (Figure 3A). This contrasts with what is observed in spreads from brains of Psf2SH0805 heterozygous larvae, where chromosomes are elongated (Figure 3B–F), sister chromatids are separated (Figure 3D), and aneuploidy is observed (Figure 3B, C, D). One or all of these defects are observed in approximately 80% of all nuclei in mitosis in the heterozygous larval brains, whereas none of these defects are observed in the wild-type control preparations. To assay the chromosome length in the mutant, X chromosomes were measured. X chromosomes were chosen for length measurements as they are readily identifiable because they are telocentric. This analysis demonstrated that chromosomes from heterozygous larvae were on average 48.9% longer (p<0.001) than wild-type chromosomes (Figure 3G). Due to the mitotic defects observed in the heterozygous larvae, it is not surprising that the quantitation of the mitotic index reveals that a there is a significant (p= 0.004) mitotic delay in heterozygous larval brains as compared to wild-type (Figure 3H). Overall, there is an average of a 6 fold increase in the number of observed mitotic figures when larvae are heterozygous for the Psf2 mutation. The introduction of a single copy of a Psf2 transgene driven by the native promoter is sufficient to rescue the mitotic defect, mitotic delay, and the lengthening of the X chromosome (Supplemental Table 1). These observations support the notion that the phenotypes observed are a result of a haploinsufficiency of Psf2.
Figure 3.
Effects of reduced dosage of Psf2 on mitotic figures in larval brains. (A) Representative micrograph of wild-type mitotic figure. (B–F) Representative micrographs of mitotic figures from Psf2/+ brain squashes showing a variety of defects including aneuploidy (B, C, D), severely mis-condensed and broken chromosomes (E, F), and premature sister chromatid separation (D arrow). (G) Quantitation of mitotic indices of Psf2/+ and wild-type. Psf2 heterozygotes show significant M phase delay compared to wild-type (p= 0.004). (H) Quantitation of X chromosome length in Psf2/+ and wild-type. X chromosome are significantly longer in Psf2/+ larval brains than in wild-type (p<0.000). Scale bar = 10μm
DNA damage
Given the severity of the defects in mitotic chromosomes and the mitotic delay we wanted to determine if DNA damage was elevated in the heterozygous larvae compared to wild type controls. In order to detect elevated levels of DNA damage we immunostained with an anti- phospho H2Av antibody as phosphorylated H2Av has been shown to be a reliable indicator of DNA damage in Drosophila (Madigan et al. 2002) (Figure 4A&B). Brain squashes and immunostaining of the relevant genotypes revealed that larvae heterozygous for the Psf2 mutation show nearly a twofold increase (p=0.009) in nuclei staining positive for phosph-H2Av suggesting that DNA damage is increased with larvae have a reduction in the dosage of Psf2 (Figure 4C).
Figure 4.
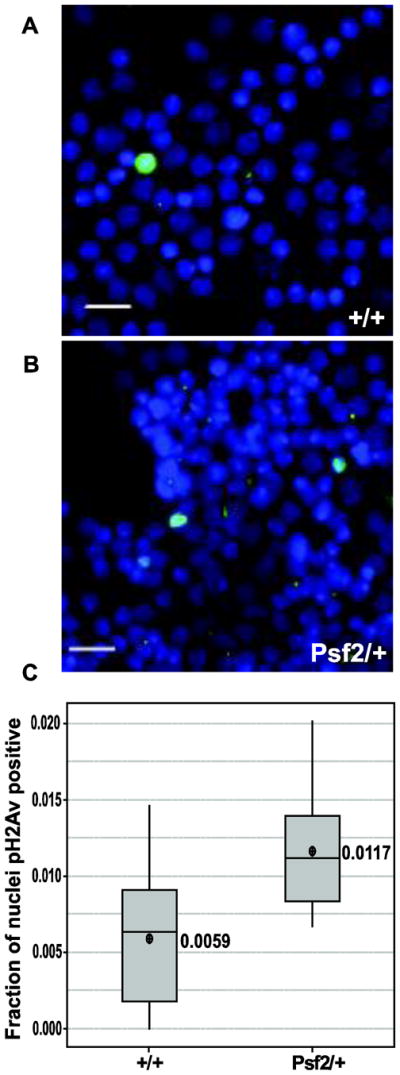
Larval brains from Psf2/+ show a significant increase in the number of nuclei that are positive of phosph-H2Av. A & B. Representative micrographs of brain squashes stained with DAPI (blue) and phospho H2Av (green) for the indicated genotypes (scale bars = 10μm). C. Boxplots of the faction of nuclei that are positive for phospho H2Av staining in the indicated genotypes
S phase
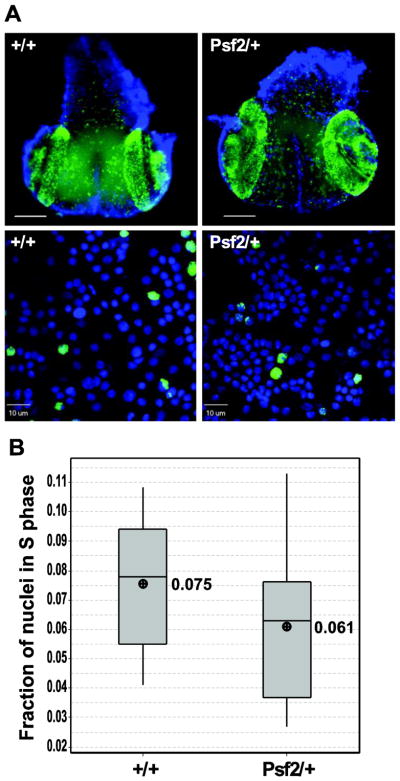
The GINS complex has been implicated in efficient DNA synthesis during S phase (MacNeill 2010). Reducing the dosage of Sld5 by half in Drosophila has been shown to double the fraction of cells in S phase in larval brains, consistent with a possible rate limiting role in S phase for the GINS complex (Gouge and Christensen 2010b). In order to test whether or not a similar reduction in Psf2 resulted in S phase delays we performed EdU (a thymidine analog) incorporation studies on larval brains either heterozygous for the Psf2 mutation or wild-type. We found no qualitative difference in EdU incorporation patterns when heterozygous whole mount brains were compared to wild-type (Figure 5, top panels). Moreover, quantitation of the S phase index through enumeration of EdU positive nuclei following brain squash revealed no significant difference in the fraction of nuclei in S phase in the heterozygous larvae as compared to the wild-type control (p=0.204)(Figure 5B).
