Abstract
Polo-like kinase 1 (Plk1) is an essential mitotic regulator and undergoes periodic phosphorylation on threonine 210, a conserved residue in the kinase’s activation loop. While phosphate-mimicking alterations of T210 stimulate Plk1’s kinase activity in vitro, their effects on cell-cycle regulation in vivo remain controversial. Using gene targeting, we replaced the native PLK1 locus in human cells with either PLK1T210A or PLK1T210D, in both dominant and recessive settings. In contrast to previous reports, PLK1T210D did not accelerate cells prematurely into mitosis, nor could it fulfill the kinase’s essential role in chromosome congression. The latter was traced to an unexpected defect in Plk1-dependent phosphorylation of BubR1, a key mediator of stable kinetochore-microtubule attachment. Using chemical genetics to bypass this defect, we found that Plk1T210D is nonetheless able to induce equatorial RhoA zones and cleavage furrows during mitotic exit. Collectively, our data indicate that K-fibers are sensitive to even subtle perturbations in T210 phosphorylation, and caution against relying on Plk1T210D as an in vivo surrogate for the natively activated kinase.
Introduction
Polo-like kinase 1 (Plk1) is an evolutionarily conserved regulator of mitosis and cell division, with essential roles in centrosome maturation, bipolar spindle assembly, chromosome congression, and cytokinesis (Archambault and Glover 2009; Petronczki et al. 2008; Randall et al. 2007). This breadth of functions is reflected in the kinase’s dynamic pattern of localization, as Plk1 initially accumulates on centrosomes in G2 phase, then associates with kinetochores in prometaphase, and finally migrates to the spindle midzone and midbody after anaphase onset. This spatial targeting is mediated by Plk1’s C-terminal Polo-box domain (PBD), a specialized phosphopeptide-binding module that also controls Plk1’s interaction with and activity towards specific substrates (Lowery et al. 2005).
A second mode of Plk1 regulation involves its mitosis-specific phosphorylation. One major phosphoacceptor, threonine 210, lies within the activation loop and plays an important role in stimulating Plk1 activity at the G2/M transition (Jang et al. 2002; Kelm et al. 2002; Lee and Erikson 1997; Qian et al. 1999). While the heteromeric Aurora A-Bora kinase complex is thought to initiate T210 phosphorylation in G2 phase (Macurek et al. 2008; Seki et al. 2008b), the kinase(s) that sustain and enhance this modification in mitosis (that is, after Bora has been ubiquitinated and destroyed (Chan et al. 2008; Seki et al. 2008a)) remain obscure. Regardless, direct substitution of T210 with aspartic acid increases Plk1’s in vitro kinase activity several-fold, while replacement with alanine or valine reduces it (Jang et al. 2002; Kelm et al. 2002; Lee and Erikson 1997; Qian et al. 1999). Consequently, Plk1T210D has been regarded as a “constitutive-active” form of the kinase and used to reach conclusions about Plk1’s in vivo roles and regulation (Deming et al. 2002; Fu et al. 2008; Kishi et al. 2009; Li et al. 2010; Lindon and Pines 2004; Loncarek et al. 2010; Macurek et al. 2008; Peschiaroli et al. 2006; Smits et al. 2000; van de Weerdt et al. 2005; van Vugt et al. 2004; Yamaguchi et al. 2005; Zhang et al. 2005; Zhou et al. 2003).
Using gene targeting and chemical genetics, we have re-examined the functional properties of Plk1T210A and Plk1T210D in both dominant and recessive settings. Whereas hemizygous expression of Plk1T210A recapitulated the breadth of defects associated with wholesale loss of Plk1 activity, hemizygous expression of Plk1T210D selectively compromised K-fiber stability, at least in part due to inadequate phosphorylation on BubR1. In contrast, we failed to find any evidence that Plk1T210D can accelerate mitotic entry or override the DNA damage checkpoint when heterozygously expressed from its native locus in the human genome. Collectively these data demonstrate that Plk1’s activation-loop phosphorylation is both essential and irreplaceable during M phase, but unlikely to be rate limiting beforehand.
