Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) is an abundant nuclear enzyme that modifies substrates by poly(ADP-ribose)-ylation. PARP-1 has well-described functions in DNA damage repair, and also functions as a context-specific regulator of transcription factors. Using multiple models, data demonstrate that PARP-1 elicits pro-tumorigenic effects in androgen receptor (AR)-positive prostate cancer (PCa) cells, both in the presence and absence of genotoxic insult. Mechanistically, PARP-1 is recruited to sites of AR function, therein promoting AR occupancy and AR function. It was further confirmed in genetically-defined systems that PARP-1 supports AR transcriptional function, and that in models of advanced PCa, PARP-1 enzymatic activity is enhanced, further linking PARP-1 to AR activity and disease progression. In vivo analyses demonstrate that PARP-1 activity is required for AR function in xenograft tumors, as well as tumor cell growth in vivo and generation and maintenance of castration-resistance. Finally, in a novel explant system of primary human tumors, targeting PARP-1 potently suppresses tumor cell proliferation. Collectively, these studies identify novel functions of PARP-1 in promoting disease progression, and ultimately suggest that the dual functions of PARP-1 can be targeted in human PCa to suppress tumor growth and progression to castration-resistance.
Keywords: prostate cancer, androgen receptor, PARP-1, PARP inhibitor, DNA damage
Introduction
Whereas the role of poly(ADP-ribose) polymerase-1 (PARP-1) in the DNA damage response is well-defined, the impact of PARP-1-mediated transcriptional regulation, especially in the context of human malignancy, is of emerging interest (1–3). PARP-1 is an abundant nuclear enzyme that catalyzes poly(ADP-ribose)-ylation (PARylation) of target proteins, utilizing nicotinamide adenine dinucleotide (NAD+) as a cofactor (1). In response to genotoxic insult, PARP-1 is recruited to sites of damage, which activates PARP-1-mediated catalytic activity (4–6). The vast majority of PARP-1 function is self-directed, and auto-modified PARP-1 recruits proteins that promote DNA repair (e.g. XRCC1), and thereby facilitates assembly and activation of the base excision repair (BER) machinery (7–9). Auto-modification also alters PARP-1 affinity for DNA and influences chromatin compaction at sites of PARP-1 action (8, 10–12). PARP-1 inhibitors have recently been developed as a means to sensitize to genotoxic insult, and are in clinical trials in combination with chemotherapy and/or radiation as well as for tumors with defects in DNA repair capacity (13–16).
Distinct from the role of PARP-1 in DNA repair, the transcriptional regulatory functions of PARP-1 are multi-fold, not universally dependent upon enzymatic activity, and are manifest through divergent functions including: enhancer binding, association with insulators, modulation of chromatin structure, and/or direct transcription factor regulation (2, 17–19). Despite these realizations, the mechanisms and impact underlying PARP-1-mediated transcriptional control in human malignancy remain largely unexplored. Roles for PARP-1 in estrogen receptor-α (ER) transcriptional activation (20), retinoic acid receptor activation via modulation of Mediator (21), serving as a co-activator for Neuron-derived orphan receptor 1 (NOR1) (22), and negatively regulating the retinoic acid receptor-thyroid hormone receptor (RXR/TR) heterodimer (23) are suggestive of a potential function for PARP-1 in nuclear receptor control and possibly hormone-dependent cancers. Additionally, recent studies identified a role for PARP-1 in mediating ETS activity, which harbors pro-oncogenic function, in prostate cancer (24) and Ewing’s sarcoma (24, 25).
Here, a critical role for PARP-1 was identified, wherein PARP-1 regulates both tumor growth and progression through transcriptional regulatory functions. Human prostatic adenocarcinoma (PCa) is dependent on androgen receptor (AR) activity for growth and survival, and is largely resistant to standard chemotherapy. As such, AR-directed therapeutics are the first-line therapeutic intervention for all patients with disseminated disease (26). Ablation of AR activity is initially effective; however, recurrent tumors arise wherein AR activity has been reactivated, as achieved through multiple mechanisms that have been reviewed extensively (26). Since there is no durable cure for this “castrate-resistant” stage of disease, there is an urgent need to develop means to sustainably suppress AR function and/or sensitize PCa cells to cytotoxic chemotherapy.
Studies herein reveal that PARP-1 is a potent modulator of both AR function and the response to DNA damage. Data shown demonstrate that PARP-1 regulates AR association with chromatin, and controls AR function in genetically-defined systems, both in vitro and in vivo. These functions of PARP-1 require enzymatic activity, and ablation of PARP-1 activity sensitizes PCa cells to both genotoxic insult and androgen depletion. Strikingly, models of castrate-resistant prostate cancer (CRPC) show marked up-regulation of PARP-1 activity, suggesting that PARP-1 may be enhanced as a function of tumor progression. Accordingly, PARP-1 suppression enhanced the anti-tumor effects of castration, delayed onset to castration resistance, and was required to maintain in vivo castrate-resistance. Moreover, primary human prostate tumor ex vivo cultures revealed a significant anti-tumor response to PARP-1 inhibition, which was correlated with diminished AR activity, thus further demonstrating the dependence of PCa on PARP-1 activity. Collectively, these studies identify a major role for PARP-1 in controlling AR signaling, and demonstrate that the dual functions of PARP-1 could be leveraged for therapeutic benefit.
Results
PARP-1 inhibition sensitizes to genotoxics and suppresses cell growth in AR-positive cells
To assess the impact of PARP-1 on the response to DNA damage in PCa cells, a clinically relevant PARP inhibitor, ABT888, was utilized (27). ABT888 sensitized both androgen deprivation-sensitive (HT, hormone therapy-sensitive) and CRPC cells to ionizing radiation (IR) and docetaxel (DCTX) (Fig. 1A), the latter in a dose-dependent manner (Supp.Fig. S1A). Unexpectedly, a reduction in cell number was observed with exposure to the PARP inhibitor alone, using doses known to sensitize cancer cells to DNA damage (compare vehicle to ABT888 alone, Fig. 1A) (28). Escalating doses of ABT888 were therefore administered and cell number assessed (Supp.Fig. S1B); although some variation was observed, each model demonstrated growth suppression that was induced by ABT888 as a single agent. Growth curve analyses further identified a role for PARP-1 in specifically controlling growth in AR-positive HT-sensitive cells (Fig. 1B, top). Inhibition of PARP-1 activity resulted not only in reduced growth of HT-sensitive PCa cells, but also cooperated with androgen deprivation to suppress cell survival (top right). Similar effects were observed in CRPC cells (Fig. 1B, middle). By contrast, no effect was seen in AR-negative PCa cells (bottom), or AR-negative immortalized prostate epithelial cells (29) (PrEC) (bottom right). Together, these data demonstrate that PARP inhibition cooperates with genotoxic insult and elicits unexpected growth-suppressive effects that are largely confined to AR-positive PCa and CRPC cells, without effecting AR-negative PCa cell growth.