Figure 5.
Larval brains from Psf2/+ do not show significant S phase delay. (A,Top) Micrographsof whole mount wandering 3rd instar larval brains showing DNA stained with DAPI (blue) and EdU incorporation (green). (A, bottom) Representative micrographs of squashed third instar wandering larval brains showing individual nuclei stained with DAPI (blue) and EdU incorporation (green) used for quantitation of S phase indices. (B) Graph showing the fraction of nuclei in S-phase from the genotypes indicated. There is no significant (p=0.204) difference in the fraction of cells in S-phase in Psf2/+ compared to wild-type. Scale bar = 10μm
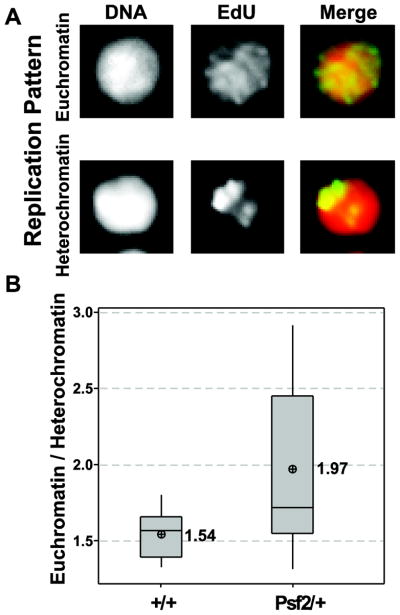
Patterns of S phase EdU incorporation have been shown to be different in cells undergoing replication of euchromatin during early/mid S phase versus cells undergoing late replication of heterochromatin (Fox et al. 2010; Schwaiger et al. 2010; Shermoen et al. 2010; Su 2010). Patterns of EdU incorporation in larval brains fall into 2 distinct categories: global incorporation during early to mid S phase (euchromatin) and restricted foci during late S phase (heterochromatin) (Figure 6A). Categorizing these patterns in heterozygous larval brains as compared to wild-type demonstrated a significant shift (p=0.043) in the relative fractions of EdU positive cells replicating euchromatin versus heterochromatin in the larvae heterozygous for the Psf2 mutation (Figure 6B). Overall, the ratio of euchromatin/heterochromatin replicating cells is 28 % higher in heterozygous mutant brains relative to wild-type.
Figure 6.
