Abstract
Unlike quantitative PCR (qPCR), digital PCR (dPCR) achieves sensitive and accurate absolute quantitation of a DNA sample without the need for a standard curve. A single PCR reaction is divided into many separate reactions that each have a positive or negative signal. By applying Poisson statistics, the number of DNA molecules in the original sample is directly calculated from the number of positive and negative reactions. The recent availability of multiple commercial dPCR platforms has led to increased interest in clinical diagnostic applications, such as low viral load detection and low abundance mutant detection, where dPCR could be superior to traditional qPCR.Here we review current literature that demonstrates dPCR’s potential utility in viral diagnostics, particularly through absolute quantification of target DNA sequences and rare mutant allele detection.
Clinical viral diagnostic approaches rely heavily on quantitative PCR (qPCR) as a method to detect and quantify viral load in patient samples. For the past twenty years fluorescence-based qPCR chemistries have revolutionized nucleic acid diagnostics and become the gold standard for viral load quantification(Mackay et al., 2002) and detection of bacterial pathogens, among myriadother applications. During qPCR, DNA is amplified until it produces a certain level of signal which is supplied through a DNA intercalating dye or sequence-specific fluorescent probe. The cycle threshold, defined as the number of amplification cycles required to reach that signal level, is used to calculate the number of DNA molecules originally present based on a standard curve(Bustin, 2004).
Though qPCRhas driven major advances in disease diagnosis, this technology has notable limitations. Quantification is based on a standard curve, which requires careful calibration and consistent source material. Additionally, the choice of signal threshold can be made by the operator, introducing subjectivity into the analysis. Due to differences in standard curve construction and potential subjectivity in analysis, interlab variation can be substantial even when using commercial kits and standardized protocols. Moreover, even within a highly trained lab the coefficient of variation for any single assay can be 20-30% or higher at lower template copy number(Lai et al., 2003; Cook et al., 2009). For example, the interassay variability for a CMV quantitation assay is considered low with a viral load coefficient of variation of 28% (Boeckh et al., 2004).
Digital PCR (dPCR) promises to remedy some of the shortcomings of qPCR by transforming the analog, exponential nature of PCR into a digital, linear signal (Vogelstein & Kinzler, 1999). Here we discuss the theoretical basis for dPCR and the currently available commercial dPCR systems. We also review current literature that demonstrates dPCR’s potential utility in viral and microbial diagnostics, particularly through absolute quantification of target DNA sequences and rare mutant allele detection.
Digital PCR
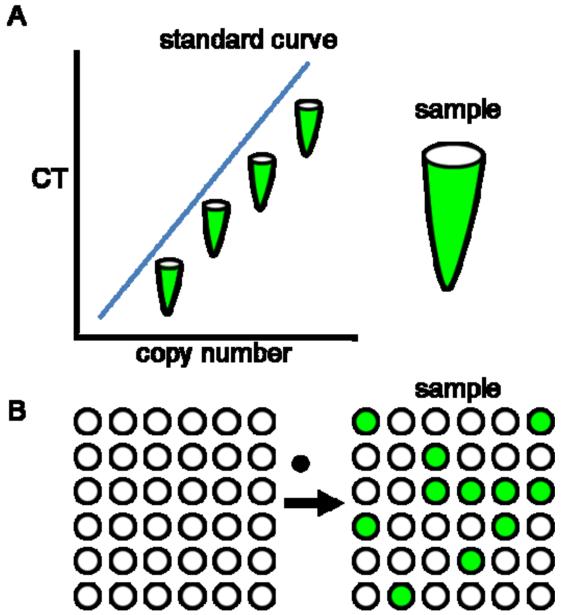
First described in the 1990s(Sidransky et al., 1992; Vogelstein & Kinzler, 1999), dPCRuses the same primers and probes as qPCR, but touts increased sensitivity and precision. These improvements are achieved by diluting the sample and partitioning it into individual reactions so that ideally each reaction contains one or no copies of the DNA of interest (Figure 1). The number of positive versus negative reactions is counted to directly calculate the number of DNA molecules in the original sample based on Poisson statistics. If the sample is not dilute, many of the individual reactions will be positive and will have contained two, three or more target molecules. In this case, simply counting the positive reactions would underestimate the true number of molecules. This underestimation can be corrected using the Poisson equation (copies per reaction = −ln (1-p) where p is the fraction of positive reactions), which calculates the average number of molecules per reaction from the observed proportion of positive reactions (Sykes et al., 1992). Using Poisson statistics, digital PCR provides absolute quantification of nucleic acids, reducing subjectivity in analysis by abrogating the need for signal thresholddetermination and standard curves.
