Abstract
Induction of cytokines by small interfering RNA (siRNA) polyplexes has been a significant concern of researchers attempting to minimize the toxicity of this promising therapy. Although cationic carriers of siRNA are known to increase cytokine levels, few systematic studies have been done to determine what properties of the carrier are important to modulate cytokines. Because branched histidine-lysine (HK) peptides are effective carriers of siRNA and their sequence can be readily modified, we selected this class of carrier to determine which sequences of the peptide were important for cytokine induction. With the use of peripheral blood mononuclear cells (PBMCs), the HK peptide with a higher number of histidines (H3K(+H)4b) in complex with siRNA induced lower levels of cytokines compared with other HK (e.g., H2K4b, H3K4b, H3K(+N)4b) siRNA nanoplexes. Notably, these peptides' siRNA polyplexes showed a similar pattern of cytokine induction when injected intravenously in a mouse model, i.e., the HK with higher content of histidines induced cytokines the least. As indicated by the pH-sensitive dye within acidic endosomes, the greater pH-buffering capacity of H3K(+H)4b compared with other HK peptides may explain why cytokine levels were reduced. In addition to buffering capacity, the size of HK polyplexes markedly influenced cytokine production.
Introduction
Development of small interfering RNA (siRNA) silencing that evades the innate immune responses of the cell has enabled this therapy to be applied to mammalian cells and clinical trials. RNA interference (RNAi) of 150 bp effectively silenced their targets in plants and lower eukaryotes, but these longer RNAi were ineffective in mammals because of activation of the innate immune system. In Drosophila melanogaster, the longer RNAi were enzymatically cleaved to 19- to 21-mer overhangs that enabled the antisense strand to combine with the RNA-induced silencing complex system. When mammalian cells were exposed to longer RNAi duplexes, however, activation of toll-like receptors (TLR) and/or intracellular enzyme sensors of RNA,1,2,3,4,5,6 resulted in stimulation of interferon (IFN) gene pathways and/or in marked inhibition of protein translation. Thus, these protective antiviral activities of the cells overshadowed the RNAi silencing activity, particularly when RNAi was larger than 30 bp. The seminal discovery by Tuschl and colleagues that siRNA of 30 bp or fewer had effective silencing ability with reduced cytokine induction has led to the use of siRNA to understand gene function in mammalian cells and development of potential treatment options.7 More recently, it has been increasingly recognized that siRNAs between 19 and 30 bp can induce cytokines, and this induction may be particularly marked when siRNAs are combined with cationic carriers.8,9,10
In addition to the cationic carriers, other factors important for induction of cytokines include the sequence patterns of the siRNA. Judge et al. determined that the sequence “UGUGU” augments cytokine induction which is in line with other studies showing that single-stranded GU-rich RNAs increased cytokines in plasmacytoid dendritic cells.8 In addition, Hornung and colleagues showed that the immunostimulatory sequence “GUCCUUCAA” embedded within siRNA also induced cytokines.11 Most siRNA that are potent inducers of cytokines, however, have no discernible sequence patterns.
For several reasons, modulation of cytokines is important in developing improved siRNA polyplexes. In some cases, because cytokines are not well tolerated by humans,3,12 siRNA polyplexes with decreased tendency to induce cytokines may be preferred. On the other hand, groups have found that siRNA polyplexes that induced cytokines coupled with siRNA silencing may have an additive to synergistic effect on the target cell. For example, together with its silencing effect, induction of cytokines in a mouse model with siRNA was demonstrated to have added benefit in reducing hepatitis B viral titers13 or in enhancing anticancer activity.14 Moreover, the therapeutic silencing effect of siRNA has in some cases been due to nonspecific induction of cytokines.15 Regardless of whether cytokines are desired or not, increased understanding of factors and greater flexibility to modulate cytokine production are required for development of siRNA polyplexes. Although siRNA sequences and modification of their bases have been examined in attempts to affect cytokine levels,16 there have been few studies that systematically examined the role of modification of carriers of siRNA on cytokine induction.17 In the study done by Kedmi et al.,17 cytokine induction primarily reflected alterations in the carrier because carrier alone and the carrier with siRNA increased levels of cytokines similarly in vitro.
To investigate the role of varying the amino acid sequences of carriers in augmenting cytokine production, we utilized histidine-rich peptide-based carriers of siRNA. Numerous groups have utilized histidine-rich carriers of nucleic acids including siRNA.18,19,20,21,22,23,24,25,26,27,28,29,30 Our group has focused on developing branched histidine-lysine (HK) peptides in which the amino acid sequences can be precisely varied with use of a peptide synthesizer.29,31,32,33 Whereas lysines within the HK polymers are responsible for binding to the negatively charged phosphates of siRNA, histidines have a role in buffering acidic endosomes, which is thought to be involved in disruption of endosomal membranes. Because pH-buffering chloroquine markedly reduced cytokines,34 presumably by affecting the interaction of single-stranded RNA with TLR7 within acidic endosomes, we were particularly interested in varying the histidine content of the peptide and the consequential effects on cytokines. In this study, we determined that siRNA polyplexes composed of peptides with higher histidine content reduced cytokine induction in vitro and in vivo.