Results and discussion
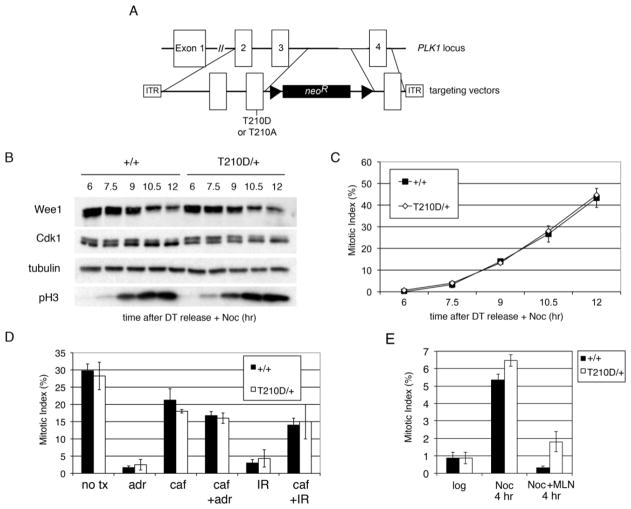
Using adeno-associated virus (AAV)-mediated gene targeting, T210A and T210D mutations were introduced into both telomerase-immortalized human retinal pigment epithelial cells (hTERT-RPE) and colorectal carcinoma (HCT116) cells (Fig. 1a and Supplementary Fig. S1a). Transgenic expression of Plk1T210D was previously reported to accelerate mitotic entry and override the G2 DNA damage checkpoint (Jackman et al. 2003; Macurek et al. 2008; Smits et al. 2000; van Vugt et al. 2004). However, once released from a double-thymidine block into nocodazole-containing medium, PLK1T210D/+ cells degraded Wee1 and dephosphorylated Cdk1 on schedule (Fig. 1b) and accumulated in mitosis with the same kinetics as their isogenic wildtype counterparts (Fig. 1c). Likewise, PLK1T210D/+ cells mounted a fully intact G2 arrest in response to either adriamycin or ionizing radiation, which induce double-stranded DNA breaks and engage the DNA damage checkpoint (Fig. 1d), and also delayed in G2 phase when challenged with the Aurora A inhibitor MLN8054 (Fig. 1e). We conclude that this mutant kinase does not advance the G2/M transition or bypass the DNA damage checkpoint when expressed from its natural genomic context.
Figure 1. PLK1T210D/+ cells enter mitosis with normal kinetics and have an intact G2 DNA-damage checkpoint.
a Schematic of PLK1 locus and gene-targeting vectors. Triangles represent FRT sites. b–c HCT116 PLK1+/+ and PLK1T210D/+ cells were synchronized by a double thymidine block and released into medium containing nocodazole. At 90-minute intervals, samples were withdrawn for immunoblotting of the indicated proteins (b) and determination of mitotic indices by Hoechst staining and fluorescence microscopy (c). Error bars indicate SEM. d G2 checkpoint assay (Papi et al. 2005). HCT116 PLK1+/+ and PLK1T210D/+ cells were pre-treated with caffeine (caf) to induce checkpoint override or left alone, challenged with adriamycin (adr) or ionizing radiation (IR) as indicated, and then cultured in the presence of nocodazole for 6 hours to trap cells escaping from G2 phase into mitosis. Mitotic indices were determined as above. e hTERT-RPE1 PLK1+/+ and PLK1T210D/+ cells were treated with nocodazole (Noc) or nocodazole plus the Aurora A kinase inhibitor MLN8054 (Noc + MLN) for 4 hr. Mitotic indices were then determined and compared to untreated cells.
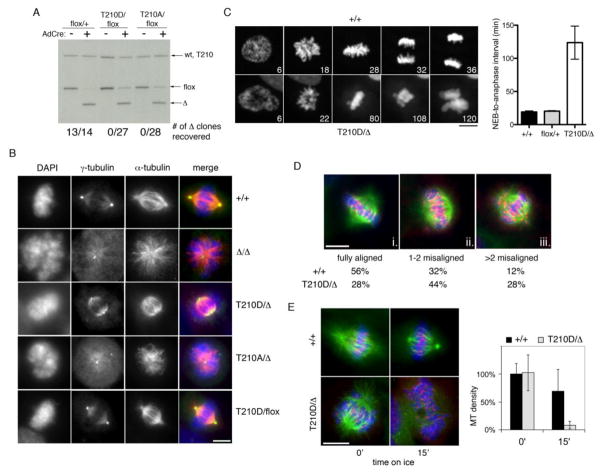
To investigate recessive phenotypes, the same gene-targeting constructs were used to modify hTERT-RPE cells bearing a PLK1 conditional-knockout allele (Burkard et al. 2007), yielding PLK1flox/T210A and PLK1flox/T210D cells. An adenovirus expressing Cre recombinase (AdCre) was then used to delete the PLK1flox allele and effect monoallelic expression of PLK1T210A and PLK1T210D (Fig. 2a and Supplementary Fig. S1b). Based on the robust catalytic activity of Plk1T210D in vitro (Jang et al. 2002; Kelm et al. 2002; Lee and Erikson 1997; Qian et al. 1999), as well as the low threshold of Plk1 activity required in this cell type (Burkard et al. 2007; Liu et al. 2006), we anticipated that one or both alleles should support cell proliferation. However, no PLK1T210D/Δ or PLK1T210A/Δ clones were recoverable by limiting dilution (Fig. 2a). To understand this result, we performed immunofluorescence microscopy and live-cell imaging experiments shortly after AdCre infection. Similar to PLK1Δ/Δ cells, PLK1T210A/Δ cells lacked mature centrosomes and often formed monopolar spindles (Fig. 2b and Table 1), resulting in a lengthy prometaphase arrest followed by mitotic slippage (Supplementary Fig. S2). In comparison nearly all PLK1T210D/Δ cells had mature centrosomes and underwent bipolar spindle assembly (Fig. 2b and Table 1), but nonetheless delayed transiently in prometaphase, often with one or more misaligned chromosomes visible (Fig. 2c), before exiting mitosis as micronucleated cells (Supplementary Fig. S2).