Figure 1.
(A) Cell number was assessed 96h after exposure to indicated treatment in indicated cell models. Treatments are: ABT888 (2.5uM), IR (ionizing radiation, 2Gy), ABT888+IR (2.5uM+2Gy), DCTX (1nM docetaxel), and ABT888+DCTX (2.5uM+1nM docetaxel). Vehicle control is set to “100%”. (B) Model systems indicated were treated with vehicle control (solid line) or 2.5uM ABT888 (dashed line), and cell number assessed at indicated time points. Cell number at the zero hour time point is set to “1”. Data reflect averages and standard deviation of at least three independent experiments, each performed with biological triplicates. Statistical significance was determined using Student’s t test. *=p<0.05, **=p<0.01, and ***=p<0.001
PARP-1 is a critical effector of AR activity
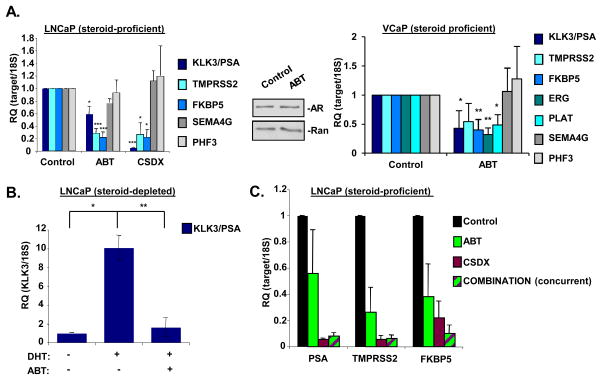
Given the specificity of PARP inhibitors for suppressing growth of AR-positive PCa and CRPC cells (but not AR-negative cells), the effect of PARP-1 on AR function was determined. Single dose (2.5uM, below IC50) exposure of ABT888 significantly reduced expression of well-characterized, prostate-specific AR target genes of clinical relevance (KLK3/PSA, TMPRSS2, and FKBP5) compared to control (Fig. 2A, left), while not altering mRNA levels of genes previously found to be refractory to PARP-1 knockdown in MCF7 cells (SEMA4G and PHF3) (30), thus demonstrating specificity. The clinically utilized AR antagonist Casodex (CSDX), served as a positive control. Observed effects on AR target gene expression were not due to alterations in AR protein levels (middle). Importantly, chromosomal rearrangements placing the coding region of ETS genes (e.g. ERG) under control of the TMPRSS2 regulatory locus occur with high frequency in PCa, and therein result in AR-dependent expression of pro-tumorigenic ETS genes (31). Therefore, the impact of PARP-1 on AR-dependent ERG gene expression was assessed in cells containing TMPRSS2:ERG fusions (VCaP cells) (31). PARP inhibition significantly suppressed expression of AR-dependent target genes including ERG and the ERG target gene PLAT, while not affecting PARP-1 refractory SEMA4G or PHF3 (Fig. 2A, right). Similar effects on AR function were observed in additional PCa models of HT-sensitive disease (Supp.Fig. S2A). PARP inhibition suppressed AR-dependent activity of an integrated ARR2(probasin)-LUC reporter, thereby confirming the ability of PARP-1 to modulate AR activity (32) (Supp.Fig. S2B). Moreover, pre-treatment of androgen-depleted cells with ABT888 prior to androgen (dihydrotestosterone, DHT) (comparison of hormone proficient and hormone deficient KLK3/PSA levels, Supp.Fig. 2C) stimulation significantly impaired androgen-dependent expression of KLK3/PSA (Fig. 2B). Thus, inhibition of PARP activity suppresses AR target gene expression.
Figure 2.
(A) Left panel: LNCaP cells were cultured in media containing complete serum, treated for 24h with either ethanol control (0.01%), ABT888 (2.5uM), or Casodex (1uM). Cells were harvested, and qPCR analyses for indicated target genes were performed. Data reflect averages and standard error of at least three independent experiments, each performed with technical triplicates. Results were normalized to 18S and control is set to “1”. Middle panel: LNCaP cells were cultured, treated, and harvested as in A, then lysed, and total protein was separated by SDS-PAGE, transferred to PVDF, and immunoblotted for indicated proteins. Representative image of at least three independent experiments is shown. Right panel: Same as left panel, except cell model is VCaP. (B) LNCaP cells were steroid deprived for 72h, then either pretreated for 30m with vehicle or ABT888 (2.5uM), stimulated with DHT (1nM, 24h) or ethanol control. Cells were harvested and qPCR analyses for KLK3/PSA were performed. Data reflect averages and standard error of at least three independent experiments, each performed with technical triplicates. Results were normalized to 18S and control is set to “1”. (C) LNCaP were cultured in media containing complete serum, treated for 24h with either ethanol control (0.01%), ABT888 (2.5uM), Casodex (1uM) or a combination of ABT888 (1.75uM) and Casodex (0.5uM). Cells were harvested, and qPCR analyses for indicated target genes were performed. Data reflect averages and standard error of at least three independent experiments, each performed with technical triplicates. Results normalized to 18S and control is set to “1”. Statistical significance was determined using Student’s t test. *=p<0.05, **=p<0.01, and ***=p<0.001
To determine whether the observed growth-suppressive function of PARP inhibition is through regulation of AR, HT-sensitive cells were exposed to: ABT888 alone, CSDX alone, or a combination thereof. Single treatment regimens elicited a decrease in AR target gene expression compared to control; however, there was no observed additive effect when used in combination (Fig. 2C). Additionally, combination treatment elicited no additive effect with regard to inhibition of cell growth (Supp.Fig. S3). Taken together, these data demonstrate that PARP enzymatic activity is required for AR function in HT-sensitive PCa cells, and that targeting PARP activity decreases AR activity in multiple models of human disease.