Results
In vivo induction of cytokines by HK siRNA polyplexes
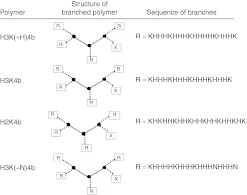
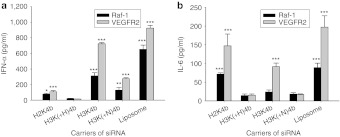
Several four-branched HK peptides (H3K(+H)4b, H3K4b, H2K4b, H3K(+N)4b (Figure 1; Table 1)) were compared for their ability to induce IFN-α, interleukin (IL)-6 or proinflammatory cytokines (IFN-γ or tumor necrosis factor (TNF)-α) in an immunocompetent Balb/c mouse model. We initially examined cytokine induction by these carriers with Raf-1 siRNA (Figure 2a,b), because this siRNA had been shown by us and others to give a potent antitumor response.29,35 In contrast to larger siRNA polyplexes which were trapped in the lungs of mice and did not silence extrapulmonary targets, we prepared smaller polyplexes that we had found gave effective silencing effects in our previous studies.33 Although there were significant variations in the cytokine responses to a particular HK siRNA nanoparticle, the HK peptide with the largest number of histidines per molecule and the highest buffering capacity had the lowest cytokine levels. Varying the charge of the HK carrier did not correlate as closely with cytokine levels, particularly for IFN-α levels. For example, siRNA polyplexes comprised of H2K4b and H3K(+N)4b, which had the highest and lowest number of lysines, respectively, both induced high levels of IFN-α. Of particular interest was the significantly lower cytokine induction by H3K(+H)4b compared with H3K4b polyplexes. Compared with H3K4b, H3K(+H)4b has one additional histidine per branch and four additional histidines per molecule. We focused primarily on the effects of HK polyplexes on IFN-α and IL-6 because the HK polyplexes did not markedly increase proinflammatory cytokines at the 6-hour time point. Notably, the HK peptides or Raf-1 siRNA administered separately induced little to no cytokines (data not shown). The amino acid composition of each peptide is given in Table 1.
Figure 1.
Schematic structure of the four primary HK peptides. “X” represents a C-terminal amide group of the 3-lysine core while “R” represents the amino acid sequences of the peptides. HK, histidine-lysine.
Table 1. Amino acid content of the HK peptides.
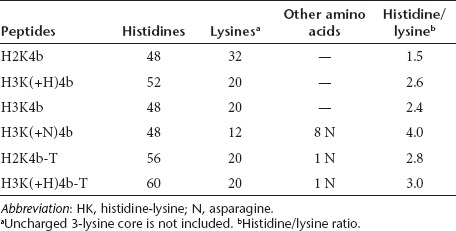
Figure 2.
Cytokine induction by carriers of Raf-1 or VEGFR2 siRNA in Balb/c mice. Several branched carriers that varied in their histidine and/or lysine content were examined for their ability to induce (a) IFN-α or (b) IL-6 in Balb/c mice. Six hours after the siRNA complexes were administered intravenously, serum cytokine levels were determined. Of the carriers of siRNA, H3K(+H)4b increased cytokine levels the least. Vehicle alone values: IFN-α, IL-6, not detectable. *P < 0.05, **P < 0.01, ***P < 0.001, H3K(+H)4b versus other HK, liposomal carriers of Raf-1 or VEGFR-2 siRNA (one-way analysis of variance with Bonferroni t-test). HK, histidine-lysine; IFN, interferon; IL, interleukin; siRNA, small interfering RNA; VEGFR2, vascular endothelial growth factor receptor-2.
Independent of the siRNA, the H3K(+H)4b carrier induced low levels of IFN-α and IL-6
In addition to Raf-1 siRNA, we examined whether the HK polymers would give similar patterns of cytokine inductions with vascular endothelial growth factor receptor-2 (VEGFR-2) siRNA (Figure 2a,b). Although most polyplexes with VEGFR2 siRNA induced significantly higher cytokine levels compared to those with Raf-1 siRNA, the patterns of cytokine induction with the various HK polymers were similar. That is, H3K(+H)4b polymer in complex with VEGFR2 siRNA induced significantly lower levels of IFN-α than other synthesized HK polymers, whereas the H3K4b polyplexes induced the highest. Independent of the siRNA, the H3(+N)K4b and H3K(+H)4b siRNA polyplexes induced very low levels of IL-6. Compared with HK siRNA polyplexes, 1,2-dioleoyl-3-trimethlyammonium propane (DOTAP) siRNA lipoplexes generally induced higher cytokine levels than HK siRNA polyplexes.
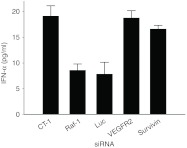
We also examined whether other siRNA in complex with the H3K(+H)4b carrier showed similarly low levels of cytokine (Figure 3). Of particular interest was the siRNA targeting survivin which has elevated number of uridines, a motif that may be immunostimulatory. Despite differences in sequence and immunostimulatory potential of the siRNAs, the H3K(+H)4b polyplexes induced low levels of IFN-α (Figure 3) and other cytokines tested (IL-6, IFN-γ, and TNF-α data not shown).
Figure 3.
Induction of cytokines with different siRNA polyplexes. In complex with H3K(+H)4b, various siRNAs (VEGR2, Raf-1, survivin, luciferase, and non-targeting control (CT-1)) were examined for their ability to increase cytokines in Balb/c mice. Independent of the siRNA, the H3K(+H)4b polyplexes did not induce high levels of cytokines. Vehicle value: IFN-α, not detectable. IFN, interferon; siRNA, small interfering RNA; VEGFR2, vascular endothelial growth factor receptor-2.
Moreover, since syntheses of these polymers are demanding and variation between batches may occur, we tested whether there were variations in cytokine induction with different synthetic batches of three of these polymers, H3K(+H)4b, H3K4b, and H2K4b. Different batches of these polymers in complex with VEGFR2 showed no significant variation in cytokine induction (data not shown). Both batches of H3K(+H)4b induced lower cytokine levels and the batches of H2K4b and H3K4b siRNA polyplexes induced higher cytokine levels.