Figure 1.
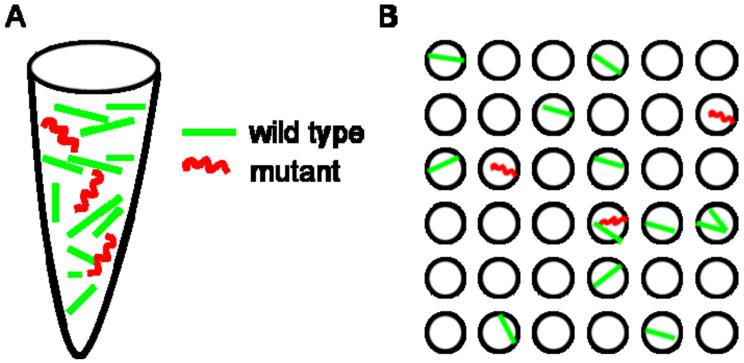
Additionally, when amplification is carried out in bulk reactions, it is difficult to quantify poorly represented target sequences in a background of more abundant species. Digital PCR increases sensitivity by isolating rare target species so they are not competing with extraneous DNA targets for primers or other reagents (Figure 2). While the concept of dPCR is a powerful one for nucleic acid analysis, the technique has been limited by technical roadblocks associated with the sheer number of reactions required for statistically significant results. The advent of multiple commercially available platforms capable of running reactions on the nano- to picoliter scalehave made dPCRa practicaltool with great potential in research and clinical settings.
Figure 2.
Digital PCR platforms
Four different dPCR platforms are currently marketed and differ mainly in their method of individual reaction partitioning (Table 1). Fluidigm Corporation and Life Technologies offer microfluidics based systems that partition sample using sophisticated chips designed with microfluidics channels that deliver nanoliter volumes of sample into individual reaction wells. These systems are limited only by the number of reactions that fit onto a single microfluidics chip (hundreds to thousands) and the cost of the consumable chips (in the hundreds of dollars) (Baker, 2012). Bio-Rad laboratories and RainDance have developed systems that divide diluted sample among many water-in-oil droplets(Kiss et al., 2008; Hindson et al., 2011). Each droplet represents a single reaction allowing simultaneous analysis of thousands (Bio-Rad) to millions (RainDance) of separate reactions(Baker, 2012).
Table 1.
Overview of Commercial Digital PCR systems. Adapted from Baker, 2012.
| Reaction partitioning format |
Vendor | Instrument and list price | Consumables and list price |
Number (and volume) of partitions |
Multiplexing capabilities |
|---|---|---|---|---|---|
| Plate format |
Fluidigm Corporation |
Biomark HD $200-250K | $400/chip | 9,180 (6 nl) partitions/chip |
5 colors |
| Life Technologies |
OpenArrayRealTime PCR System $140K and QuantStudio 12K Flex instrument $90-190K |
$150/plate | 3, 072 (33 nl) partitions/plate |
2 colors | |
| Droplet format |
Bio-Rad Laboratories |
QX100 ddPCR system $89K | $3/sample 8samples/chip |
20,000 (1nl) droplets/sample |
2 colors |
| RainDance | RainDrop Digital PCR $100K | $10-30 sample 8samples/chip |
1,000,000 (5 pl) droplets/sample |
2 colors |
In addition to commercially available dPCR systems, several labs are developing simpler dPCR systems with the goal of making this technology practical in resource limited settings. For example, the SlipChip platform relies simply on the movement or “slipping” of two plates to reproducibly and precisely deposit discreet volumes suitable for parallel compartmentalization of nucleic acids(Shen et al., 2010). Any system capable of dividing one bulk PCR reaction into many discrete reactions is suitable for dPCR, whose utility derives simply from the ability to identify the amplification of a single nucleic acid template in many separate reactions.
Applications of dPCR
While dPCR promises more sensitive and accurate nucleic acid detection, its use has been mainly limited to research applications. For example, Tadmor and colleagues used digital PCR instead of classical phage enrichment to identify virus-bacteria interactions in uncultured bacteria. The group targeted phage-like elements with degenerate primers and targeted bacterial small subunit ribosomal RNA genes with universal “all bacterial” primers in a microfluidics dPCR platform to identify previously unknown, uncultured bacteria in the termite hindgut (Tadmor et al., 2011).