PARP-1 modulates AR-chromatin interaction
As PARP enzymatic activity was shown to influence AR activity, the underlying impact on chromatin association was assessed. Using chromatin tethering assays, which have previously been demonstrated to differentiate between nuclear and chromatin-associated proteins (33), DHT-induced AR association with chromatin compared to cells cultured in androgen-depleted conditions was determined (Fig. 3A, compare lanes 4 and 5). The DHT-stimulated AR occupancy of chromatin was markedly reduced in cells that were pre-treated with a PARP inhibitor (compare lanes 5 and 6). To determine if AR is PARylated in response to DHT, immunoprecipitations followed by immunoblot were performed. AR was not PARylated (either in the presence or absence of androgen), whereas PARP-1 automodification was suppressed by PARP inhibition (Supp.Fig. S4A), indicating that regulation of AR by PARP-1 is not due to PARylation of AR. To assess whether the functional interplay observed in the tethering assay held true at sites of known AR function, chromatin immunoprecipitation (ChIP-qPCR) was used to assess recruitment of AR and PARP-1 to the regulatory loci of AR target genes. As expected, DHT stimulation induced AR occupancy at the KLK3/PSA enhancer (Fig. 3B, top left), the KLK3/PSA promoter (top right), and a critical androgen responsive element (ARE) that controls expression of TMPRSS2 (bottom left). Strikingly, PARP-1 was associated with each region as a function of AR occupancy. By contrast, DHT-induced occupancy of both AR and PARP-1 was markedly suppressed in cells pre-treated with ABT888 (Fig. 3B), thus suggesting that PARP-1 function may be critical for AR and PARP-1 residence at key sites of AR function. To determine if AR and PARP-1 co-occupy sites of chromatin, ChIP-reChIP was performed. reChIP signals of AR/PARP-1 were not discernable from IgG controls (Supp.Fig. S4B), indicating that while AR and PARP-1 occupy the same AR target gene loci, they do so in separate protein complexes. To eliminate the possibility that these observations represent random binding of either AR or PARP-1, ChIP assays to assess the residency of these proteins in a genomic region devoid of coding genes were assessed. Neither PARP-1 nor AR binding was detectable (Fig. 3B, bottom right). Previous studies demonstrated that PARP-1-dependent transcriptional regulation can be manifest through the ability of PARP-1 to act as an exchange factor with linker histone H1 at active promoters (34); however, histone H1 occupancy was not altered as a function of DHT stimulation or PARP inhibition at both the KLK3/PSA promoter and enhancer (Fig. 3C), suggesting that the function of PARP-1 at AR-dependent loci is independent of histone H1 regulation. To determine whether PARP-1 inhibition altered the transcriptional competency for activation near AR target gene regulatory loci, ChIP assays to quantify histone H3K4 di-methylation (H3K4me2) (35) and histone H3K4 tri-methylation (H3H4me3) were performed. DHT induced the enrichment of both H3K4me2 and H3K4me3, while PARP inhibition suppressed accumulation of these DHT-induced chromatin modifications (Fig. 3D and E, respectively). To determine the effect of PARP inhibition on pioneering factor occupancy (36, 37), ChIP assays were performed for GATA2 and FOXA1. GATA2 occupancy increased upon DHT stimulation, and this occupancy was attenuated upon PARP inhibition (Fig. 3F), distinct from what was observed with FOXA1 (Supp.Fig. S5), suggesting that PARP enzymatic activity is required for the hierarchical programming for transcriptional competency of AR target gene loci. GATA2 has previously been demonstrated to be required for AR recruitment to chromatin (38), and may play a role in more aggressive PCa (39), therefore the role of PARP enzymatic activity in regulating GATA2 occupancy is likely of strong translational significance. To further examine the relative state of chromatin at sites of AR action after PARP inhibition, MNase protection assays were performed. Amplicons spanning ±400 bases from the transcriptional start site of KLK3/PSA demonstrated significant differences in MNase sensitivity between DHT stimulated and PARP inhibited cells (Fig. 3G), as such that PARP inhibition renders the chromatin of a clinically relevant AR target gene less accessible, thus less transcriptionally competent, which not only results in less AR binding (along with PARP-1, GATA2, and active histone marks), but ultimately leads to diminished AR transcriptional output. Together, these data suggest that PARP-1 is required for AR occupancy on chromatin at sites of AR function in response to ligand and exerts AR-modulatory functions dependent on its enzymatic activity. These data support a model wherein PARP-1 function is critical for GATA2 binding, selective histone modification, and promoting an open chromatin structure to allow transcriptional activity of AR.
Figure 3.
(A) LNCaP cells were steroid deprived for 72h, then either pretreated for 30 min with vehicle or ABT888 (2.5uM), stimulated with DHT (1nM, 1h) or ethanol control. Cells were then harvested, lysed, and differentially centrifuged as described in the material and methods section, resulting in a soluble fraction (GAPDH serves as control) or a chromatin-tethered fraction (histone H4 serves as control). Immunoblots were performed for the indicated proteins. A representative image of at least three independent experiments is shown. (B, C, D, and E) LNCaP cells were steroid-deprived for 72h, pretreated for 30m with vehicle or ABT888 (2.5uM), and then stimulated with DHT (10nM, 1h) or ethanol control. Cells were fixed, lysed, and chromatin immunoprecipitation (ChIP) for AR and PARP-1 (B), histone H1 (C), di-methylated lysine 4 of histone H3 (D), tri-methylated lysine 4 of histone H3 (E), or GATA2 (F) was performed. DNA was purified from immunoprecipitates (IPs) and utilized in qPCR reactions for indicated genomic loci. Data is representative of at least three independent experiments, each performed with technical triplicates, and depicted as the averages of immunoprecipitated signals to input signals and standard error. Statistical significance was determined using Student’s t test. *=p<0.05, **=p<0.01, and ***=p<0.001. (G). LNCaP treated as in A. MNase protection assays were performed as described in the Materials and Methods section, from which DNA was purified and utilized in qPCR reactions for amplicons spanning 400bp on either side of the transcriptional start site (TSS) of KLK3/PSA. Data are representative of at least three independent experiments performed in technical triplicate, depicted as the averages of MNase sensitive:protected DNA and standard deviation. Grey box indicates amplicons that are statistically significant between DHT treated samples, and ABT pretreated->DHT treated samples by Student’s t test.