Similar cytokine induction with mouse splenocytes and human PBMCs
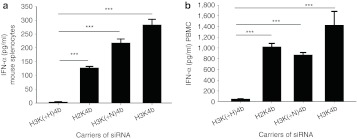
To investigate further the in vivo results and the mechanisms of cyokine induction, we carried out in vitro experiments using mouse splenocytes and human peripheral blood mononuclear cells (PBMCs). siRNA polyplexes were prepared in isotonic medium which we previously determined gave effective silencing results in vitro.29,31 Even with likely differences in their surface coating by proteins and size of polyplexes (Table 2), the polyplexes induced similar cytokine patterns in splenocytes and the mouse experiments in vivo. For example, in both mouse splenocytes and in vivo mouse models, H3K(+H)4b siRNA polyplexes induced low levels of IFN-α while H2K4b and H3K4b induced significantly higher levels (Figure 4a). An interspecies similarity also was found for cytokine induction between the human and mouse cells. Thus, transfected PBMCs with H2K4b siRNA polyplexes induced more IFN-α than did H3K(+H)4b polyplexes (Figure 4b).
Table 2. Size and zeta potential measurements.
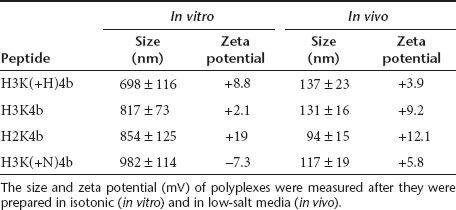
Figure 4.
In vitro comparison of HK carriers of siRNA for their ability to induce IFN-α. Six hours after transfection of (a) mouse splenocytes or (b) human PBMCs with different HK carriers in complex with VEGFR2 siRNA, medium levels for IFN-α were measured by ELISA. Vehicle alone values for IFN-α were 4.1 and 5.2 pg/ml for splenocytes and PBMCs, respectively. ***P < 0.001, H3(+H)K4b versus H2K4b, H3K(+N)4b, H3K4b (one-way analysis of variance, Bonferroni multiple comparison test). ELISA, enzyme-linked immunosorbent assay; HK, histidine-lysine; IFN, interferon; PBMC, peripheral blood mononuclear cell; siRNA, small interfering RNA.
The endosomal pH-buffering agent, chloroquine, inhibited IFN-α production
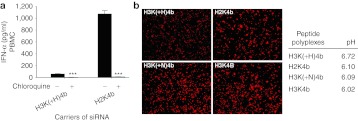
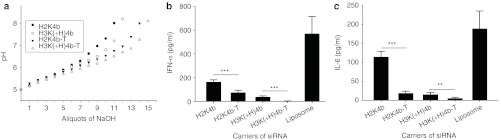
Further corroboration of our findings was that addition of chloroquine, which increases endosomal pH, markedly reduced cytokine induction of the H2K4b polyplex (Figure 5a). This suggested that greater buffering capacity of H3K(+H)K4b carrier may also reduce cytokine induction, by inhibiting endosomal acidification. To test this directly, we compared several HK siRNA polyplexes for their ability to inhibit endosomal acidification in human PBMCs. On the basis of decreased fluorescence of the endosomal pH-sensitive dye, the H3K(+H)4 polyplex was most effective in preventing endosomal acidification in PBMCs (Figure 5b) or MDA-MB-435 cells (Supplementary Figure S1). The pH of endosomes of PBMCs incubated with the H3K(+H)4b polyplexes had an average pH of 6.7 while H2K4b, H3K(+N)4b, and H3K4b polyplexes had pH values of 6.1, 6.09, and 6.02, respectively (Figure 5b).
Figure 5.
Endosomal buffering reduces cytokines produced by HK siRNA polyplexes in PBMCs. (a) In the presence or absence of chloroquine (100 µmol/l), levels of IFN-α were examined after exposure of PBMC to different carriers of siRNA. After transfection of human PBMCs, medium levels for IFN-α levels were measured by ELISA. Vehicle alone value: IFN-α was 2.8 pg/ml. ***P < 0.001, chloroquine-treated cells versus untreated cells (t-test). (b) The endosomal pH dye, pHrodo Dextran, indicates that H3K(+H)4b siRNA polyplexes more effectively buffer the endosomes of PBMCs compared with other HK polyplexes. Three hours after the cells were incubated with dye and varied HK polyplexes (H3(+H)K4b versus H2K4b, H3K(+N)4b, H3K4b), the cells were fixed and images were captured with a fluorescent microscope (×100). The diminution of fluorescent intensity indicates that the H3K(+H)4b siRNA buffers the endosomal pathway more than HK siRNA polyplexes. From a pH titration curve with the pH-sensitive dye, the average pH values of endosomes in PBMCs incubated with the varied HK polyplexes are given. See “Materials and Methods” for more details. ELISA, enzyme-linked immunosorbent assay; HK, histidine-lysine; IFN, interferon; PBMC, peripheral blood mononuclear cell; siRNA, small interfering RNA.
To account for its greater buffering capacity, we examined if the uptake of the H3K(+H)4b polyplex compared with the other HK polyplexes. We found no significant difference of uptake by the H3K(+H)4b siRNA polyplexes at early time points compared with other HK polyplexes (Supplementary Figure S2). The intracellular uptake of the four HK polyplexes was elevated in 80% or more of IFN-α–inducing cell populations isolated from PBMCs (monocytes, plasmacytoid dendritic cell (pDC)-enriched). Moreover, the uptake and fluorescence level of HK polyplexes was particularly high in the pDC-enriched population, consistent with marked increased IFN-α levels associated with some HK polyplexes (Supplementary Figure S2a). Similar to uptake in immune cells, the uptake of HK polyplexes was also high in non-immune MDA-MB-435 cells. Notably, although H3K4b polyplexes induced significantly higher cytokine levels than H3K(+H)4b polyplexes, the uptake in the several cell populations or cell lines of these two HK polyplexes were similar.