Larval brains from Psf2/+ show significant shift in EdU incorporation pattern. (A) Representative micrographs of EdU incorporation patterns for (global) euchromatin and (punctate) heterochromatin as indicated. (B) Graph showing a significant increase in the ratio of euchromatin/heterochromatic patterns of EdU incorporation in Psf2/+ individual compared to wild-type (p=0.043)
Endoreplication
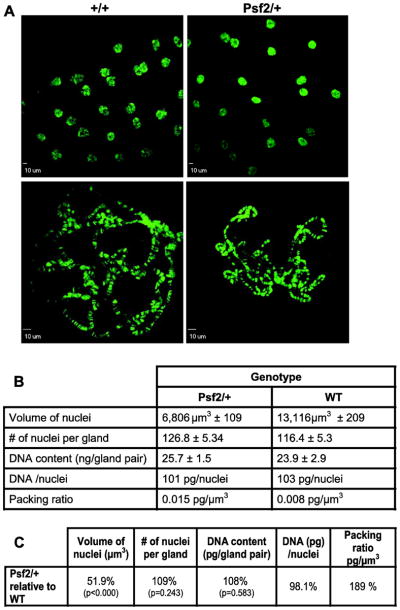
The presence of mitosis in the normally cycling cells of the larval brain complicates the analysis of whether or not the defects observed in mitotic chromosomes are the function of a discrete role for Psf2 in metaphase chromosome biology or are the result of defects in S phase being manifested in M phase. Drosophila salivary glands provide a tissue where repeated rounds of DNA replication occur without being followed by mitosis, thereby allowing the examination of large interphase polytene chromosomes for replication defects and/or chromosome packaging defects. Imaging of salivary glands from the respective genotypes suggested that nuclei from the heterozygous larvae were slightly smaller (Figure 7, top micrographs). Squashed and spread polytene chromosomes from these same genotypes suggested that these smaller nuclei were a consequence of under-replicated polytene chromosomes (Figure 7A, bottom micrographs). However, attempts to quantify the relative DNA content in chromosome spreads using imaging techniques were confounded by the high variability in apparent chromosome size and quality of spreads that were obtained from mutant larvae. We addressed this issue in two ways. First, we sought to use the volume of the nuclei without squashing as a surrogate for DNA content. Second, we dissected salivary glands from numerous individual larvae, extracted the DNA, and quantitated the DNA spectroscopically. Curiously, the average nuclear volume reduction relative to wild-type in the mutant is 48.1% (p<0.001), whereas there is no significant change in the DNA content of salivary glands in heterozygous larvae relative to wild-type (p=0.583)(Figure 7B&C). This result was not due to differences in the number of nuclei between the genotypes, as the number of nuclei per gland was not significantly different (p=0.243). Nor was this result a failure of the spectroscopic method to detect differences in DNA content as we have used this technique to detect under replication of DNA in a PCNA mutant line and have shown that spectroscopic measurements of 2nd instar larvae is consistent with lower ploidy at this earlier developmental stage (Chmielewski and Christensen 2011)(Supplemental table 2). These data sets allow us to calculate average packaging ratios for the DNA in the nuclei from the respective genotypes. These calculations show that the DNA packaging ratio (pg/μm3) in the heterozygous mutant larva polytene nuclei is 189% higher relative to wild-type (Figure 7C). The addition of a single extra copy of Psf2 is sufficient to restore the packing ratio of these nuclei back to wild type levels (Supplemental table 1). In summary, Psf2 appears to be dispensable for endoreplication of salivary glands but required for proper packaging of polytene chromosomes.
Figure 7.
Normal dosage of Psf2 is not required for endoreplication of salivary gland nuclei but is required for normal chromosome compaction. (A, Top) Representative micrographs of intact polytene chromosomes within whole salivary glands. (A, Bottom) Micrographs of polytene chromosome spreads showing smaller apparent size in Psf2/+ compared to wild-type. (B) Table of average values for different measures as indicated for the respective genotypes. (C) Table showing the phenotype of Psf2/+ compared to wild-type for the measures indicated
Unfortunately defects in DNA packaging in salivary glands are not informative to the extent that we cannot discern whether or not these defects have the potential to lead to subsequent mitotic chromosome defects. In fact, developmental modulation in the regulation of condensation has not been reported in Drosophila salivary gland chromosomes but has been reported in the polyploid nurse cells of the ovary (Dej and Spradling 1999). Several lines of evidence suggest that the polyploid nurse cells undergo a mitosis-like transition between stages 5 and 6 where nurse cell chromosomes transition from a highly condensed 5-blob stage to a decondensed stage which is fully manifested by stage 7 (Dej and Spradling 1999; Reed and Orr-Weaver 1997; Edgar and Orr-Weaver 2001). During this transition, like in M phase, homologous chromosomes separate before sister chromatids and the newly dissociated chromatids resemble mitotic chromosomes (Dej and Spradling 1999). Defects in this transition have been noted for a hypomorphic mutation in MCM10, a protein that is implicated in DNA replication and chromatin remodeling, and in mutants in condensin II (Apger et al. 2010; Hartl et al. 2008). Although we were unable to detect defects in this transition, we noted that 35% of the ovarioles from PSF2/+ females displayed apoptosis at stage 7, a sign of failure to pass the previtellogenic checkpoint. The prevalence of this defect was abrogated by the re-introduction of a single good copy of Psf2 (Supplemental table 1). In wild-type, we only observed apoptosis in 5% of the ovarioles (Figure 8).
Figure 8.
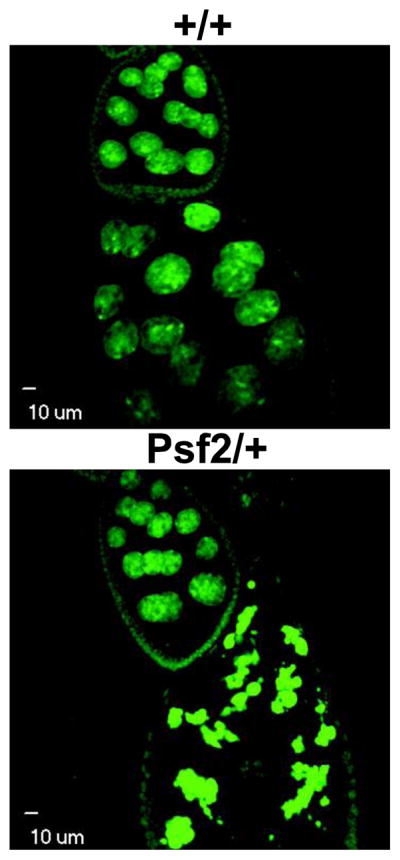
Confocal micrographs of Drosophila egg chambers and nurse cell nuclei. Representative images of stage 9 egg chambers from wild-type and Psf2/+. There is an increase in the number of apoptotic stage 9 egg chambers in Psf2/+ females
With respect to ovaries it should also be noted that eggs laid by Psf2/+ females did not exhibit any defects in chorion deposition indicating that chorion gene amplification is likely not impaired by haploinsufficiency of Psf2 (data not shown).