PARP-1 activity is increased in CRPC, and regulates castration-resistant AR activity
The lethal stage of PCa, CRPC, arises due to resurgent AR activity despite maintenance of AR-directed therapeutics (26). Remarkably, analyses of PARP-1 activity in cell line models of CRPC revealed that PARP-1 is highly PARylated in CRPC cells, as compared to HT-sensitive PCa (Fig. 4A, compare lanes 1–3 vs. lanes 4 and 5). Noting that auto-modification accounts for much of PARP-1 activity (40), these findings suggested that PARP-1 function may be enhanced in CRPC. This postulate was confirmed by ELISA, which demonstrated that global PAR levels are significantly elevated in CRPC cells (Fig. 4B) compared to HT-sensitive LNCaP cells. Given the ability of AR to remain active after androgen deprivation in CRPC cells, the impact of enhanced PARP-1 activity on AR function was determined under conditions of hormone depletion. Whereas Casodex was incompletely effective at suppressing AR activity in CRPC cells (Supp.Fig. S6), suppression of PARP activity attenuated AR target gene expression (Fig. 4C, top). Importantly, PARP inhibition suppressed expression of UBE2C, a key regulator of cell cycle progression in castration-resistant prostate cancer cells (38). Consistent with observations in HT-sensitive cells, no changes in AR protein were apparent (Fig. 4C, bottom). Although the present findings identify PARP-1 activity as a major effector of AR-chromatin occupancy, AR function, and AR-dependent cell growth in prostate cancer, PARP inhibitors show modest impact on other PARP family members, especially PARP-2 (27). Therefore, additional means were used to address the specificity of PARP-1 for modulation of AR function. Validated shRNAs directed against PARP-1 (or control) (30) were introduced into CRPC cells, and AR target gene expression was assessed. Moderate PARP-1 knockdown (Supp.Fig. S7A, middle top) resulted in reduced PARP-1 activity as determined by immunoblotting for total PAR (right); in addition, decreased AR target gene expression was observed in CRPC cells deprived of androgen (left) without altering AR protein levels (Supp.Fig. S7A, middle bottom). Additionally, combining AR knockdown with pharmacologic PARP inhibition did not result in a cooperative effect with respect to inhibition of cell growth (Supp.Fig. S7B), suggesting that the growth inhibitory effects of PARP inhibition are likely attributed to suppressed AR function in CRPC cells. Moreover, ChIP analyses showed that AR is recruited to loci regulating KLK3/PSA and TMPRSS2 expression in the absence of hormone, and that PARP-1 co-occupied these regions (Fig. 4D). Within each region, suppression of PARP activity resulted in depletion of both AR and PARP-1 on chromatin. Combined, these observations demonstrate that PARP-1 activity is up-regulated in models of CRPC, and demonstrate that CRPC-associated, ligand-independent AR function and chromatin occupancy is dependent on PARP-1 enzymatic activity.
Figure 4.
(A) Indicated model systems were cultured in media containing complete serum, harvested, lysed, and total protein was separated by SDS-PAGE, transferred to PVDF, and immunoblotted for indicated proteins. Representative image of at least three independent experiments is shown. (B) PAR ELISA was performed on whole cell lysates of indicated model systems. Data reflect averages and standard deviation of at least three independent experiments, each performed with technical and biological triplicates. (C) Upper panel: indicated model systems were steroid deprived for 72h, then either treated for 30 min with vehicle or ABT888 (2.5uM). Cells were harvested and qPCR analyses were performed for indicated target genes. Data reflect averages and standard error of at least three independent experiments, each performed with biological triplicates. Results were normalized to 18S and control is set to “1”. Lower panel: indicated model systems were treated as above, then harvested, lysed, and total protein was separated by SDS-PAGE, transferred to PVDF, and immunoblotted for indicated proteins. Representative image of at least three independent experiments is shown. (D) C4-2 cells were steroid deprived for 72h, then treated for 30m with vehicle or ABT888 (2.5uM). Cells were fixed, lysed, and ChIP for AR and PARP-1 was performed. DNA was isolated from IPs and utilized in qPCR reactions for indicated genomic loci. Data are representative of at least three independent experiments, each performed with technical triplicates, and depicted as the average of immunoprecipitated signal to input signal and standard error. Statistical significance was determined using Student’s t test. *=p<0.05, **=p<0.01, and ***=p<0.001.
PARP-1 is required for AR function in genetically-defined systems and in vivo models
To assess PARP-1-regulated AR function in defined genetic systems, mouse embryonic fibroblasts (MEFs) isolated from wild-type and Parp-1−/− (41) mice were transfected with plasmids encoding β-galactosidase (to normalize for transfection efficiency), AR, and the ARR2-LUC reporter (for determination of AR activity). Both basal and ligand-induced AR reporter activity was reduced in Parp-1−/− MEFs when compared to wild-type MEFs (Fig. 5A, left). To ensure specificity, a reporter specific for E2F1 function was also assessed. There was no discernable difference of reporter activity in wild-type versus Parp-1−/− MEFs transfected with a 3xE2F reporter construct (Supp.Fig. S8). Parallel studies were performed wherein Parp-1−/− MEFs were transfected with plasmid encoding either a wild-type or a catalytically inactive allele of PARP-1 (E988A) (42). Expression of wild-type PARP-1 in Parp-1−/− MEFs restored AR activity, whereas the catalytic mutant failed to support AR function (Fig. 5A, middle). Expression of transfected PARP-1 alleles was confirmed by immunoblot (right). Together, these data demonstrate that PARP-1 promotes basal and ligand-induced AR activity, and further establish the requirement of PARP-1 catalytic activity for AR function. To determine whether the observed regulation of AR by PARP-1 can be targeted in vivo by another PARP inhibitor (Olaparib), short-term (4h) treatments were initially performed using human tumor xenografts. Upon tumor development, mice were treated with Olaparib, Casodex, or control for 4 hrs prior to sacrifice, and gene expression was analyzed (Fig. 5B, top). Tumors exposed to PARP inhibition showed decreased expression of KLK3/PSA, TMPRSS2, and FKBP5 as compared to control, whereas responses to Casodex were varied and target gene-dependent (Fig. 5B, bottom), which is consistent with the observations in Supp.Fig. S3, in which there was no additive effect in vitro when PARP inhibition was combined with Casodex. To further these observations, parallel xenograft studies were performed wherein host animals were treated with ABT888 instead of Olaparib. PARP inhibition resulted in significant decreases in AR target gene expression (Fig. 5C), and importantly, strongly suppressed PCa tumor growth in vivo (Fig. 5D) (tumor volumes depicted in Supp.Fig. S9). PARP inhibition significantly suppressed tumor doubling compared to control to a degree strikingly similar to castration alone. When castration and PARP-1 inhibition were combined, there was a significant lengthening of the duration of tumor doubling compared to castration alone (50%). This is significant, as these findings indicate that PARP-1 inhibition not only cooperates with castration to limit tumor doubling in vivo, but also suppresses the transition to CRPC. Additionally, in castrated animals treated with the PARP inhibitor, 20% of the tumors never achieve a full doubling through the duration of the study (60 days). To examine the in vivo effect of PARP inhibition on CRPC cell tumor growth, C4-2 xenografts were established. Castration plus PARP inhibition significantly decreased tumor volume compared to castration alone (Fig. 5E, top panel), and this decreased tumor volume was associated with attenuated AR target gene expression (Fig. 5E, bottom panel). Together, these data demonstrate that suppressing or silencing PARP-1 results in decreased AR-dependent target gene expression, AR function, and ability of AR to promote tumor progression both in vitro and in vivo. These observations predict that PARP-1 could serve as a viable therapeutic target in AR-positive PCa, as PARP inhibitors not only suppressed tumor growth in cooperation with castration, but also delayed the onset of castration resistance.