Figure 2. Monoallelic PLK1T210D expression is inadequate for chromosome congression.
a hTERT-RPE1 cells of the indicated genotypes were infected with AdCre and processed for Southern blotting (to confirm excision of the PLK1flox allele in the bulk culture) and limiting dilution (to assess clonal viability post-excision). b Three days after AdCre infection, cells were fixed, stained with antibodies to γ-tubulin (green) and α-tubulin (red), and examined microscopically. Scale bar represents 10 microns. c Cells expressing histone H2B-GFP were infected with AdCre and followed by timelapse microscopy 48 to 72 hours later. Time after chromosome condensation is indicated. At least 20 cells were scored for each line. Error bars indicate SEM. d Two days after AdCre infection, cells were treated with monastrol and then released into MG132 for 1 hour. Chromosome alignment was scored by CREST (red) and α-tubulin (green) immunofluorescence microscopy. At least 50 cells of each genotype were scored. e K-fiber stability assay. Cells were incubated on ice for 15 minutes before fixation and staining with CREST (red) and α-tubulin (green) antibodies. Microtubule density was scored as the fraction of pixels with intensity greater than 2X above background. Error bars indicate SEM.
Table 1.
T210 phosphorylation is required for spindle bipolarity
| unseparated centrosomes | monopolar spindles | |
|---|---|---|
| +/+ | 4% | 4% |
| Δ/Δ | 60% | 52% |
| T210D/Δ | 16% | 14% |
| T210A/Δ | 30% | 24% |
| as + 3-MB | 60% | 60% |
| as/wt + 3-MB | 4% | 4% |
| as/T210D + 3-MB | 2% | 2% |
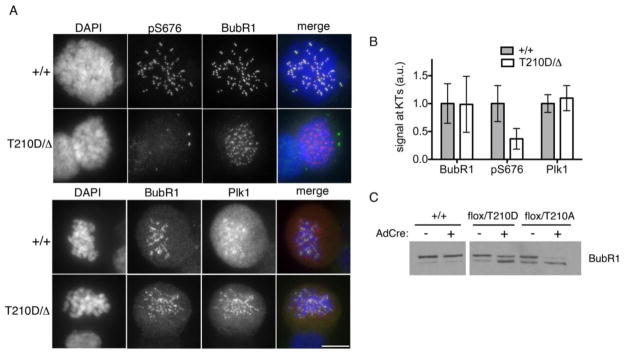
To evaluate chromosome congression in more detail, we fixed cells after transient treatment with monastrol and analyzed them by immunofluorescence microscopy. Consistent with our timelapse studies, the frequency of chromosome alignment errors was markedly higher in PLK1T210D/Δ cells as compared with PLK1+/+ cells (Fig. 2d). We also tested the integrity of kinetochore-bound microtubules (also called K-fibers), which normally resist cold-induced depolymerization (Rieder 1981). However, the K-fibers of PLK1T210D/Δ cells were extremely labile (Fig. 2e). We then asked if the T210D mutation affects the phosphorylation of known Plk1 substrates at the kinetochore. One leading candidate in this regard is BubR1, whose phosphorylation by Plk1 is important in stabilizing kinetochore-microtubule interactions (Elowe et al. 2007; Matsumura et al. 2007). Remarkably, despite proper targeting of both Plk1 and BubR1 to kinetochores, BubR1’s phosphorylation on S676 was sharply reduced in PLK1T210D/Δ cells (Fig. 3a–b). We also observed a prominent (though incomplete) increase in BubR1’s electrophoretic mobility (Fig. 3c), which normally is retarded by Plk1-dependent phosphorylation on S676 and other sites (Elowe et al. 2007; Lenart et al. 2007; Matsumura et al. 2007).