Digital PCR has significantly advanced research capabilities but itspotential for clinical application has been investigated only to a limited degree, partly because devices that are practical, both in cost and dynamic range of detection, are just now becoming commercially available. As commercial systems gain wider use, dPCR could become a standard diagnostic approach for nucleic acid quantition. Two areas where dPCR has shown potential clinical diagnostic utility are absolute quantification of target DNA sequences and rare mutant allele detection.
Absolute quantification
Digital PCR provides a sensitive method for the direct measure of viral nucleic acid, providing the absolute number of copies/ml without the need for a standard curve.White and colleagues utilized the FluidigmdPCR system to quantify GB Virus Type-C (GBV-C), an occult RNA virus associated with HIV-1 infection(White et al., 2012). Co-infection of HIV-1 patients with GBV-C has been suggested to lead to a decrease in the temporal progression to AIDS(Bhattarai & Stapleton, 2012; Gretch, 2012). Therefore, tracking the presence of GBV-C early in infection could provide the information needed for a more comprehensive patient prognosis. White and colleagues compared quantification of GBV-C isolated from transfected cells lines using standard qPCRanddPCR;theyfound that dPCRhad an average coefficient of variation (CV, measure of precision) of 11.7±2.2% for viral load testingwhile standard qPCR had an average CV of 25.8±4.9%. Using dPCR they could detect between 3 and 10 DNA molecules/microliter, a level that could not be detected by traditional qPCR in parallel experiments.
The second comparison of viral qPCR and dPCR was carried out by Henrich and colleagues on HIV-1 quantitation. They found that serial dilutions of HIV-1 or human CCR5 DNA amplicon standards quantitated by droplet digital PCR (ddCPR, Bio-Rad) matched expected nominal copy numbers. When they ran the same assay on patient samples they found that both ddPCR and qPCR had similar sensitivity but ddPCR enumerated 10-40% fewer DNA copies compared with qPCR. While the reason for this discrepancy is speculative, the authors offer that error could be introduced into the qPCR assay by the spectrophotometric determination of the DNA concentration of the standards (Henrich et al., 2012).
A study by Kiss et. al. validated the RainDancedPCR platform for sensitive absolute quantification. This system utilizes millions of picoliter droplets to divide a PCR reaction into millions of separate negative or positive outputs. They detected adenovirus at starting template concentrations as low as 1 template molecule/167 droplets or 92 molecules/ul(Kiss et al., 2008).
The move towards absolute quantification of viral load is driven not only by commercially available dPCR systems but by simple systems suitable for point-of-care and resource limited settings. Shen and colleagues used a SlipChip system to show that absolute quantification of HIV and HCV could be achieved within a large dynamic range (Shen et al., 2011). They utilized a rotational SlipChip, a microfluidic platform that manipulates liquid samples from microliter to picoliter scales through the relative movement of different plates without requiring complex control systems. They validated their assay with viral RNA from two HIV patients. Their measurements had good agreement with measurements from the standard clinical assay, and achieved a dynamic range of 3-fold (0.5 log10) resolution from1.7 × 102 to 2.0 × 107 molecules/ml, with a lower detection limit of 40 molecules/ml. They also validated a multiplex SlipChip with a five-plex panel to simultaneously detect HIV and HCV along with a negative and positive control with a dynamic range of 1.8×103 to 1.2×107 molecules/ml.
Validation of dPCR for clinical viral diagnostics is still in its infancy. However, these initial studies demonstrate the potential clinical utility of dPCR for rapid, sensitive, and accurate quantification of viral load in patient samples.
Rare mutant detection
A second important application of dPCR in viral diagnostics, which has been most well-studied thus far in the oncology field, is the detection of rare point mutants in a background of wild type sequences. Wang and colleagues used a RainDancedPCR platform for the sensitive and quantitative detection of mutations in the KRAS oncogene, one of the most common oncogenic alterations in a range of human cancers(Pekin et al., 2011). In oncology diagnostics, somatic mutations in tumor DNA are used as highly specific biomarkers to distinguish cancer cells from their normal counterparts. Current qPCRTaqMan assays and pyrosequencing cannot detect less than 1-10% mutant genes in a non-mutated DNA background(Pekin et al., 2011). Digital PCR (on the RainDance platform) improves mutant detection by compartmentalizing genomic DNA (gDNA)into millions of picoliter droplets at a concentration of less than one genome equivalent per droplet with two TaqMan probes, one for mutant (green) and one for wild type (red). The ratio of green to red fluorescent reactions determines the ratio of mutant to wild type genes. Wang et. al. quantified mutations in codons 12 and 13 of the KRAS oncogene in gDNA fom several different human cell lines and were able to detect 1 mutant in a background of 200,000 wildtypeKRAS genes (0.005% mutant) by analyzing 106 droplets(Pekin et al., 2011).