Position effect variegation
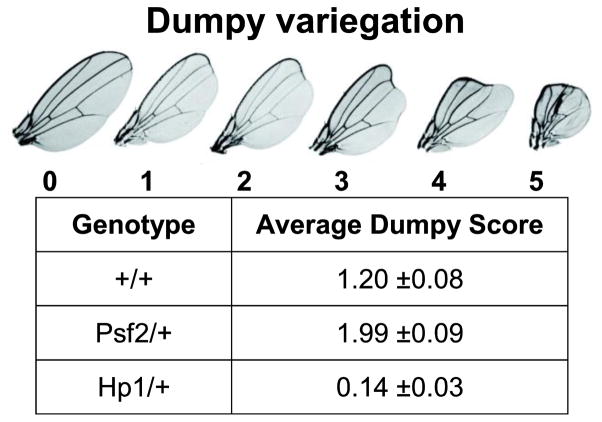
Measurements of position effect variegation (PEV) in Drosophila have been used to assess the status of heterochromatin silencing. Genes placed in proximity to regions of heterochromatin within the Drosophila genome display variegating expression and expansions or contractions of the adjacent heterochromatic domain leads to corresponding decreases or increases in the expression of these genes (Elgin 1996). It has been well documented that DNA replication associated proteins play a role in heterochromatic silencing in Drosophila (Shareef et al. 2001; Pak et al. 1997; Apger et al. 2010). This literature combined with our observations of chromosome condensation defects in Psf2/+ larvae suggested that Psf2 may also play a role in heterochromatic silencing. To test this hypothesis we utilized a previously described variegating allele of dumpy as a reporter for PEV (Apger et al. 2010). We found that Psf2SH0805/+ flies had a significantly higher (p=0.008) average dumpy score as compare to wild-type controls, while Hp1/+ flies had a significantly lower average score (Figure 9). These results suggest that the Psf2SH0805 mutant allele is a dominant enhancer of variegation.
Figure 9.
Psf2SH0805 is a dominant enhancer of variegation. Using a variegating allele of dumpy (see scoring rubric for phenotype) PEV was measured and average “dumpy” scores are reported in the table. Hp1 mutation suppresses variegation of the reporter compared to wild-type whereas Psf2 mutation enhances PEV compared to wild-type
Discussion
Although a late comer to the cadre of DNA replication factors our understanding of the GINS complex has benefited from careful biochemical and structural analysis (Aparicio et al. 2006; MacNeill 2010; Moyer et al. 2006). These studies have placed the GINS complex as an integral member of the CMG complex (Cdc45-MCM-GINS) which is involved in performing and coordinating helicase function during DNA replication. Much of the early characterization of the GINS has concentrated on its role in DNA replication; however there are an increasing number of clues that this complex or its subunits may have roles outside of DNA replication. In Drosophila, for instance, only approximately 5% of the GINS complex in embryo extracts is found in the CMG complex, leaving the possibility that 95% of the GINS complex members present may participate in other functions within the organism, although this may simply represent maternal loading (Moyer et al. 2006). The GINS complex members have also been shown to be present at high levels (100,000 units/cell) in untransformed human fibroblasts (Aparicio et al. 2009). These observations combined with work in budding yeast that point to the fact that only one GINS complex may be required for each DNA replication fork (Gambus et al. 2006), leaves open the real possibility that the GINS complex and/or its members have separable roles in other cellular processes. Indeed, other roles are beginning to be defined. Work in fission yeast has suggested that the GINS complex is important for chromosome segregation and that a subset of human Psf2 localizes to the mitotic spindle and the midzone during cytokinesis (Huang et al. 2005). Separate studies suggest that members of the GINS complex are important for development. Psf2 knockdown impairs eye development in Xenopus, and normal Psf1 dosage is required for proper stem cell proliferation and embryonic development in mice (Walter et al. 2008; Ueno et al. 2005; Ueno et al. 2009). This study presents data to support the idea that the proper dosage of Psf2 is required for a number of cellular processes related to chromosome biology (Table 1). These results lend support to the hypothesis that members of the GINS complex may have roles outside of DNA replication. We show that a haploinsufficiency of Psf2 in Drosophila has no significant impact on S phase progression in normally cycling cells in the larval brain or on endoreplication in salivary glands. These observations imply that the core GINS complex function with respect to its established role in DNA replication may be largely intact, despite the lowered levels of a Psf2. Instead the phenotypes we observe are consistent with a role for Psf2 in establishing or maintaining proper chromosome condensation during S phase. In our analysis of polyploid salivary gland nuclei we show that despite a normal DNA content, DNA is more tightly packaged when Psf2 levels are reduced. Moreover, the condensation defects we observe likely also have impacts on transcriptional regulation, as we have found that reduced dosage of Psf2 enhances position effect variegation. All this being said it is a real possibility that the role of Psf2 in the condensation dynamics of chromosomes is indirect. Specifically the observation that a reduction in Psf2 results in elevated levels of DNA damage suggest that the condensation and cell cycle defects observed may in fact be a secondary effect of the DNA damage. Increases in DNA damage have previously been associated with defects in chromosome condensation in Drosophila (Krause et al. 2001).