Figure 3. Plk1T210D is unable to phosphorylate BubR1 in vivo.
a Nocodazole-arrested cells were stained with pS676-BubR1 (green) and total BubR1 (red) antibodies (top panels) or total BubR1 (green) and Plk1 (red) antibodies (bottom panels). Scale bar represents 10 microns. b BubR1 and Plk1 signals were quantified from at least 50 kinetochores in 6 cells of each genotype. c Lysates from nocodazole-arrested cells were resolved by SDS-PAGE and immunoblotted with BubR1-specific antibodies.
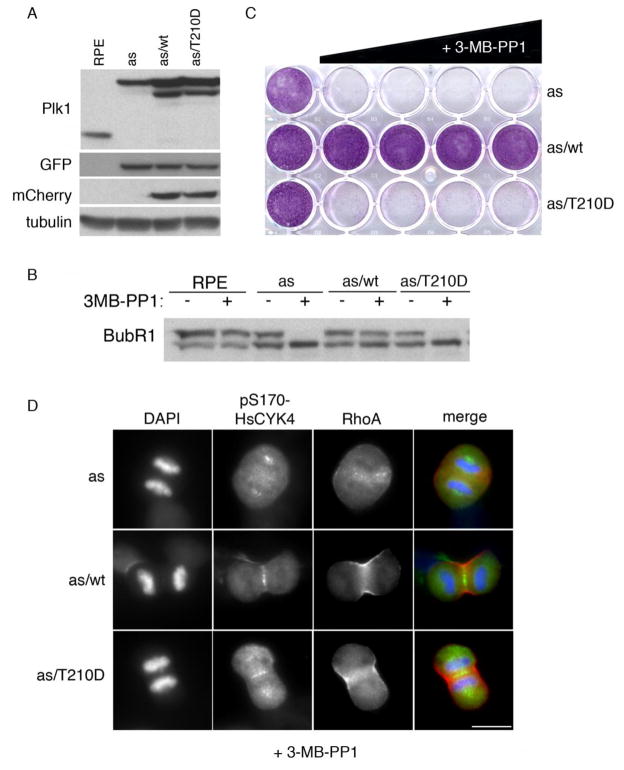
To clarify if BubR1’s residual phosphorylation should be attributed to Plk1T210D, as opposed to traces of wildtype Plk1 lingering after Cre-mediated recombination, we employed an orthogonal chemical-genetic approach. Plk1wt and Plk1T210D were expressed alongside an analog-sensitive form of the kinase (Plk1as) in PLK1Δ/Δ cells (Fig. 4a). Treatment with the bulky purine analog 3-MB-PP1 was then used to selectively inhibit Plk1as and unmask the functionality of Plk1T210D. Remarkably, BubR1’s phosphorylation-dependent mobility shift disappeared almost completely in analog-treated Plk1as/T210D cells (Fig. 4b), resulting in highly unstable K-fibers that were unable to complete chromosome bi-orientation and satisfy the spindle checkpoint (Table 2 and Supplementary Fig. S3a–d). Importantly, these phenotypes occurred despite the fact that the in vitro kinase activity of Plk1T210D was similar to that of wildtype Plk1 isolated from prometaphase cells (Supplementary Fig. S3e), or about 6- to 12-fold higher than uninhibited Plk1as (Burkard et al. 2007). Consistently, Plk1as/T210D cells proliferated only in the absence of 3-MB-PP1, whereas Plk1as/wt cells were insensitive to this inhibitor (Fig. 4c). Leveraging the rapid kinetics of this system, we then asked whether Plk1T210D is able to trigger cytokinesis, which depends on anaphase-specific phosphorylation of the centralspindlin component HsCYK-4 (Burkard et al. 2009). Briefly, Plk1as/wt and Plk1as/T210D cells were synchronized in prometaphase by monastrol block and release, then treated with 3-MB-PP1 as they entered anaphase. Robust HsCYK-4 phosphorylation and induction of equatorial RhoA zones, were observed in both cell lines (Fig. 4d and Table 3). Further, Plk1as/T210D cells formed cleavage furrows at a similar rate as Plk1as/T210D cells (Table 3). Thus, the T210D substitution does not interfere with Plk1’s ability to initiate cell division, in contrast to its effects on chromosome congression.