Modification of HK carriers with increased histidine content reduced cytokine induction in vivo
Because HK peptides with higher buffering capacity, H3K(+H)4b, in complex with siRNA induced lower amounts of cytokines, we synthesized analogs of H3K(+H)4b and H2K4b with additional histidines (Table 1) and greater buffering capacity (Figure 6a). A histidine-rich “tail” (T) containing eight histidines was inserted off the core of these two HK peptides, where it should not interfere with binding of the HK polymer with the siRNA. Both modified H2K4b and H3K(+H)4b peptides with the histidine-rich tails and thus higher buffering capacity induced lower levels of cytokines in mice compared with their parent peptides (Figure 6b,c). For example, H2K4b-T and H3(+H)4b-T reduced IFN-α levels by more than 80% compared with the H2K4b and H3(+H)4b parent peptides. Levels of IL-6, TNF-α, and IFN-γ were also reduced significantly with incorporation of histidine-rich tails within HK peptides (Figure 6, Supplementary Figure S3). In addition, the histidine-rich polyplex analogues had similar uptake levels as their parent HK polyplexes (Supplementary Figure S2).
Figure 6.
Histidine-rich tails on HK peptides reduced cytokine levels in vivo. Two peptides, H2K4b-T and H3K(+H)4b-T, with eight additional histidines attached to the C-terminal lysine core, were compared with the parent peptides, H2K4b and H3K(+H)4b. (a) Before testing their ability to induce cytokine in vivo, the buffering capacity of these histidine-rich HK peptides and their parent HKs were tested. These peptides in complex with VEGFR-2 siRNA were injected into Balb/c mice with cytokine measurements measured 6 hours later. (b,c) H2K4b-T and H3K(+H)4b-T polyplexes induced cytokines (IFN-α (in b); IL-6 (in c)) significantly less than H2K4b and H3K(+H)4b polyplexes. Vehicle alone values: IFN-α 2.1; IL-6, 4.3 pg/ml. **P < 0.01; ***P < 0.001, H2K4b versus H2K4b-T; H3K(+H)4b versus H3K(+H)K4b-T (t-test). The titration curves represent the mean of two independent experiments. HK, histidine-lysine; IFN, interferon; IL, interleukin; siRNA, small interfering RNA; VEGFR2, vascular endothelial growth factor receptor-2.
Cytokine induction occurred primarily by macropinocytosis in vitro
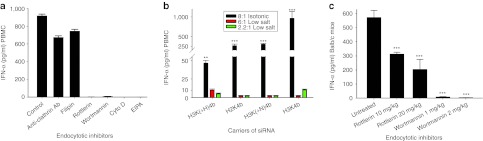
Because TLR7 has been reported to be localized within acidic endosomes, we investigated the roles of the particular endosomal pathways in augmenting IFN-α by HK polymers. We selected H2K4b as a carrier of siRNA since this polyplex effectively induced IFN-α. Although inhibition of clathrin- and calveolin-mediated endocytosis did not affect IFN levels, inhibition of macropinocytosis by wortmannin and rottlerin greatly reduced cytokine levels exposed to the HK polyplexes (Figure 7a). Other macropinocytosis inhibitors including cytochalasin D, 5-(N-ethyl-N-isopropyl)-amiloride (EIPA), and DMA (5′-(N,N-Dimethyl)amiloride) also markedly inhibited IFN-α induced by the HK polyplexes in PBMC.
Figure 7.
Inhibitors of macropinocytosis reduced cytokine induction in vitro and in vivo. (a) Inhibitors of endosomal pathways (clathrin antibodies, fillipin, rottlerin, wortmannin, cytochalasin D, or EIPA) were added to PBMC cells before addition of the large H2K4b siRNA polyplexes formed in isotonic growth medium. While inhibition of the clathrin- and calveolae-mediated endocytosis did not markedly reduced cytokine, inhibition of macrocytosis markedly inhibited cytokine induction. (b) In contrast to larger HK polyplexes formed in isotonic media, smaller HK polyplexes formed in low-salt medium induced significantly less cytokines from PBMCs. Vehicle alone: IFN-α was 2.8 pg/ml. **P < 0.01, ***P < 0.001, HK polyplexes formed in isotonic medium versus HK polyplexes formed in low salt (one-way analysis of variance with Bonferroni t-test). (c) In vivo. Forty-five minutes before H3K4b siRNA polyplexes, two inhibitors of macropinocytosis, rottlerin (10 or 20 mg/kg) or wortmannin (1 or 2 mg/kg), were injected intraperitoneally into mice. Six hours later, mice were euthanized and cytokines in serum were measured. ***P < 0.001, untreated versus macrocytosis inhibitors. Ab, antibody; EIPA, 5-(N-ethyl-N-isopropyl)-amiloride; HK, histidine-lysine; IFN, interferon; PBMC, peripheral blood mononuclear cell; siRNA, small interfering RNA.