Figure 5.
(A) Left: Indicated MEFs were transfected as described in Material and Methods. Cells were then stimulated for approximately 36 hours with 1nM DHT or ethanol control and relative luciferase activity determined. Normalized AR activity in the absence of ligand in the wild type MEFs was set to “1”. Data shown reflects the mean of at least nine independent biological replicates ±SE. Middle: PARP-1−/− MEFs were transfected as above, with the addition of either wild type PARP-1 or a PARP-1 catalytic domain point mutant allele, then treated, processed, harvested, and analyzed as above. *=p<0.05, **=p<0.01, and ***=p<0.001 Right: Parp-1 −/− MEFs were transfected as before, except cells were harvested, lysed, and total protein was separated by SDS-PAGE, transferred to PVDF, and immunoblotted for indicated proteins. (B) Upper panel: Schematic: VCaP xenografts were established for 4 weeks prior to treatment, at which time the mice received a single dose of either Casodex or Olaparib (100mg/kg), tumors were harvested 4 hours later and qPCR analyses were performed for indicated target genes. Data reflect average and standard deviation of at least three independent xenograft tumors, each performed with technical triplicates. Results were normalized to β-actin and control is set to “1”. (lower panel). (C) Tumors were established as in B, except mice were treated with ABT888 (100mg/kg twice daily). 72h later, tumors were harvested and qPCR analyses were performed for indicated target genes. Data reflect average and standard deviation of at least three independent experiments, each performed with technical triplicates. Results were normalized to 18S and control is set to “1”. (D) Upper panel: VCaP xenograft tumors were established as in B and C. Treatment was initiated when tumors reached 150mm3, and consisted of: control, castration alone (Cx), ABT888 (100mg/kg twice daily)(PARPi), and castration + ABT888 (Cx+PARPi.) Tumor volumes were assessed three times each week. The cumulative incidence plot depicts the percent of tumors in each treatment group that have doubled in volume, as a function of time. Each treatment group is significantly different than the control group. The combined treatment group is significantly different than the individual treatment groups as determined by with log-rank (Mantel-Cox) analysis. Lower panel: Median time elapsed before tumor volume doubled for each treatment group. (E) Upper panel: C4-2 xenografts were established as in B. Treatment was initiated when tumors reached 150mm3, and consisted of: castration alone (Cx) as control, and castration + ABT888 (Cx+PARPi). Tumor volumes were assessed daily until animals were sacrificed. Lower panel: tumors from upper panel were excised, homogenized in Trizol, cDNA was generated, and qPCR for the indicated mRNA was performed. Data are presented as mean ± SD of at least three xenografts from each treatment group. Statistical significance was determined using Student’s t test. *=p<0.05, **=p<0.01, and ***=p<0.001.
PARP-1 promotes tumor cell proliferation in primary human disease
As the above studies identified PARP-1 as a potent mediator of AR activity, PCa growth and progression, the impact of PARP inhibition on primary human tumor growth was assessed using a novel ex vivo culture system. Tumor tissue was obtained immediately upon resection at radical prostatectomy, and placed into the explant system as described (43, 44). Explant specimens retain all salient features of the original tumor, including AR expression, tumor and normal tissue histoarchitecture, stromal/tumor cell interaction, proliferative rate, and can be used to monitor response to treatment(s). As depicted in Fig. 6A, ex vivo culture of primary tumor tissue obtained at radical prostatectomy was subdivided, cultured on a matrix, and subjected to targeted therapy. To assess efficacy of suppressing PARP activity, immunohistochemistry for total PAR was performed after ABT888 treatment. Global PAR levels in ABT888 treated tumor cells were significantly decreased compared to control (Fig. 6A, bottom). While no observable alteration in histoarchitecture was observed after ABT888 exposure, the Ki67 index of the tumor cell population was markedly decreased (Figs.6B and 6C). This observed decrease in proliferative index was associated with diminished AR target gene expression (Fig. 6D). Taken together, these data demonstrate that PARP-1 can be targeted in human PCa, and that suppressing PARP activity results in decreased primary tumor cell proliferation.
Figure 6.
(A) Upper panel: Schematic of explant assay is depicted as described in the Materials and Methods sections. Lower panel: Representative images of explant tissues that were treated with either control or 2.5uM ABT888, then were formalin fixed and paraffin embedded. Tissue from these paraffin blocks was cut with a microtome and placed on microscope slides. Slides were then utilized to determine relative levels of PAR in these explant tissues by immunohistochemistry using standard techniques. (B) Same as in A, but tissue slides were either stained with hematoxylin & eosin (top) or stained for Ki67 via immunohistochemistry using standard techniques. (C) Quantification of Ki-67 (as a percent of tumor cells that are nuclear positive) as depicted in B from three explants harvested at indicated time points treated with either control or 2.5uM ABT888. Data are presented as mean ± SD of three explant assays from three distinct prostatectomy specimens. (D) Explants were performed as in A, tissue was homogenized in Trizol, cDNA was generated, and qPCR for the indicated mRNA was performed. Data are presented as mean ± SD of at least three distinct explant tissue per treatment group. Statistical significance was determined using Student’s t test. ***=p<0.001.
Discussion
Although the role of PARP-1 in the DNA damage response is well understood, the impact of PARP-1-mediated transcriptional effects on tumor cell behavior remains poorly elucidated. The present study illuminates the dual roles of PARP-1 as supporting AR activity and resistance to genotoxic insult in PCa. Key findings demonstrate that (i) PARP-1 inhibition sensitizes human PCa cells to genotoxic insult, independently yields cytostatic effects in AR-positive cells, and cooperates with androgen depletion to suppress cell proliferation; (ii) PARP-1 is recruited to sites of AR action, PARP-1 enzymatic activity is critical for AR occupancy on chromatin and for AR activity in both HT-sensitive and CRPC cancer cells; (iii) PARP-1 activity is enhanced and required in CRPC, wherein AR activity is sustained in castrate conditions; (iv) ablation of PARP-1 activity in vivo is sufficient to suppress AR function, decrease tumor growth, delay the onset to castration resistance, and is required to maintain castrate-resistance; (v) PARP-1 inhibition suppresses proliferation of primary human tumor cells. Together, these findings identify the dual functions of PARP-1 as critical in the DNA damage response, as well as supporting AR signaling, pro-tumorigenic phenotypes, and tumor progression.