Another example of rare mutant detection by dPCRis the detection of low abundance epidermal growth factor receptor (EGFR) mutations in tumor tissue and plasma (Yung et al., 2009; Wang et al., 2010). Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors retard the progression of some lung cancers. Responsiveness to these inhibitors is associated with the presence of activating mutations in the EGFR kinase domain. Therefore, Yung and colleagues investigated dPCR analysis (Fluidigm platform) for detection of the two most common EGFR mutations in tumor tissues and plasma of lung cancer patients. Direct sequencing was commonly used in early studies, but this technique only detected mutant sequences greater than 30% of the total genetic content(Yung et al., 2009). Using dPCR, they were able to identifymutant sequences that were not detected by traditional sequencing methods. In these samples mutant sequence constituted 2-14% of the total DNA. Similarly, Wang and colleagues utilized dPCR (Fluidigm platform) to detect and quantitaterare (0.02%-9.26% abundance) drug-sensitizing EGFR mutations in tumor DNA.
These studies distinguish dPCR as a powerful tool for identifying low abundance mutant alleles in a background of high abundance wild type sequence. While these data focus on oncology diagnostic applications, the principles demonstrated here translate to virology diagnostic applications where detection of low abundance mutant sequences, such as those mediating antiviral resistance, can significantly impact treatment outcome.
Potential applications and limitations ofdPCR
Digital PCR’s potential for sensitive and accurate quantitation of nucleic acids could offer significant improvements over current viral diagnostic procedures, particularly in detecting very low viral loads. Clinical significance of low levelviremia has not been well established, partly because the typical lower limit of 95% detection is around 40-60 copies/ml for typical viral assays.(Widdrington et al., 2011; Waggoner et al., 2012). At this level, viral load is detectable but not realiablyquantitated, resulting in a large number of patients with ongoing but unquantifiable or undetectable levels of viremia. One CMV studysuggests that increases in viral load even at very low levels were clinically meaningful(Waggoner et al., 2012). Other studies on very low levelviremia in HIV infected patients suggest that low level detection of HIV-1 viral load could be useful in predicting subsequent suboptimal viral control in patients on retroviral therapy (Widdrington et al., 2011; Doyle & Geretti, 2012; Doyle et al., 2012).Therefore, if future work indicates that dPCR assays have greater sensitivity and precision than qPCR assays at low viral loads, clinical treatment and outcome could be improved in situations where patient management relies on low-level viral load detection. Moreover, just as dPCR has been utilized to identify low abundance oncogenic mutations, it could be adapted to identify low frequency virus variants, e.g. emerging drug resistant mutants of CMV, HIV, or HBV in patients on antiviral therapy. As mentioned above, sequencing techniques, which are often employed for drug resistance mutant detection, can not detect less than 1-10% mutant genes in a wild type DNA background. Allele specific digital PCR has the potential to detect very low abundance, emerging drug resistance mutations for applications where only a few key mutations need to be monitored.
Another application of dPCRcould be the detection of chromosomally integrated viral genomes. Human herpes virus 6 (HHV-6) can integrate into human chromosome telomere regions, causing complications in the interpretation of HHV-6 PCR testing because normal PCR assays detect HHV-6 infections and integrated DNA. One study estimated that about half of all HHV-6 positive cerebrospinal fluid samples were due to detection of integrated HHV-6 rather than actual infection (Ward et al., 2007).A current assay for integration involves detection of the ratio between cellular DNA and HHV-6 DNA. However, with the variation inherent in current qPCR assays, the ratio of cell DNA to viral DNA can range from 0.5 to 2.0 or worse. The precision and reproducibility of dPCR, which does not require a standard curve for quantitation, may improve such ratio-based chromosomal integration assays.
Despite dPCR’s potential, there may be limitations to utilizing this technology in a clinical diagnostic setting. Some of the commercial dPCR platforms have a relatively small number of partitions that can only be scaled up using multiple costly, one-time-use microfluidics chips. Also, studies need to investigate whether the sensitivity of dPCR assays exceeds current clinical qPCR assay sensitivity. Theoretically, dPCR should be more sensitive and more precise at low virus levels using the same qPCRtaqman primers and probes, but practical issues such as limits on template input volume and master mix compatibility on the dPCR platform need to be considered.The digital platforms also add another layer of complexity to any assay, potentially slowing workflow and introducing error during sample pipetting and transfer, depending on the dPCR system used.