Table 1.
Summary of the phenotypes observed in flies heterozygous for the Psf2 mutation
| Psf2/+ | |
|---|---|
| Viability | Semi-lethal |
| Mitosis | delayed |
| Mitotic figures | Aberrant and aneuploidy |
| Mitotic chromosome size | longer |
| DNA damage | higher |
| S phase length | normal |
| Ratio of S phase nuclei with euchromatic pattern | higher |
| DNA replication of salivary glands | normal |
| Chromosome compaction of polytene chromosomes | higher |
| Ovary apoptosis | higher |
| Position effect variegation | enhanced |
One central question that arises out of our observations is whether or not the phenotypes we have observed are a result of lowered levels of the GINS complex or of Psf2 specifically. Put another way, is reduced dosage of the GINS complex responsible for the phenotypes observed or are these phenotypes the result of just Psf2 reduction? Given that the GINS complex has been convincingly shown to be heterotetrameric with a 1:1:1:1 stoichiometry of Sld5:Psf1:Psf2:Psf3 (MacNeill 2010), we would expect that knockdowns of other GINS complex members would result in similar phenotypes because the entire pool of the heterotetrameric complex would be reduced. However, our previous and ongoing work with Sld5 mutants suggests that there are significant differences. Haploinsufficiency of Sld5, unlike Psf2, leads to both an M and S phase delays in Drosophila (Gouge and Christensen 2010b). Chromosome defects in animals heterozygous for a Sld5 null mutation include polycentric chromosomes and telomere fusions (Gouge and Christensen 2010b). These defects are not observed in the Psf2 mutant. In addition, aneuploidy is observed in the Psf2 mutant but not Sld5 mutants. Haploinsufficiency of Sld5 does not result in an increase in DNA damage as evidenced by phospho H2Av staining like it does in flies lacking a dose of Psf2 (our unpublished data; Figure 4). We have also recently found that while salivary gland nuclei in larva heterozygous for the Sld5 mutation have normal DNA packaging ratios they contain nearly 2 times the DNA of wild type controls (our unpublished data). This is in stark contrast to the increased packaging ratios and normal DNA content we have observed in Psf2 mutant salivary glands. Taken all together the differences in phenotypes between Psf2 and Sld5 knockdowns suggest that these proteins have roles outside of the GINS complex or that there are alternate versions of the GINS complex in vivo.
Precisely how or if Psf2 functions directly in chromosome condensation remains a compelling question. Our observation that haploinsufficiency of Psf2 results in an increase in the fraction of S phase nuclei with euchromatic patterns of DNA replication in larval brain tissue suggests that proper levels of Psf2 may be required for the transition of DNA replication origin usage from early to late origins. This defect in replication timing may result in aberrant chromosome condensation. This observation combined with the slight (though not significant) decrease in cells in S phase (Figure 5) suggests that the role of Psf2 in S phase may not be completely separate from a role in condensation. There is precedence for this as defects in replication timing have been linked to chromosome condensation defects in Drosophila mutated for Orc2 (Loupart et al. 2000). Alternatively the role of Psf2 in maintaining chromosome condensation may be more direct through interaction with factors involved in modulating chromatin structure. There are a number of proteins that have been predicted, or shown, to interact with Psf2 that have established roles in chromosome biology. These partners include Mcm10 which has been shown to interact with Hp1 and is a suppressor of PEV (Apger et al. 2010; Christensen and Tye 2003; Muramatsu et al. 2010), Nurf-38; a member of the NURF complex involved in chromatin remodeling (Hazbun et al. 2003; Badenhorst et al. 2002), and Survivin; which is implicated in chromosome structure (Huang et al. 2005; Ruchaud et al. 2007).
Our characterization of the phenotypes associated with haploinsufficiency of Psf2 in Drosophila represents a foundation for future investigation of the possible roles of the GINS complex. A careful examination of the similar and differing phenotypes associated with haploinsufficiency of other GINS complex members may lead to clues that allow us to begin to understand the potentially separate roles for the different complex members.
Materials and Methods
Fly husbandry / stocks
Fly stocks (Psf2SH0805 /CyO Flybase ID: FBti0026079)( w 1118; +/+; +/+ Flybase ID: FBst0006326)( Df(2L)M24F-B/SM1 Flybase ID: FBst0000744) and (Hp15 Flybase ID: FBst0006234, In(1)w[m4h]; Su(var)205[5] / In(2L)Cy, In(2R)Cy, Cy[1]) were obtained from the Szeged stock center and the Bloomington Fly stock center. Psf2SH0805 line was backcrossed >7 times to w 1118; +/+; +/+ and rebalanced prior to analysis. The dumpy variegating line (w; SM1 / dp[w18] T(2:3) 3L^2R 2L^3L) and a Gla,dpov line (Gla, dp[Olv-12]/ Cy, Roi) were kindly provided by R. MacIntyre (Cornell University). Psf2SH0805 P element insertion was confirmed by PCR (Data not shown). All fly stocks were maintained on Drosophila K12 media (US Biological # D9600-07B) at room temperature unless otherwise noted.