Since size of the polyplex may be important for induction, we investigated whether smaller polyplexes (<200 nm) modulated cytokine levels (Supplementary Table S1). By lowering the ionic strength of the solution in which HK polyplexes were formed, polyplexes of reduced size were made, similar to those of what other investigators have reported.36,37 Although smaller siRNA polyplexes and larger polyplexes were equally as effective in silencing their luciferase targets in MDA-MB-435-Luc cells (Supplementary Figure S4), in marked contrast to findings with larger polyplexes, the smaller polyplexes did not induce cytokines in PBMCs (Figure 7b). Moreover, the smaller HK polyplexes entered primarily through the clathrin-mediated pathway in MDA-MB-435 cells (Supplementary Figure S5). Thus, the cytokine pattern induced by the small and large HK siRNA polyplexes in PBMC was consistent with the different pathways of uptake in MDA-MB-435-Luc cells.
Macropinocytosis inhibitors reduced cytokine levels in vivo
For in vivo studies, delineation of endosomal pathways is not always possible because specific inhibitors are lacking for most pathways. Nevertheless, we determined that mice injected with HK siRNA polyplexes had significantly lower cytokine induction, when the mice were first treated intraperitoneally with the macropinocytosis inhibitors, rottlerin, and wortmannin (Figure 7c).38,39,40,41,42 In the absence of these inhibitors, induction of cytokines by these polyplexes occurred even though the polyplexes were smaller than 100 nm (Table 2). It is likely that the size of these polyplexes when they reached their target in vivo may be different from their measurements before injection.
Discussion
Although TLR3 has been implicated in induction of cytokines by “naked” siRNA,10 cytokine induction by siRNA (delivered by nonviral carriers) is primarily through its interaction with TLR7 in mice and with TLR7/8 in humans. TLR7(8) is located in acidic endosomes and presumably, in all endosomal pathways (including the clathrin-mediated, caveolin, non-caveolin, and macropinocytosis), but this has not yet been validated by investigators. TLRs are differentially expressed in macrophages, plasmacytoid, and myeloid dendritic cells, and in particular, TLR7 is highly and constitutively expressed in pDC and lymphocytes. Moreover, uptake of siRNA lipoplexes into pDC or monocytes isolated from human PBMCs has been found to induce high levels of IFN-α.8,43 In addition to TLR, cytosolic receptors such as retinoic acid inducible receptor-I and oligoadenylate synthetase may be activated by siRNA, which results in cytokine induction. Notably, since changes in the buffering capacity of HK polyplexes are likely to have their primary effect on the interaction of TLR7 with siRNA within acidic endosomes, such modifications will likely not affect the ability of siRNA to induce cytokines by cytosolic receptors.44
Despite the limited number of studies to define the essential factors for modulating cytokines by carriers of siRNA, there have been two commonly used methods to reduce cytokines induced by nonviral siRNA polyplexes. The most common approach is modification of the sense strand with 2-O methyl bases which has been shown to interact with TLR7 and to reduce cytokine levels of siRNA polyplexes.16 Although this modification thus far appears safe and effective in preclinical studies, its safety has not been fully established in clinical trials, and other options need to be explored. The use of chloroquine is a second approach that can effectively suppress cytokine levels induced by siRNA complexes in vitro.34 Although the exact mechanism that reduces cytokine induction is not known, it has been postulated that the alteration of the pH of endosomes by chloroquine may affect the interaction of siRNA with the TLR7 receptor.8 Once unprotonated chloroquine traverses the endosomal membrane into the acidic environment of endosomes, chloroquine becomes protonated, accumulates, and buffers these vesicles. An alternative but not mutually exclusive mechanism is that chloroquine may enable lysis of endosomes and escape of siRNA from the endosomes,45 thereby reducing the likelihood of siRNA interaction with TLR7. Because cationic carriers of siRNA enter by endosomal pathways, accumulation of siRNA within the acidic endosomes plays a role in activating cytokines and chloroquine inhibits this induction. By affecting pH, release of siRNA from endosomes, and/or interactions between siRNA and TLR7, chloroquine can effectively suppress cytokine levels induced by siRNA in vitro. Nevertheless, chloroquine did not inhibit cytokine levels in vivo (data not shown), probably due to lack of sufficient endosomal accumulation.46
Based on data obtained with chloroquine in vitro, we compared several HK peptides that varied in their potential to affect the pH of endosomes. Interestingly, addition of a single histidine to each branch of H3K4b, forming H3K(+H)4b, resulted in a marked decrease in cytokine induction in vitro and in vivo. While polyplexes prepared with the H3K(+H)4b peptide did not induce the levels of cytokines significantly, the polyplexes with fewer histidines such as H2K4b, H3K4b, and H3K(+N)4b did induce cytokines. Although these peptide complexes varied in their sequence patterns and their lysine content, these variations could not explain the proclivity of these particles to induce cytokines. Moreover, HK peptides with greater histidine content than H2K4b or H3K(+H)4b (e.g., H2K4b-T or H3K4b-T) in complex with siRNA induced correspondingly lower cytokine levels. Thus, there appears to be a tipping point in the buffering capacity between the peptides that induce cytokines and those that do not based on their histidine content.
In addition to buffering endosomes, we have examined other mechanisms such as uptake that might explain variation in cytokine production. Because IFN-α was markedly stimulated by at least some HK polyplexes in vitro, we were particularly interested in examining uptake of polyplexes in pDCs or monocytes, two cell populations producing high levels of this cytokine.8,43 We found no significant differences in uptake of the different HK polyplexes formed in isotonic media of these cell populations (Supplementary Figure S2), suggesting that uptake was not a primary factor in cytokine variation. Nevertheless, both cell populations exhibited high uptake of the HK polyplexes, providing a “necessary but not sufficient” rationale as to why IFN-α was strikingly stimulated by some HK polyplexes. Similar to the immune cell populations, we observed high uptake rates of the HK polyplexes in the MDA-MB-435 cells. Although other mechanisms, such as altered stability of siRNA polyplexes within endosomes, may be possible for induced cytokines by some HK polyplexes, our data supports the primary role of buffering capacity of the HK polyplexes in cytokine variation.