Figure 4. Comparing the early and late mitotic competencies of Plk1T210D via chemical genetics.
a PLK1Δ/Δ cells expressing Plk1wt or Plk1T210D (fused to mCherry) and/or GFP-tagged Plk1as were processed for Western blotting with the indicated antibodies. b Cells were grown for 6 days in the presence of 0, 0.078, 0.313, 1.25, 5, or 20 μM 3MB-PP1 before fixation and staining with crystal violet. c Cells were treated with nocodazole ± 10 μM 3MB-PP1 for 12 hours, then harvested for Western blotting. d Cells were treated with 3-MB-PP1 30 minutes after monastrol washout, then fixed and stained with antibodies to pS170-HsCYK-4 (green), RhoA (red), or DAPI (blue). Scale bar represents 10 microns.
Table 2.
Plk1T210D is inadequate for chromosome congression
| fully aligned | 1–2 chromosomes misaligned | >2 misaligned | |
|---|---|---|---|
| as/wt + 3-MB | 70% | 24% | 6% |
| as/T210D + 3-MB | 36% | 36% | 28% |
Table 3.
Plk1T210D is effective in triggering cytokinesis
| RhoA zone | − | + | + |
|---|---|---|---|
| furrow ingression | − | − | + |
| as + 3-MB | 80% | 14% | 6% |
| as/wt + 3-MB | 2% | 2% | 96% |
| as/T210D + 3-MB | 8% | 8% | 84% |
Numerous studies have used Plk1T210D as a proxy for the mitotically activated form of Plk1, with strong expression of this kinase variant reported to cause precocious entry into mitosis (Jackman et al. 2003) and override of the G2 arrest caused by DNA damage (Macurek et al. 2008; Smits et al. 2000; van Vugt et al. 2004). We were therefore surprised to find that replacing the native PLK1 locus with PLK1T210D did not accelerate the G2/M transition in a dominant manner, either in the presence or absence of DNA damage. Thus, we have no evidence to support the notion that T210 phosphorylation is rate limiting for mitotic entry, but do not exclude the possibility that this modification could have a subtle role in G2/M transit that becomes more obvious if Plk1 is overexpressed. Rather, using two orthogonal approaches, we found that the T210D substitution severely diminishes Plk1’s ability to support BubR1 phosphorylation, K-fiber stability, chromosome congression, and cell proliferation. These observations contrast with an earlier report that Plk1T210D reconstitutes the cell cycle profile of Plk1 RNAi-depleted cells with similar efficiency to wildtype Plk1 (van de Weerdt et al. 2005). This discrepancy is likely explained by technical differences in the rate and extent of Plk1 inactivation. While gene deletion and allele-specific chemical inhibition revealed similar T210-linked defects in prometaphase cells, only the latter allowed us to circumvent these defects and demonstrate Plk1T210D’s competence to trigger cytokinesis once the spindle checkpoint had been satisfied.
Plk1 plays a crucial role in stabilizing kinetochore-microtubule attachments, as evidenced by its role in generating the 3F3/2 phosphoepitope, which marks kinetochores that are not experiencing spindle-generated tension (Ahonen et al. 2005; Wong and Fang 2005). While the identity of the 3F3/2 phosphoantigen(s) remains to elucidated, it is clear that Plk1 phosphorylates a growing list of kinetochore-specific substrates, including CENP-U/PBIP, CLIP-170, NudC, Bub1, and BubR1 (Elowe et al. 2007; Kang et al. 2006; Li et al. 2010; Matsumura et al. 2007; Nishino et al. 2006; Qi et al. 2006). Furthermore, Plk1’s own recruitment to kinetochores depends on its phosphorylation- and PBD-dependent interaction with several of these factors (Kang et al. 2006; Nishino et al. 2006; Qi et al. 2006). Here we found that, despite normal kinetochore targeting, Plk1T210D was unable to phosphorylate BubR1 on S676 or other sites that contribute to its mitotic upshift on SDS-PAGE. Importantly, direct mutation of BubR1’s phosphorylation sites has similar effects on K-fiber stability and chromosome congression (Elowe et al. 2007; Matsumura et al. 2007). Thus, loss of this post-translational modification is already sufficient to explain the major T210D-linked phenotypes found in our study. Nevertheless, we do not exclude the possibility that other Plk1 substrates may also share BubR1’s stringent requirement for T210 phosphorylation. Given the paucity of validated phosphospecific antibodies, further efforts at identifying such substrates will require quantitative phosphoproteomic profiling before and after allele-specific inhibition in Plk1as/T210D cells (Oppermann et al. 2012). Nonetheless, our results already show that Plk1T210D cannot support the phosphorylation of at least one critical substrate, and thus is not a faithful surrogate of the natively activated kinase.