The observed recruitment of PARP-1 to sites of AR function and the ability of PARP-1 inhibitors to reduce AR occupancy on chromatin is consistent with the observation that PARP-1 can interact with histones and alter chromatin accessibility (11, 19, 45–50). It is enticing to speculate that up-regulation of PARP-1 activity in CRPC cells may assist in the development of inappropriate AR activity and concomitant transition to CRPC; this postulate is strongly supported by data herein, which demonstrate that PARP inhibitors are highly effective in CRPC models. Recent findings in both PCa and Ewing’s sarcoma indicate that similar effects are observed upon activation of ERG (24, 25). This function of the PARP-1/ERG interaction is both a transcriptional regulatory, as well as a DNA damage effect, similar to the functions of PARP-1 described herein. These observations provide the first evidence that PARP-1 function is a requisite for AR-chromatin association, AR activity, and AR-dependent cellular proliferation, and unveil a critical new understanding of AR regulation in PCa cells.
Chemo- and radiosensitization by PARP inhibition suggests that PARP-1 may be important for promoting resistance to these modalities. This is of significant clinical relevance, as PCa is largely resistant to genototoxic chemotherapies, and as of yet there is no means to sensitize PCa to the combination of irradiation and AR-directed therapy, which is the standard of care for management of locally advanced disease (26, 51). That PARP-1 inhibition can sensitize PCa to DCTX could be of clinical significance, as DCTX is currently utilized after the disease has transitioned to the CRPC stage, wherein treatment options are limited. As it has been shown that DCTX affords only a modest (~2–3 month) survival advantage (52, 53), utilizing PARP-1 inhibitors as an adjuvant to DCTX may serve as a means to increase the therapeutic window in advanced disease.
The ability of PARP-1 to promote aberrant ligand-independent AR activity was remarkable, as the vast majority of lethal, late-stage prostate cancers remain dependent on AR for growth and survival (26, 54). PARP-1 inhibitors proved effective at enhancing the effect of androgen depletion on cell survival, suggesting the provocative hypothesis that PARP-1 inhibitors could be assessed clinically as a means to sensitize PCa to AR-directed therapeutics. The PARP-1 inhibitors used in this study are currently being evaluated in clinical trials for multiple tumor types, have favorable toxicity profiles (55–58) and show pre-clinical and clinical efficacy (14, 55, 59). The data herein bring forth important new understanding for PARP-1 inhibitors, and demonstrate that the transcriptional regulatory functions of PARP-1 may significantly promote clinically important transcriptional events in PCa. Currently, endeavors to design clinical trials combining ABT888 and AR-directed therapy (abiraterone) (NCI 9012 also designated as NCT01576172), as well as ABT888 and IR, are underway.
The cooperation between castration and PARP-1 inhibition in vivo holds significance. Mimicking human disease, xenografts utilized re-activate AR despite continued therapeutic targeting of AR. In vivo data demonstrate that combining PARP-1 inhibition and HT may result in slowing evolution of the tumor to the lethal CRPC stage, and may therefore serve a benefit over AR-directed strategies alone. The combination of castration and PARP-1 inhibition appears to not only target ligand-dependent AR activity, but also aberrant, ligand-independent, CRPC-associated AR activity, as was determined in model systems of CRPC under castrate conditions. This potentially is of considerable clinical significance, in that the vast majority of mortality associated with PCa occurs after the disease has undergone the transition to castration-resistance, as the data suggest that PARP-1 inhibition may serve benefit for tumors that have already undergone the transition to CRPC.
Together, the data support a new paradigm in regard to PARP-1 function in human malignancy, whereby the dual functions of PARP-1 in DNA damage repair and AR-regulation can be leveraged to suppress pathways critical for PCa cell survival and progression. Mechanistically, PARP-1 enzymatic activity was determined to be required for AR transcriptional function. This is of importance, given the well-understood role of AR at all stages of PCa. Based on the data, PARP-1 inhibition may afford therapeutic benefit in several contexts: (i) as a means to radiosensitize in primary disease, as well as in salvage and palliative radiotherapy, (ii) as a single agent in primary disease, (iii) in combination with existing AR-directed therapeutics, potentially increasing the time to development of CRPC, and (iv) in combination with docetaxel (or other DNA damage-inducing therapeutics) in late-stage, metastatic CRPC that is, by definition, refractory to other AR-directed strategies. These data provide evidence that PARP-1 is a potentially novel therapeutic target in PCa based on its role in DNA damage repair, and its heretofore unknown role in regulating the function of AR.
Materials and Methods
Cell culture and treatments
LNCaP, C4-2, and VCaP cells were maintained in IMEM supplemented with 5% FBS (heat-inactivated fetal bovine serum). LAPC4 cells were maintained in IMDM supplemented with 10% ΔFBS. PC3, DU145, Parp-1+/+ MEFs, and Parp-1−/− MEFs were maintained in DMEM supplemented with 10% FBS. 22Rv1 cells were maintained in RPMI supplemented with 10% FBS. LNCaP-abl cells were maintained in phenol red-free IMEM supplemented with 5% charcoal dextran treated serum (CDT). PrEC cells were maintained in PrEBM Prostate Epithelial Cell Basal Medium, supplemented with PrEGM SingleQuots (Clonetics). All media supplemented with 2mM L-glutamine and 100 units/ml penicillin/streptomycin. ABT888 was obtained from Alexis Biochemicals and dissolved in H2O and utilized at indicated concentrations. For steroid depleted conditions, cells were plated in appropriate phenol red-free media supplemented with 5–10% CDT. Casodex and DHT were dissolved in ethanol. Cell lines were not cultured longer than six months after receipt from original source or ATCC.
Cell growth assays
Cells were seeded at equal densities, treated as indicated, and harvested at indicated time points. At the time of harvest, cell number was determined using trypan blue exclusion and a hemocytometer.
Gene expression analysis
Cells were seeded at equal density in steroid-proficient (fetal bovine serum, FBS) or depleted (charcoal dextran-treated fetal bovine serum, CDT) conditions as indicated, treated as specified, and RNA was isolated using Trizol, and cDNA generated using Superscript III (Invitrogen). Quantitative PCR was performed with primers described in Supplemental Table 1 and using an ABI Step One machine and PowerSybr according to manufacturer’s specifications.
Chromatin Immunoprecipitation (ChIP) and re-ChIP analysis
Cells were cultured in media containing CDT for 72 hrs and treated as indicated. ChIP analyses and qPCR were performed as previously described (60) using primers described in Supplemental Table 1. ReChIP was performed as ChIP, except after first IP, samples were eluted at 37°C for 30 minutes, supernatant was collected and subjected to the second IP, then processed as ChIP samples.