Psf2 containing transgenic flies were made by amplifying genomic DNA using primers: CACCCGTATATAGTTTAAAATTGAGATCACTTTTG and CTGAGAAAACAGAGAGTTACTGTTCGG. This product includes 2000bp upstream of the translation start site of Psf2 and removes the stop codon for use in C terminal fusions. The PCR product was cloned into pENTR/D TOPO and sequence verified. This genomic Psf2 construct was then cloned into the pTWF vector for use in germline transformation (BestGene Inc.). A transgenic fly line containing a non-lethal insertion into the 3rd chromosome was identified and used to show that the lethality of Psf2SH0805 could be rescued by the transgene.
Antibodies/Western blots
Western Blot Analysis was performed on 0–2hr embryos collected from WT and Psf2/+ females on grape agar plates. Embryos were then ground in 1X SDS (1X SDS, 0.5M DDT) 100mg embryos/ml and immediately boiling for 10 minutes followed by centrifugation for 4 minutes. Samples were loaded into a 12.5% SDS page gel. Samples were run in 1X Hepes running buffer at 100V for 1 hour followed by transfer to Immobilon transfer membrane (Millipore). Transfer was run semi-dry for 4 hours at 33mAmps. Membrane was blocked overnight at 4°C in 5% BSA. After block, membrane was incubated at 4°C with rabbit anti-Psf2 (Moyer et al. 2006) and mouse anti-tubulin DM1a 1:1000 (Sigma) primary antibodies diluted 1:2000 and 1:20000 respectively in 5% BSA. After incubation membrane was washed briefly 3X with wash buffer followed by 5X 10 minutes washes. Membrane was then incubated with HRP conjugated secondary antibodies goat anti-rabbit and goat anti-mouse (Millipore), 1:5000 and 1:20000 respectively, at room temperature for 1 hour. Membrane was washed and developed with Immobilon™ Western Chemiluminescent HRP substrate (Millipore).
Phospho H3 immunostaining was carried out on embryos collected as above. Embryos were fixed with methanol/EGTA, prepared, and stained as in (Kellum and Alberts 1995). Embryos were incubated (1:500) with rabbit anti-Phospho H3(Millipore) at room temperature on a rotator for 5 hours followed by washes and incubation for 1 hour with 1:1000 goat anti-rabbit secondary antibody conjugated to fluorescein (Millipore). Embryos were then washed and stained with DAPI and mounted in Vectashield™ for visualization using an Olympus IX81 Motorized Inverted Microscope with Spinning Disk Confocal controlled by the SlideBook™ software.
Larval Brain squashes / Mitotic index / H2Av index / EdU incorporation
These protocols were carried out as previously described (Apger et al. 2010; Gouge and Christensen 2010b; Gouge and Christensen 2010a). For mitotic indices 3rd instar wandering larva were harvested and dissected, incubated in hypotonic solution (Sodium Citrate 0.5%) for 10 min, and then fixed in Acetic acid : Methanol : Water 11:11:2 for 30 seconds. Brains were then squashed, mounted in Vectashield® with DAPI, and visualized. Mitotic index determination were performed on these squash preparations by selecting 10 random well populated fields of view for each of 10 brain preparations using a 60X objective (total of 100 images). Total nuclei were counted for each field and this was divided by the total number of mitotic figures observed in each field to generate the fraction of cells in mitosis. Statistical analysis was performed using Minitab™.
Phospho-H2Av indices were carried out as for mitotic indices except that after hypotonic solution incubation slides were incubated in 3% BSA for 30 minutes, rinsed with PBS and then covered with 100μl of (1:500 dilution) primary antibody (Rockland Inc. Anti-Histone H2AvD pS137 (RABBIT) Antibody - 600-401-914) in 3% BSA and incubated at 4°C for 12–14hrs or overnight in a humidified chamber. Following this the coverslip is gently removed and the slide is rinsed 5X in PBS. The slides are then incubated with 100μl (1:500 dilution) of secondary antibody (Invitrogen, anti-rabbit Alexafluor 488) in 3% BSA for 1 hour at room temp. Following incubation the slides are again rinsed 5X with PBS and stained with 3μg/ml DAPI for 10 minutes and washed to remove excess stain using PBS. 7μl of Vectashieldtm; H1000 is pipetted onto the air-dried slide and a coverslip added, edges are sealed with nail polish and slides are stored at 4°C until imaging. Slides were imaged and images analyzed as for mitotic indices above.
EdU (5-Ethynyl-2′-deoxyuradine) incorporation analysis was performed as published with the exception that brains were squashed in 11:11:2 Methanol: acetic acid: water fixative (Gouge and Christensen 2010a). IX81 Motorized Inverted Microscope with Spinning Disk Confocal controlled by the SlideBooktm; software. S phase indices were determined as for mitotic indices above except that EdU positive cells were scored. Euchromatin/Heterochromatin indices were obtained in a similar fashion as S phase indices using the same images. EdU incorporation dispersed throughout the entire area of the nuclei was scored as Euchromatin replication. EdU incorporation that appeared as small blobs or that were localized to one side of a nucleus was scored as heterochromatin.