Materials and Methods
Cell culture
HEK293, HeLa, and Phoenix cells were grown in Dulbecco’s modified Eagle (DME) medium. Human retinal pigment epithelial cells were grown in DME: Ham’s F12 medium (1:1) with 2.5 mM L-glutamine. HCT116 cells were grown in McCoy’s 5A medium. All media were supplemented with 10% fetal bovine serum and 100 units/ml penicillin-streptomycin. Double-thymidine synchronization and DNA-damage checkpoint assays were carried out as previously described (Papi et al. 2005).
Gene targeting and retroviral transgenesis
PLK1-specific sequences were amplified from a genomic BAC (RP11-1149D11) using PfuTurbo polymerase (Stratagene) and modified at the T210 codon using site-directed mutagenesis (QuikChange, Stratagene). All constructs were fully sequenced to verify their integrity. AAV-mediated gene targeting was carried out as described (Berdougo et al. 2009). High-titer adenovirus expressing Cre recombinase (AdCre) was purchased from the Baylor University Vector Development Laboratory and used at a MOI of 200 plaque-forming units per cell. mCherry-Plk1 fusions were cloned into pQCXIP (Clontech) and cotransfected with pVSV-G into Phoenix cells using FuGene (Roche) to generate retroviral stocks, which were then used to transduce PLK1-null cells reconstituted with EGFP-Plk1as (Burkard et al. 2007).
Immunofluorescence and immunoblotting
For most experiments, cells were fixed with 4% paraformaldehyde. For γ-tubulin and active RhoA, cells were fixed with ice-cold methanol and 10% trichloroacetic acid, respectively. After permeabilization in PBS-T for 10 minutes, cells were blocked in PBS-T + 10% goat serum for 30 minutes, incubated with primary and Alexa 488- and 594-conjugated secondary antibodies for 1 to 2 hours each, counterstained with 4,6-diamidino-2-phenylindole (DAPI), and mounted in Prolong Plus (Invitrogen). Cells were imaged on a Nikon TE2000 microscope equipped with a 100x 1.4 NA oil objective and Hammamatsu ORCA ER camera. Images were acquired using MetaMorph (Molecular Devices). Extracts were prepared by lysing cells on ice in HB2 buffer (50 mM HEPES, [pH 7.5], 0.5% NP-40, 10% glycerol, 100 mM NaCl, 10 mM Na pyrophosphate, 5 mM β-glycerophosphate, 50 mM NaF, 0.3 mM Na3VO4, 1 mM DTT, 1 mM PMSF, and 1× complete protease inhibitor cocktail [Roche]), followed centrifugation at 10,000 × g for 15 minutes at 4°C.
Antibodies
The following antibodies were used: α-tubulin (1:5000, Santa Cruz and Chemicon); γ-tubulin (1:200, Sigma); Plk1 (1:500, Santa Cruz); EGFP (1:1000, Invitrogen), RhoA (1:200, Santa Cruz), BubR1 (1:300); cyclin B1 (1:2000, Cell Signaling); cdc2 (1:1000, Cell Signaling); phospho-Histone H3 (1:1000, US Biologicals); Wee1 (Santa Cruz, 1:1000); DsRed (1:1000, Clontech); CREST (1:5000). pS676-BubR1 was a gift of Erich Nigg (Biozentrum). pS170-CYK4 (1:500) was used as previously described (Burkard et al. 2009).
Live-cell microscopy
Cells were imaged on a Nikon TE2000 microscope equipped with 10x, 40x, and 60x long working distance objectives, a temperature-controlled stage enclosure and CO2 enrichment system (Solent Scientific). Image acquisition was automated using Metamorph. H2B-GFP expressing cells infected with AdCre were followed by timelapse microscopy between 48 and 72 hours post-infection. Images were taken every 2 minutes over a 6-hour time course.
K-fiber stability assay
Cells were incubated on ice for 0–15 minutes before fixation with PTEMF (20 mM PIPES pH 6.8, 4% paraformaldehyde, 0.2% Triton X-100, 10 mM EGTA, 1 mM MgCl2) and staining with CREST and -tubulin antibodies. Microtubule density was scored as the fraction of pixels with intensity greater than twice the background.
In vitro kinase assays
mCherry-tagged versions of Plk1 (wildtype, T210A, or T210D) were immunoprecipitated from nocodazole-arrested cells with DsRed-specific antibodies, washed three times in HB2, twice in kinase buffer [20 mM Tris (pH 7.4)/10 mM MgCl2/50 mM KCl/1 mM DTT], and then incubated in kinase buffer plus 5 μg casein, 1 μM ATP and 5 μCi [γ–32P]ATP [3000Ci/mmol, 10 mCi/ml] for 0–20 minutes at 30°C. Reactions were stopped by addition of sample buffer. 32P incorporation was quantified on a PhosphorImager (Fuji-Film Medical Systems USA, Stamford, CT).