MNase protection assay
These assays were performed as described previously (45). Briefly, cells were cross-linked with formaldehyde, nuclei were collected, and chromatin was isolated. Chromatin was divided into two aliquots, one receiving MNase treatment. Chromatin was incubated overnight at 65°C to reverse crosslinks, then treated with proteinase K and RNase H. DNA was extracted and precipitated, and used in qPCR reactions with primers described in Supplemental Table 1 and using an ABI Step one machine and PowerSybr according to manufacturer’s specifications.
Antibodies Immunoprecipitation and Immunoblotting
Protein isolation, immunoprecipitation, and immunoblotting were performed as previously described (61), using antisera described in the Supplemental Materials and Methods.
PAR ELISA
Total cell lysates were analyzed for PAR utilizing the HT Colorimetric PARP/apoptosis Assay (Trevigen) per manufacturers’ instructions.
Reporter assays
Parp-1+/+ and Parp-1−/− MEFs were transfected in serum free media as described in the Supplemental Materials and Methods, and treated with DHT or control for ~36h post-transfection. Relative luciferase activity was assessed using the Promega Luciferase Assay Kit and Galacto-Star reagent (Applied Biosystems) was used to detect β-galactosidase activity.
Chromatin tethering
LNCaP cells were steroid-deprived for 72h and treated as indicated. At harvest, plates were harvested and processed as previously described (62). Supernatant was removed and SDS sample buffer was added. The insoluble, chromatin tethered fraction was re-suspended in SDS sample buffer, sonicated briefly, and then both the supernatant and insoluble fraction were denatured at 95°C for 5m. After denaturation, samples were separated by SDS-PAGE, transferred to PVDF, and immunoblotted as per usual.
Expression vectors and shRNA retroviruses
Retroviral shRNA constructs were generously provided by W. Lee Kraus (UT Southwestern) and were previously described (30). Plasmid constructs used in reporter assays were previously described (63). Wild type and catalytic deficient PARP-1 expression plasmid constructs were previously described (46).
Xenograft analysis
Four-week-old male Balb C nu/nu mice were purchased from Charles River, Inc. VCaP (2 × 106 cells), were re-suspended in 100μl of saline with 50% Matrigel (BD Biosciences) and were implanted subcutaneously into the flank of the mice. All tumors were staged for four weeks before starting the drug treatment. For assessment of in vivo AR target gene expression, tumors from mice treated with a single dose of Olaparib (100mg/kg via intraperitoneal injection) or ABT888 (100 mg/kg via oral gavage), and harvested 4 or 72 hours after treatment, respectively. Total RNA was extracted by Trizol. For long-term xenograft growth studies, treatment was initiated when tumors were approximately 150 mm3, with tumor volumes balanced between all treatment groups. Mice allocated to the castration group were surgically castrated on the first day of treatment initiation (day 0). Mice allocated to the ABT888 groups were given 100mg/kg of ABT888 via oral gavage twice per day (with treatments separated by 10–12 hours) for five consecutive days each week, starting on the first day of treatment initiation until tumor harvest (i.e. 0–4, 7–11, 14–18, etc). Tumor volume (via caliper measurement) and mouse weight were assessed three times weekly. No mice lost more than 5% of their initial body weight. Mice were sacrificed once their tumors reached a size of approximately 800 mm3. All procedures involving mice were approved by the University Committee on Use and Care of Animals (UCUCA) at the University of Michigan and conform to their relevant regulatory standards. Changes in tumor volume within each treatment group were depicted using cumulative incidence plots per tumor volume doubling, with statistical differences among treatment groups assessed with the log-rank (Mantel-Cox) analysis.
Human prostate tumor ex vivo explant cultures
Human prostate ex vivo explant cultures were performed as previously described (43). Briefly, fresh tissue was obtained from a pathologist immediately following radical prostatectomy. The de-identified specimen was processed under a laminar flow hood using sterile technique and transported to the lab in RPMI on ice. The Thomas Jefferson University IRB has reviewed this procurement protocol and determined this research to be in compliance with federal regulations governing research on de-identified specimens and/or clinical data [45 CFR 46.102(f)]. The following procedures were performed under sterile tissue culture conditions. Veterinary dental sponges (Novartis Cat# 96002) were placed in 12 well plates and soaked in 500uL media (IMEM supplemented with 5% heat inactivated FBS, hydrocortisone, insulin from bovine pancreas, and 100 units/mL penicillin-streptomycin) and appropriate treatment (either vehicle control or 2.5uM ABT888) for 5–10 minutes at 37°C. Tissue was placed into the lid of a 10cm plate, dissected into 1mm3 pieces with a scalpel. Three pieces of tissue were placed on each sponge using sterile tweezers/forceps. Plates were placed in an incubator at 37°C and 5% CO2. Media was replaced every day with appropriate treatment. Tissue was harvested at indicated time points and fixed in 4% formalin. Formalin fixed paraffin embedded blocks were cut with a microtome, and slides underwent standard hematoxylin and eosin staining, as well as standard Ki67 immunohistochemistry and scored by a clinical pathologist (Dr. McCue).
Supplementary Material
Statement of significance.
These studies introduce a paradigm shift with regard to PARP-1 function in human malignancy, and suggest that the dual functions of PARP-1 in DNA damage repair and transcription factor regulation can be leveraged to suppress pathways critical for pro-malignant phenotypes in prostate cancer cells by modulation of the DNA damage response and hormone signaling pathways. The combined studies highlight the importance of dual PARP-1 function in malignancy and provide the basis for therapeutic targeting
Acknowledgments
Financial Support This work was supported by NIH grants (R01 CA099996-09, and R01 ES016675-11 to K.E.K.), Commonwealth of Pennsylvania grant (SAP# 41000548782 to K.E.K.), a Prostate Cancer Foundation Creativity award (to K.E.K and A.D.P.) a Prostate Cancer Foundation Young Investigator Award (F.Y.F.), grants from the National Health and Medical Research Council of Australia (627185; W.D.T & L.M.B), a Cancer Council of South Australia Senior Research Fellowship (L.M.B.), Predoctoral Fellowships from the DOD (PC094195 to M.J.S. and PC094596 to M.A.A.), and The Kimmel Cancer Center was supported by the NIH/NCI Cancer Center Core grant P30-CA-56036.
The authors thank members of the K. Knudsen laboratory for input and commentary. The authors specifically acknowledge S. Balasubramaniam, C.E.S. Comstock, A.K. McClendon, and R.S. Schrecengost. The authors thank Elizabeth Schade for artistic assistance. Also, we express gratitude to W. Lee Kraus for providing retroviral constructs.