Polytene chromosomes spreads and DNA compaction analysis
Polytene chromosome spreads were performed as previously described (Apger et al. 2010). DNA Compaction methods and analysis were performed as described previously (Chmielewski and Christensen 2011). Briefly, for salivary gland whole mounts to acquire DNA volume, salivary glands from third instar wandering larva are dissected out in 100μl of 1x PBS, fixed in 4% formaldehyde, and stained with DAPI. After staining with DAPI, glands where transferred to a slide prepared with Vaseline® to serve as a support for the coverslip and mounted in Vectashield® Mounting Medium (Cat. No. H-1000, Vector Laboratories). Montage images where acquired using 20X magnification on an Olympus IX81 Motorized Inverted Microscope with Spinning Disk Confocal equipped with Slidebooktm. Areas for polytene nuclei were established using Adobe® Photoshop® element CS5 software. To determine DNA content per salivary gland pair, salivary glands were dissected out of wandering 3rd instar larvae in 150μl of HyQ Graces’s Unsupplemented Insect Cell Culture Medium (Cat. No. 30610.01, HyClone, Logan, UT). Once dissected, glands pairs were transferred to a PCR tube (Fisher, Cat. No. 14230225) prefilled with 3–5x 1mm glass beads (Biospec Products, Inc., Cat. No. 11079110) along with 300 micron glass beads (Sigma, 212–300 microns) and 25μl of squishing buffer (20μg/ml proteinase K, 10mM Tris-Base, 25mM NaCl, and 1mM EDTA). Glands were vortexed to facilitate the break-up of the salivary gland then incubated at 37°C 30 minutes 85°C for 10 minutes vortexed again for 15 seconds and spun at 12,000 rpm for 2 minutes. DNA content was determined using the Qubit® dsDNA HS Assay Kit (Invitrogen, Cat. No. Q32854) along with the Qubit® 2.0 Florometer (Invitrogen, Cat. No. Q32866) following manufactures instructions.
Ovary Analysis
Ovary analysis was performed on virgin females of WT and Psf2/+ lines fed for three days on yeast paste (1:1 active dry yeast to dH2O). Ovaries were then dissected in 1X PBS (0.137M NaCl, 2.68mM KCl, 1.44mM Na2HPO4, 1.76mM KH2PO4) then fixed with 4% formaldehyde solution in 1X PBX (1X PBS with 1% triton X-100) for 20 minutes. Ovaries were then stained with DAPI solution (1μg/ml) for 5 minutes. Ovaries were washed in 1X PBX 4X for 5 minutes, 1X for 1hr, and 3X for 10 minutes. Ovaries were mounted in 20μl of Vectashield® Mounting Media. Images were captured as above.
PEV analysis
The dpov allele was introgressed into the Psf2SH0805 and Hp1 mutant lines using standard genetic crosses. w; dpov, Psf2SH0805/CyO flies and w; Hp15/CyO were then crossed to w; Gla, dpov/Cy. The resulting F1 flies with the respective genotypes: w; dpov, Psf2SH0805/Gla,dpov and w; dpov, Hp15/Gla,dpov were crossed to the dumpy variegating line w; Cy / dpw18 T(2:3). This cross was incubated at 25°C as dumpy variegation is temperature sensitive (R. MacIntyre, personal comm.). The adult non-Glazed-eye progeny from these crosses were then scored for wing morphology (see text) and compared with Gla, dpov individuals acting as sibling controls. Greater than 500 flies from each genotype were scored.
Supplementary Material
Acknowledgments
The authors would like to thank Michael Reubens and Chad Hunter for productive discussions concerning the experimental methodologies, Tom Fink of the East Carolina University Imaging Core for his support, and Michael Botchan for again generously sharing reagents. We’d also like to thank Divya Devadasan for working out the H2Av staining protocol in the lab and Paul Hagar for suggestions on methods. Lastly we’d like to thank East Carolina University Department of Biology for the Next Step scholarship awarded to J.C and the National Institute of Health for funding (1R15GM093328-01) awarded to T.W.C.