Chemicals
3-MB-PP1 was a gift of Chao Zhang and Kevan Shokat (UCSF) and used at 10 μM. Caffeine (5 mM), thymidine (2.5 mM), MLN8054 (3 μM), monastrol (100 μM), adriamycin (0.5 μM), and nocodazole (0.2 μM) were also used.
Supplementary Material
a Southern blot analysis confirmed recovery of clones with each of the desired genotypes. B. RT-PCR analysis confirmed monoallelic expression of PLK1T210A and PLK1T210D after AdCre infection.
a Cells of the indicated genotypes were infected with AdCre and harvested 3 days laster. Nocodazole was added to the media 12 hours prior to cell harvest. Lysates were probed with an antibody against Plk1. b Cells were collected 0, 1, 2, or 3 days after AdCre infection and scored based on nuclear morphology. At least 3 sets of 100 cells were counted for each timepoint. Error bars represent SEM.
a Chromosome alignment assay. Cells were released from a nocodazole block into medium containing 3-MB-PP1 and MG132. Two hours later, cells were fixed and stained with DAPI (blue), CREST antisera (red) and α-tubulin antibodies (green). At least 50 cells were scored per sample. Scale bar represents 10 microns. b Cells were treated with 3-MB-PP1 and nocodazole, then stained for S676-phosphorylated and total BubR1 as in Fig. 3a. c Cold stability of K-fibers in 3-MB-PP1 treated Plk1as/wt and Plk1as/T210D cells was determined as in Fig. 2e. d Timecourse of mitotic arrest and slippage after allelic-specific inhibition in Plk1as/T210D cells. Percentages of mitotic and micronucleated cells were determined from three sets of 100 cells each. Error bars represent SEM. e Wildtype, T210D, or T210A Plk1 were immunoprecipitated from Plk1as/wt, Plk1as/T210D, and Plk1as/T210A cells using mCherry-specific antibodies, then incubated with casein and [γ-32P]ATP for the times indicated. Samples were resolved by SDS-PAGE and either processed for Coomassie staining and phosphorimager detection (top and middle rows) or blotted to detect Plk1 (bottom row).
Acknowledgments
We thank Sabine Elowe and Erich Nigg for kindly providing the pS676-BubR1 antibody. This study was supported by grants from the American Cancer Society (114520-RSG-08-093-01-CCG) and National Institutes of Health (R01GM094972) to P.V.J.
References
- Ahonen LJ, Kallio MJ, Daum JR, Bolton M, Manke IA, Yaffe MB, Stukenberg PT, Gorbsky GJ. Polo-like kinase 1 creates the tension-sensing 3F3/2 phosphoepitope and modulates the association of spindle-checkpoint proteins at kinetochores. Curr Biol. 2005;15:1078–1089. doi: 10.1016/j.cub.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Terret ME, Jallepalli PV. Functional dissection of mitotic regulators through gene targeting in human somatic cells. Methods Mol Biol. 2009;545:21–37. doi: 10.1007/978-1-60327-993-2_2. [DOI] [PubMed] [Google Scholar]
- Burkard ME, Maciejowski J, Rodriguez-Bravo V, Repka M, Lowery DM, Clauser KR, Zhang C, Shokat KM, Carr SA, Yaffe MB, Jallepalli PV. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009;7:e1000111. doi: 10.1371/journal.pbio.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci U S A. 2007;104:4383–4388. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EH, Santamaria A, Sillje HH, Nigg EA. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008;117:457–469. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J Biol Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindall DJ, Chen J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–1082. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002;277:44115–44120. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- Kang YH, Park JE, Yu LR, Soung NK, Yun SM, Bang JK, Seong YS, Yu H, Garfield S, Veenstra TD, Lee KS. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Kelm O, Wind M, Lehmann WD, Nigg EA. Cell cycle-regulated phosphorylation of the Xenopus polo-like kinase Plx1. J Biol Chem. 2002;277:25247–25256. doi: 10.1074/jbc.M202855200. [DOI] [PubMed] [Google Scholar]
- Kishi K, van Vugt MA, Okamoto K, Hayashi Y, Yaffe MB. Functional dynamics of Polo-like kinase 1 at the centrosome. Mol Cell Biol. 2009;29:3134–3150. doi: 10.1128/MCB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- Li H, Liu XS, Yang X, Wang Y, Turner JR, Liu X. Phosphorylation of CLIP-170 by Plk1 and CK2 promotes timely formation of kinetochore-microtubule attachments. EMBO J. 2010;29:2953–2965. doi: 10.1038/emboj.2010.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol. 2004;164:233–241. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lei M, Erikson RL. Normal cells, but not cancer cells, survive severe Plk1 depletion. Mol Cell Biol. 2006;26:2093–2108. doi: 10.1128/MCB.26.6.2093-2108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr Biol. 2010;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery DM, Lim D, Yaffe MB. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Matsumura S, Toyoshima F, Nishida E. Polo-like kinase 1 facilitates chromosome alignment during prometaphase through BubR1. J Biol Chem. 2007;282:15217–15227. doi: 10.1074/jbc.M611053200. [DOI] [PubMed] [Google Scholar]
- Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee LY. NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol. 2006;16:1414–1421. doi: 10.1016/j.cub.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Oppermann FS, Grundner-Culemann K, Kumar C, Gruss OJ, Jallepalli PV, Daub H. Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol Cell Proteomics. 2012;11:O111–012351. doi: 10.1074/mcp.O111.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M, Berdougo E, Randall CL, Ganguly S, Jallepalli PV. Multiple roles for separase auto-cleavage during the G2/M transition. Nat Cell Biol. 2005;7:1029–1035. doi: 10.1038/ncb1303. [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell. 2006;23:319–329. doi: 10.1016/j.molcel.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian YW, Erikson E, Maller JL. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol Cell Biol. 1999;19:8625–8632. doi: 10.1128/mcb.19.12.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall CL, Burkard ME, Jallepalli PV. Polo kinase and cytokinesis initiation in mammalian cells: harnessing the awesome power of chemical genetics. Cell Cycle. 2007;6:1713–1717. doi: 10.4161/cc.6.14.4501. [DOI] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Du H, Jang CY, Yates JR, 3rd, Fang G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008a;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008b;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- van de Weerdt BC, van Vugt MA, Lindon C, Kauw JJ, Rozendaal MJ, Klompmaker R, Wolthuis RM, Medema RH. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Mol Cell Biol. 2005;25:2031–2044. doi: 10.1128/MCB.25.5.2031-2044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MA, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Wong OK, Fang G. Plx1 is the 3F3/2 kinase responsible for targeting spindle checkpoint proteins to kinetochores. J Cell Biol. 2005;170:709–719. doi: 10.1083/jcb.200502163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Goto H, Yokoyama T, Sillje H, Hanisch A, Uldschmid A, Takai Y, Oguri T, Nigg EA, Inagaki M. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J Cell Biol. 2005;171:431–436. doi: 10.1083/jcb.200504091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Fletcher L, Muschel RJ. The role of Polo-like kinase 1 in the inhibition of centrosome separation after ionizing radiation. J Biol Chem. 2005;280:42994–42999. doi: 10.1074/jbc.M505450200. [DOI] [PubMed] [Google Scholar]
- Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
a Southern blot analysis confirmed recovery of clones with each of the desired genotypes. B. RT-PCR analysis confirmed monoallelic expression of PLK1T210A and PLK1T210D after AdCre infection.
a Cells of the indicated genotypes were infected with AdCre and harvested 3 days laster. Nocodazole was added to the media 12 hours prior to cell harvest. Lysates were probed with an antibody against Plk1. b Cells were collected 0, 1, 2, or 3 days after AdCre infection and scored based on nuclear morphology. At least 3 sets of 100 cells were counted for each timepoint. Error bars represent SEM.
a Chromosome alignment assay. Cells were released from a nocodazole block into medium containing 3-MB-PP1 and MG132. Two hours later, cells were fixed and stained with DAPI (blue), CREST antisera (red) and α-tubulin antibodies (green). At least 50 cells were scored per sample. Scale bar represents 10 microns. b Cells were treated with 3-MB-PP1 and nocodazole, then stained for S676-phosphorylated and total BubR1 as in Fig. 3a. c Cold stability of K-fibers in 3-MB-PP1 treated Plk1as/wt and Plk1as/T210D cells was determined as in Fig. 2e. d Timecourse of mitotic arrest and slippage after allelic-specific inhibition in Plk1as/T210D cells. Percentages of mitotic and micronucleated cells were determined from three sets of 100 cells each. Error bars represent SEM. e Wildtype, T210D, or T210A Plk1 were immunoprecipitated from Plk1as/wt, Plk1as/T210D, and Plk1as/T210A cells using mCherry-specific antibodies, then incubated with casein and [γ-32P]ATP for the times indicated. Samples were resolved by SDS-PAGE and either processed for Coomassie staining and phosphorimager detection (top and middle rows) or blotted to detect Plk1 (bottom row).