Even with marked difference in the sizes of polyplexes prepared for use with in vitro and in vivo studies, their propensity for inducing varied levels of cytokines was quite similar (Figures 2,4). While some HK siRNA polyplexes of larger particle size (e.g., H2K4b, H3K(+N)4b, and H3K4b) induced cytokines in vitro, similar in make-up HK polyplexes but smaller in size were determined to markedly induce cytokines in vivo. For the studies in vivo, it is possible that the surfaces of the polyplexes are altered sufficiently in whole blood to affect their size and/or the endosomal pathway by which they enter their target cells. Indeed, several lines of evidence support the notion that despite their small size before intravenous injection, a portion of the HK polyplexes increased their size in vivo before reaching their target cells such as the pDC. First, results in vitro demonstrated that only large polyplexes induced cytokines whereas smaller siRNA polyplexes were significantly less effective at inducing cytokines. Second, there was a close correlation of induced cytokines observed between studies in vitro and in vivo suggesting that the HK polyplexes injected intravenously were altered and their uptake was similar to their endosomal uptake in vitro. Third, preliminary data indicates a marked reduction of cytokines with pegylated H2K4b siRNA polyplexes that may be due in part to their ability to maintain integrity and repel proteins from altering their surface. This would be consistent to what we previously reported that pegylated HK polyplexes were more stable in vitro and in vivo and effectively silenced their targets in vivo.33,47 Fourth, inhibitors of macropinocytosis for in vitro and in vivo experiments minimized induction of IFN-α, suggesting larger polyplexes may induce cytokines. Since in vivo conditions may alter not only the size but the stability of polyplexes, it is likely that free siRNA and polymer are released within the blood stream after injection of the polyplex. Nevertheless, siRNA or HK polymers by themselves induced low levels of cytokines.
On the basis of what other investigators have suggested,3 a critical level of siRNA within endosomes is necessary to induce cytokines. Consequently, it is perhaps not surprising that larger siRNA polyplexes may induce cytokines more effectively. In contrast, smaller H2K4b polyplexes (<200 nm) when entering endosomes such as the clathrin-mediated pathway may not induce cytokines because of the reduced siRNA payload (Figure 7b). Thus, reduction of cytokines might extend to small non-buffering siRNA complexes (lipoplexes or polyplexes) predominantly entering via the clathrin pathway, but this has not been investigated. If these smaller complexes entering the clathrin-mediated pathway induce cytokines, then pH-buffering H2K4b polyplexes would be expected to induce lower levels of cytokines. An alternative mechanism that may explain the variability in cytokine induction is that TLR7 is differentially expressed (or its signal pathways activated) in endosomal pathways. Differentially expressed TLR7 appears less likely since larger size polystyrene-lysine complexes comprised of TLR-9 agonist oligonucleotides also increased cytokine production more than smaller complexes.48 Nevertheless, these mechanisms for cytokine induction require further study.
Despite the many studies demonstrating sequence-specific silencing of siRNA therapy, a recent study suggested that at least some siRNA therapeutic effects were due to nonspecific off-target effects from cytokines.10,15 More important, the investigation by Kleinman et al. underscored that greater understanding of the factors modulating cytokine induction by siRNA polyplexes is essential for development of an improved siRNA carrier. In summary, this study indicates that minor modifications in the sequence of the HK carrier and changes in the particle size of the polyplexes have marked effects on cytokine induction in the target cell. We anticipate that these findings for HK carriers may have broader implications for other pH-buffering and non-pH buffering carriers of siRNA.
Materials and Methods
Cell lines
Human PBMCs. Human PBMCs from normal healthy donors (BRT Laboratories, Baltimore, MD.) were separated from whole blood cell products by density gradient centrifugation in Lymphocyte Separation Medium (ICN Biomedicals, Aurora, OH). The cells were frozen in RPMI-1640 (Invitrogen, Grand Island, NY) containing 20% human AB serum (Gemini Bio-Products, Woodland, CA) and 10% dimethylsulfoxide (Sigma, St Louis, MO) by using an automated cell freezer (Gordinier Electronics, Roseville, MI) and stored in the vapor phase of liquid nitrogen until used.
Splenocytes. The spleen was isolated from mice and teased apart in Dulbecco's modified Eagle's medium. After centrifugation, the single cell suspension was treated with ACK-lysing buffer (8.29 g/l NH4Cl, 1 g/l KHCO2, 37.2 mg/l EDTA) for 10 minutes to lyse red blood cells. After centrifugation, cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum until they were cultured in 24-well plates for polyplex and cytokine experiments.
MDA-MB-435 cells. MDA-MB-435 cells, a human cancer cell line (ATCC, Manassas, VA) were stably transfected with luciferase and the cells were maintained in Dulbecco's modified Eagle's medium/10% fetal calf serum.
Synthesis of HK peptides. The biopolymer facility at the University of Maryland (College Park, MD) synthesized the polymers as previously described.31 The branched HK polymers were synthesized through attaching the four-branches to the 3-lysine core. The following schematic represents the HK polymers:
where R = (KHKHHKHHKHHKHHKHHKHK) for H2K4b and H2K4b-T; R = (KHHHKHHHKHHHKHHHK) for H3K4b; R = (KHHH KHHHKHHHHKHHHK) for H3K(+H)4b and H3K(+H)4b-T; R = (KH- HHKHHHKHHHNHHHN) for H3K(+N)4b); R = (KNKNNKNNKNN KNNKNNKNK) for N2K4b. Both H2K4b-T and H3K(+H)4b-T had a histidine-rich tail represented by X = (HHHHNHHHH) while for other HK peptides the X represented a C-terminal amide group. Polymers were first analyzed by HPLC (Beckman, Fullerton, CA) and were not purified further if HPLC showed that the polymers were at least 95% pure. The polymers were further analyzed by Voyager MALDI-TOF mass spectroscopy (Applied Biosystems, Foster City, CA) and amino acid analysis (AAA Laboratory Service, Boring, OR).