References
- Aparicio T, Guillou E, Coloma J, Montoya G, Mendez J. The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res. 2009;37 (7):2087–2095. doi: 10.1093/nar/gkp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio T, Ibarra A, Mendez J, Aparicio T, Ibarra A, Mendez J. Cdc45-MCM-GINS, a new power player for DNA replication. Cell Division. 2006;1:18. doi: 10.1186/1747-1028-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apger J, Reubens M, Henderson L, Gouge CA, Ilic N, Zhou HH, Christensen TW. Multiple Functions for Drosophila Mcm10 Suggested Through Analysis of Two Mcm10 Mutant Alleles. Genetics. 2010;185 (4):1151–1165. doi: 10.1534/genetics.110.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci U S A. 1995;92 (25):11791– 11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16 (24):3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YP, Wang G, Bermudez V, Hurwitz J, Chen XS. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc Natl Acad Sci U S A. 2007;104 (31):12685– 12690. doi: 10.1073/pnas.0705558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielewski JP, Christensen TW. Novel Method for Determining Chromosome Compaction and DNA Content of Salivary Gland Nuclei in Drosophila. Drosophila Information Services. 2011;94 [Google Scholar]
- Christensen TW, Tye BK. Drosophila MCM10 interacts with members of the prereplication complex and is required for proper chromosome condensation. Mol Biol Cell. 2003;14 (6):2206– 2215. doi: 10.1091/mbc.E02-11-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126 (2):293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105 (3):297– 306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Elgin SC. Heterochromatin and gene regulation in Drosophila. Current Opinion in Genetics & Development. 1996;6 (2):193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Fox D, Gall J, Spradling A. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24 (20):2294– 2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8 (4):358– 366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gouge CA, Christensen TW. Detection of S phase in multiple tissues utilizing the EdU labeling technique. Drosophila Information Services. 2010a;93 [Google Scholar]
- Gouge CA, Christensen TW. Drosophila Sld5 is essential for normal cell cycle progression and maintenance of genomic integrity. Biochem Biophys Res Commun. 2010b;400 (1):145– 150. doi: 10.1016/j.bbrc.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T, Smith H, Bosco G. Chromosome alignment and transvection are antagonized by condensin II. Science. 2008;322 (5906):1384– 1387. doi: 10.1126/science.1164216. [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmström L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu C-H, Snydsman BE, Bradley P, Muller EGD, Fields S, Baker D, Yates JR, III, Davis TN. Assigning Function to Yeast Proteins by Integration of Technologies. Molecular Cell. 2003;12 (6):1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Huang HK, Bailis JM, Leverson JD, Gomez EB, Forsburg SL, Hunter T. Suppressors of Bir1p (Survivin) identify roles for the chromosomal passenger protein Pic1p (INCENP) and the replication initiation factor Psf2p in chromosome segregation. Mol Cell Biol. 2005;25 doi: 10.1128/MCB.25.20.9000-9015.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108 (4):1419–1431. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. MITOTIC CHROMOSOME CONDENSATION. Annual Review of Cell and Developmental Biology. 1996;12 (1):305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Krause SA, Loupart ML, Vass S, Schoenfelder S, Harrison S, Heck MM. Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol Cell Biol. 2001;21 (15):5156–5168. doi: 10.1128/MCB.21.15.5156-5168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A, Labib K, Gambus A. A key role for the GINS complex at DNA replication forks. Trends in Cell Biology. 2007;17 (6):271–278. doi: 10.1016/j.tcb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Loupart M-L, Krause S, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10 (24):1547–1556. doi: 10.1016/s0960-9822(00)00844-7. [DOI] [PubMed] [Google Scholar]
- MacNeill SA. Structure and function of the GINS complex, a key component of the eukaryotic replisome. Biochem J. 2010;425 (3):489–500. doi: 10.1042/BJ20091531. [DOI] [PubMed] [Google Scholar]
- Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30 (17):3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103 (27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S, Hirai K, Tak YS, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24 (6):602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obama K, Ura K, Satoh S, Nakamura Y, Furukawa Y. Up-regulation of PSF2, a member of the GINS multiprotein complex, in intrahepatic cholangiocarcinoma. Oncol Rep. 2005;14 (3):701–706. [PubMed] [Google Scholar]
- Oh S, Kingsley T, Shin H, Zheng Z, Chen H, Chen X, Wang H, Ruan P, Moody M, Hou S. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics. 2003;163 (1):195– 201. doi: 10.1093/genetics/163.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91 (3):311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Pflumm MF. The role of DNA replication in chromosome condensation. Bioessays. 2002;24 (5):411–418. doi: 10.1002/bies.10092. [DOI] [PubMed] [Google Scholar]
- Reed BH, Orr-Weaver TL. The Drosophila gene morula inhibits mitotic functions in the endo cell cycle and the mitotic cell cycle. Development. 1997;124 (18):3543– 3553. doi: 10.1242/dev.124.18.3543. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8 (10):798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Kohler H, Oakeley E, Stadler M, Schübeler D. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 2010;20 (6):771– 780. doi: 10.1101/gr.101790.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12 (6):1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A, McCleland M, O’Farrell P. Developmental control of late replication and S phase length. Curr Biol. 2010;20 (23):2067– 2077. doi: 10.1016/j.cub.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T. Heterochromatin replication: better late than ever. Curr Biol. 2010;20 (23):R1018– 1020. doi: 10.1016/j.cub.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17 (9):1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, Zhang H, FlyBase C. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37 (Database issue):D555– 559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Itoh M, Kong L, Sugihara K, Asano M, Takakura N. PSF1 is essential for early embryogenesis in mice. Mol Cell Biol. 2005;25 (23):10528–10532. doi: 10.1128/MCB.25.23.10528-10532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno M, Itoh M, Sugihara K, Asano M, Takakura N. Both alleles of PSF1 are required for maintenance of pool size of immature hematopoietic cells and acute bone marrow regeneration. Blood. 2009;113 (3):555–562. doi: 10.1182/blood-2008-01-136879. [DOI] [PubMed] [Google Scholar]
- Walter BE, Perry KJ, Fukui L, Malloch EL, Wever J, Henry JJ. Psf2 plays important roles in normal eye development in Xenopus laevis. Mol Vis. 2008;14:906–921. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.