Footnotes
Disclosure: The University of Michigan has filed a patent linking ETS fusions in Ewing’s sarcoma and prostate cancer as markers of sensitivity to PARP inhibitors and DNA-PKcs inhibitors. A.M.C. and J.C.B. are named as inventors.
References
- 1.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342 ( Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin RC, Gill DM. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980;255:10493–10501. [PubMed] [Google Scholar]
- 6.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 7.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 9.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata N, Ueda K, Kagamiyama H, Hayaishi O. ADP-ribosylation of histone H1. Identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J Biol Chem. 1980;255:7616–7620. [PubMed] [Google Scholar]
- 11.Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci U S A. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 13.Kummar S, Chen A, Ji J, Zhang Y, Reid JM, Ames M, et al. Phase I study of PARP inhibitor ABT-888 in combination with topotecan in adults with refractory solid tumors and lymphomas. Cancer Res. 71:5626–5634. doi: 10.1158/0008-5472.CAN-11-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 15.Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol. 2008;8:363–369. doi: 10.1016/j.coph.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 17.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 19.Tulin A, Chinenov Y, Spradling A. Regulation of chromatin structure and gene activity by poly(ADP-ribose) polymerases. Curr Top Dev Biol. 2003;56:55–83. doi: 10.1016/s0070-2153(03)01007-x. [DOI] [PubMed] [Google Scholar]
- 20.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 21.Pavri R, Lewis B, Kim TK, Dilworth FJ, Erdjument-Bromage H, Tempst P, et al. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 22.Ohkura N, Nagamura Y, Tsukada T. Differential transactivation by orphan nuclear receptor NOR1 and its fusion gene product EWS/NOR1: possible involvement of poly(ADP-ribose) polymerase I, PARP-1. J Cell Biochem. 2008;105:785–800. doi: 10.1002/jcb.21876. [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto T, Kakizawa T, Hashizume K. Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol Cell Biol. 1999;19:2644–2649. doi: 10.1128/mcb.19.4.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenner JC, Feng FY, Han S, Patel S, Goyal SV, Bou-Maroun LM, et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. doi: 10.1158/0008-5472.CAN-11-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15:4792–4798. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donawho CK, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 28.Liu SK, Coackley C, Krause M, Jalali F, Chan N, Bristow RG. A novel poly(ADP-ribose) polymerase inhibitor, ABT-888, radiosensitizes malignant human cell lines under hypoxia. Radiother Oncol. 2008;88:258–268. doi: 10.1016/j.radonc.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 29.Berger R, Febbo PG, Majumder PK, Zhao JJ, Mukherjee S, Signoretti S, et al. Androgen-induced differentiation and tumorigenicity of human prostate epithelial cells. Cancer Res. 2004;64:8867–8875. doi: 10.1158/0008-5472.CAN-04-2938. [DOI] [PubMed] [Google Scholar]
- 30.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9:200–206. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burd CJ, Petre CE, Morey LM, Wang Y, Revelo MP, Haiman CA, et al. Cyclin D1b variant influences prostate cancer growth through aberrant androgen receptor regulation. Proc Natl Acad Sci U S A. 2006;103:2190–2195. doi: 10.1073/pnas.0506281103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lake RJ, Geyko A, Hemashettar G, Zhao Y, Fan HY. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 35.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28:3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 40.Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981;256:4135–4137. [PubMed] [Google Scholar]
- 41.Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- 42.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem. 1995;270:3247–3254. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 43.Centenera MM, Gillis JL, Hanson AR, Jindal S, Taylor RA, Risbridger GP, et al. Evidence for Efficacy of New Hsp90 Inhibitors Revealed by Ex Vivo Culture of Human Prostate Tumors. Clin Cancer Res. 2012;18:3562–3570. doi: 10.1158/1078-0432.CCR-12-0782. [DOI] [PubMed] [Google Scholar]
- 44.Dean JL, McClendon AK, Hickey TE, Butler LM, Tilley WD, Witkiewicz AK, et al. Therapeutic response to CDK4/6 inhibition in breast cancer defined by ex vivo analyses of human tumors. Cell Cycle. 2012:11. doi: 10.4161/cc.21195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langelier MF, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 285:18877–18887. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- 48.Mathis G, Althaus FR. Release of core DNA from nucleosomal core particles following (ADP-ribose)n-modification in vitro. Biochem Biophys Res Commun. 1987;143:1049–1054. doi: 10.1016/0006-291x(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 49.Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Wacker DA, Ruhl DD, Balagamwala EH, Hope KM, Zhang T, Kraus WL. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol Cell Biol. 2007;27:7475–7485. doi: 10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwok Y, Yovino S. Update on radiation-based therapies for prostate cancer. Curr Opin Oncol. 22:257–262. doi: 10.1097/CCO.0b013e3283378c84. [DOI] [PubMed] [Google Scholar]
- 52.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 53.Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr, Jones JA, Taplin ME, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 54.Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 21:315–324. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 56.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 57.Kummar S, Kinders R, Gutierrez ME, Rubinstein L, Parchment RE, Phillips LR, et al. Phase 0 clinical trial of the poly (ADP-ribose) polymerase inhibitor ABT-888 in patients with advanced malignancies. J Clin Oncol. 2009;27:2705–2711. doi: 10.1200/JCO.2008.19.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forster MD, Dedes KJ, Sandhu S, Frentzas S, Kristeleit R, Ashworth A, et al. Treatment with olaparib in a patient with PTEN-deficient endometrioid endometrial cancer. Nat Rev Clin Oncol. doi: 10.1038/nrclinonc.2011.42. [DOI] [PubMed] [Google Scholar]
- 59.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 60.Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, et al. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest. 120:4478–4492. doi: 10.1172/JCI44239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knudsen KE, Arden KC, Cavenee WK. Multiple G1 regulatory elements control the androgen-dependent proliferation of prostatic carcinoma cells. J Biol Chem. 1998;273:20213–20222. doi: 10.1074/jbc.273.32.20213. [DOI] [PubMed] [Google Scholar]
- 62.Sever-Chroneos Z, Angus SP, Fribourg AF, Wan H, Todorov I, Knudsen KE, et al. Retinoblastoma tumor suppressor protein signals through inhibition of cyclin-dependent kinase 2 activity to disrupt PCNA function in S phase. Mol Cell Biol. 2001;21:4032–4045. doi: 10.1128/MCB.21.12.4032-4045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schiewer MJ, Morey LM, Burd CJ, Liu Y, Merry DE, Ho SM, et al. Cyclin D1 repressor domain mediates proliferation and survival in prostate cancer. Oncogene. 2009;28:1016–1027. doi: 10.1038/onc.2008.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.