Preparation of liposomes. As previously described,49,50 we prepared liposomes that were composed of DOTAP/cholesterol (1:1 mol:mol ratio) (Avanti, Birmingham, AL). The lipids were hydrated and then sonicated until clear with a Branson 1210 bath sonicator (Branson Ultrasonics, Danbury, MA) in the presence of argon. The liposomes were then extruded through a 50-nm pore size polycarbonate membrane with a LipsoFast-Basic extruder (Avestin, Ottawa, Ontario, Canada).
siRNA sequences. Except for the control (CT-1) RNAi (Qiagen, Hilden, Germany), RNAi were obtained from Dharmacon (Lafayette, CO). Their sequences are as follows: (i) Raf-1, target sequence: 5′-UGU CCA CAU GGU CAG CAC C-3′, sense: 5′-UGU CCA CAU GGU CAG CAC CdTdT-3′, antisense: 5′-GGU GCU GAC CAU GUG GAC AdTdT-3′ (ii) VEGFR2, target sequence: 5′-GCA AAT ACA ACC CTT CAG A-3′, sense: 5′-GCA AAU ACA ACC CUU CAG AdTdT-3′, antisense: 5′-UCU GAA GGG UUG UAU UUG CdTdT-3′ (iii) survivin, target sequence: 5′-GGC AGU GUC CCU UUU GCU A-3′, sense: 5′-GGC AGU GUC CCU UUU GCU A dTdT-3′, antisense: 5′-UAG CAA AAG GGA CAC UGC C dTdT-3′ (iv) luciferase, target sequence: 5′-CUG CAC AAG GCC AUG AAG A-3′, sense: 5′-CUG CAC AAG GCC AUG AAG A dTdT-3′, antisense: 5′-UCU UCA UGG CCU UGU GCA G dTdT-3′ (v) CT-1, sense: 5′-CAG UUG CGC AGC CUG AAU G dTdT-3′, antisense: 5′-CAU UCA GGC UGC GCA ACU GdTdT-3′. For uptake experiments with flow cytometry, the siRNA targeting luciferase was labeled on the 5′ sense strand with the fluorescent Dye547 (immune cells) or fluorescein (MDA-MB-435 cells) (Dharmacon). The siRNAs were reconstituted with 1× Dharmacon buffer (60 mmol/l KCl, 6 mmol/l HEPES, 0.2 mmol/l MgCl2, pH 7.4) at concentrations between 1 and 2 mg/ml.
Cytokine measurements. A multiplex assay method was used to measure levels of IL-6, IFN-γ, and TNF-α in serum and medium. Briefly, a 96-well filter plate (Millipore, Billerica, MA) was wetted with assay buffer (200 µl) (Upstate, Lake Placid, NY), subjected to a vacuum, and then standard/sample/control (25 µl) were added to the appropriate wells. After 25 µl of a mixture containing requested cytokines which had been conjugated to beads were added to each well, the plate was placed on a shaker at 4 °C overnight, washed twice, and then a biotin-labeled mixture containing requested cytokines (25 µl) was added to each well. The plate was then placed on a shaker at room temperature for 60 minutes with phycoerythrin (25 µl) later added to each well for an additional 30 minutes. After the plate was washed, the plate was analyzed by using the Luminex 100 System and the final concentrations were calculated with Bio-Plex software (Bio-Rad Laboratories, Hercules, CA).
Levels of mouse (PBL Biochemical Laboratories, Piscataway, NJ) or human IFN-α (R&D Systems, Minneapolis, MN) were measured with an ELISA kit as described by the manufacturer.
Uptake experiments with flow cytometry. Intracellular uptake of polymer–siRNA complex in MDA-MB-435 or monocytes from human PBMCs was measured by flow cytometry. Twenty-four hours before the treatment, cells were plated in 24-well plate. The polymer siRNA polyplex were formed in Opti-M at the ratio of 8:1, at room temperature for 30 minutes. Then, the polymer–siRNA (luciferase siRNA conjugated to fluorescein (MDA-MB-435 cells) or Dye547 (PBMCs)) polyplexes were added to the cell culture medium. At several time points (1, 2, and 3 hours), transfected cells were harvested, fixed with 4% formalin, and resuspended in FACS buffer for analysis. Results from the fluorescently labeled monocyte population of PBMCs were then acquired using a FACSCantoII (BD Biosciences, Franklin Lakes, NJ) and analyzed using FlowJO software (TreeStar, Ashland, OR) on flow cytometer. To isolate pDC within the monocyte population, carboxyfluorescein conjugated anti-human ILT7/CD85g monoclonal antibody (1:100 dilution) (R&D Systems) was incubated with polyplex-treated PBMCs at room temperature for 1 hour.