Digital PCR is unlikely to supplant qPCR in the short term, but instead will be a complementary approach in certain applications. Digital PCR has the potential to improve inter- and intra-lab variation. Currently qPCR assays base quantitation on a standard curve, which can vary from lab-to-lab or even run-to-run.Digital PCR requires no standard curve to quantitate nucleic acid molecules, so is a more direct, accurate method of quantitation. The sensitivity of dPCR is limited only by the number of individual reactions that are run simultaneously on a sample, so dPCR should provide the ability to detect below one viral copy per milliliter sample using already established quantitative PCR protocols for many viruses, including CMV, HIV and HCV. The absolute quantitation provided by dPCR could also improve specialty assays such as those for viral chromosomal integration. Digital PCR’s sensitive and reproducible nucleic acid detection demonstrated thus far in research settings could translate well to a diagnostics setting as commercially available, high throughput dPCR systems become more accessible.
Acknowledgements
This work was funded in part by NIH U19 AI96111.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker M. Digital PCR hits its stride. Nat Meth. 2012;9:541–544. [Google Scholar]
- Bhattarai N, Stapleton JT. GB virus C: the good boy virus? Trends Microbiol. 2012;20:124–130. doi: 10.1016/j.tim.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, et al. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol. 2004;42:1142–1148. doi: 10.1128/JCM.42.3.1142-1148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA. A-Z of Quantitative PCR. International University Line; La Jolla, CA: 2004. [Google Scholar]
- Cook L, Atienza EE, Bagabag A, Obrigewitch RM, Jerome KR. Comparison of methods for extraction of viral DNA from cellular specimens. Diagn Microbiol Infect Dis. 2009;64:37–42. doi: 10.1016/j.diagmicrobio.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Doyle T, Geretti AM. Low-level viraemia on HAART: significance and management. Curr Opin Infect Dis. 2012;25:17–25. doi: 10.1097/QCO.0b013e32834ef5d9. [DOI] [PubMed] [Google Scholar]
- Doyle T, Smith C, Vitiello P, Cambiano V, Johnson M, Owen A, et al. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy. Clin Infect Dis. 2012;54:724–732. doi: 10.1093/cid/cir936. [DOI] [PubMed] [Google Scholar]
- Gretch D. Advocating the Concept of GB Virus C Biotherapy Against AIDS. Clin Infect Dis. 2012 doi: 10.1093/cid/cis591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods. 2012;186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss MM, Ortoleva-Donnelly L, Beer NR, Warner J, Bailey CG, Colston BW, et al. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KK, Cook L, Wendt S, Corey L, Jerome KR. Evaluation of real-time PCR versus PCR with liquid-phase hybridization for detection of enterovirus RNA in cerebrospinal fluid. J Clin Microbiol. 2003;41:3133–3141. doi: 10.1128/JCM.41.7.3133-3141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11:2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Digital PCR on a SlipChip. Lab Chip. 2010;10:2666–2672. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, et al. Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load. J Am Chem Soc. 2011;133:17705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, et al. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- Tadmor AD, Ottesen EA, Leadbetter JR, Phillips R. Probing individual environmental bacteria for viruses by using microfluidic digital PCR. Science. 2011;333:58–62. doi: 10.1126/science.1200758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J, Ho DY, Libiran P, Pinsky BA. Clinical significance of low cytomegalovirus DNA levels in human plasma. J Clin Microbiol. 2012;50:2378–2383. doi: 10.1128/JCM.06800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ramakrishnan R, Tang Z, Fan W, Kluge A, Dowlati A, et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin Chem. 2010;56:623–632. doi: 10.1373/clinchem.2009.134973. [DOI] [PubMed] [Google Scholar]
- Ward KN, Leong HN, Thiruchelvam AD, Atkinson CE, Clark DA. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J Clin Microbiol. 2007;45:1298–1304. doi: 10.1128/JCM.02115-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Quake SR, Curr K. Digital PCR provides absolute quantitation of viral load for an occult RNA virus. J Virol Methods. 2012;179:45–50. doi: 10.1016/j.jviromet.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Widdrington J, Payne B, Medhi M, Valappil M, Schmid ML. The significance of very low-level viraemia detected by sensitive viral load assays in HIV infected patients on HAART. J Infect. 2011;62:87–92. doi: 10.1016/j.jinf.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15:2076–2084. doi: 10.1158/1078-0432.CCR-08-2622. [DOI] [PubMed] [Google Scholar]