In vitro cytokine and silencing studies. Human PBMCs were obtained from human donors, and 1 × 106 cells were placed in each well of a 24-well plate with 10% human AB serum (total volume-500 µl). Similarly, mouse splenocytes (1 × 106) were seeded into each well with 10% fetal calf serum. Unless otherwise indicated, the HK peptides and siRNAs were prepared with the weight ratio of 8:1 (N:P ratio≈10:1) in isotonic growth medium (Opti-MEM; 2 µg siRNA/well) at room temperature for 30 minutes. In some experiments, small polyplexes formed in low-salt conditions (2 mmol/l KCl, 6.6 nmol/l MgCl2) and with different peptide to DNA ratios (2.2:1 or 6:1) were also tested.36,37 After 6 hours, the supernatant was collected and cytokines were quantified as described above. For silencing studies, similar ratios of HK to siRNA were prepared (2.2:1, 6:1, and 8:1), and the polyplexes were incubated with luciferase-expressing MDA-MB-435 cells for 48 hours. Untransfected control and transfected cells were lysed and luciferase activity was then determined.
To determine the role of a specific endosomal pathway in cytokine induction or in silencing, various inhibitors of endosomal pathways (rottlerin, 5 µg/ml; wortmannin, 5 nmol/l; cytochalastin D, 1 µmol/l; Filipin, 2.0 µg/ml; EIPA, 50 µmol/l; and anti-human clathrin antibody (1:125 vol/vol dilution in cell culture medium)) were added to the well 30 minutes before the carrier siRNA complexes. The data for the in vitro siRNA experiments are presented as the mean and SD of four experiments.
Effect of HK siRNA polyplexes on endosomal pH. Human PBMC (1 × 106 cells/well) were placed in each well of a 24-well plate 24 hours before starting the transfection experiment. HK VEGFR2 siRNA polyplexes were prepared in isotonic growth medium at a ratio of 8:1, as described previously. The endosomal pH indicator, pHrodo Dextran (cat. no. P10361, 30 µg/ml; Invitrogen) (excitation, 560; emission, 585) was added 30 minutes before the polymer siRNA polyplex was placed into the wells. Three hours later, the cell culture medium was removed, the cells were washed three times and then fixed with 4% formaldehyde for 5 minutes. Images were then captured on the fluorescent microscope (Nikon TE2000; Nikon, Tokyo, Japan) and maximal intensity of discreet endosomal fluorescence was measured with MetaMorph software (Molecular Device, Sunnyvale, CA) and reported as an average ± SD (n = 25). Similarly, a pH curve was made by adding the pHrodo Dextran to the cells for 3 hours followed by fixing the cells with 4% formaldehyde and 0.05% Triton-100 for 5 minutes. Then 400 µl of different pH-adjusted 0.1 mol/l HEPES solutions (pH 7.4, pH 7.0, pH 6.5, pH 6.0, pH 5.5) were placed in each well for 30 minutes and the maximal intensity of individual endosomal fluorescence (n = 25) was measured as described above. From the pH versus fluorescence data fitted with SigmaPlot 11.0 software (Systat Software, San Jose, CA), the endosomal pH values of PBMCs transfected with different HK siRNA were calculated.
In addition to measuring the intracellular fluorescence individually, each well was read by a Victor3 fluorescence plate reader (Perkin Elmer, Waltham, MA) at 575 excitation. As a control for uptake, total intracellular uptake was measured 3 hours after addition of the various HK peptides in complex with a rhodamine-labeled siRNA.
In vivo experiments. Balb/c (six mice per group) were treated by injection with siRNA (40 µg) in complex with one of several carriers: H2K4b, H2K4b-T, H3K4b, H3K(+H)4b, H3K(+H)4b-T, H3K(+N)4b, and DOTAP/cholesterol liposomes. The ratios of HK polymers to DNA was 4:1 (wt:wt) while that of the liposomes to DNA was 4.3:1; the complexes were formed in a low-salt medium (10 mmol/l KCl, 0.33 nmol/l MgCl2). Six hours after injection of six mice per group with the test formulations, serum samples were collected and cytokine levels were measured (data presented as mean ± SD). In experiments with macrocytosis inhibitors, mice were injected with either rottlerin (10 or 20 mg/kg) or wortmannin (1 or 2 mg/kg) intraperitoneally, 45 minutes before injecting the HK siRNA polyplexes intravenously; dosages were based on previous studies38,39,40,41,42 with the optimal dosages then empirically derived. All experimental procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by University of Maryland School of Medicine Institutional Animal Care and Use Committee.
Measurement of particle size/zeta potential of peptide–siRNA complexes. After the HK siRNA polyplexes were formed as described in the above in vitro or in vivo experiments, their size was determined by measurement of light scattering at a 90° angle on a Zetasizer (Malvern, Westborough, MA). The size is reported as the average size obtained from unimodal analysis carried out with software provided by the instrument manufacturer. The zeta potential was also measured on the Zetasizer. Each particle size data point represents the mean ± SD of three measurements whereas the zeta potential represents the mean of two measurements.
Statistical analysis. All results, reported as means ± SD, represent at least three separate data points for the in vitro studies and six data points for the in vivo experiments unless indicated otherwise. Results were analyzed using a t-test or a one-way analysis of variance followed by Bonferroni t-test; P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL Figure S1. H3K(+H)4b siRNA polyplexes more effectively buffer the endosomes of MDA-MB-435 cells compared with other HK polyplexes. Figure S2. Elevated uptake of HK peptide polyplexes in immune (human monocytes, pDC) and MDA-MB-435 cells. Figure S3. Histidine-rich tails on HK peptides reduced levels of IFN-γ and TNF-α in vivo. Figure S4. Silencing of luciferase by small and large HK polyplexes in MDA-MB-435-Luc cells. Figure S5. Uptake of small HK polyplexes is mediated primarily by clathrin endocytosis. Table S1. Size of HK polyplexes prepared for in vitro experiments.
Acknowledgments
The authors thank Pamela Talalay for her helpful suggestions and careful reading of this manuscript. We also thank the Biopolymer lab at the University of Maryland School of Medicine for synthesizing the peptides in this study. This work was supported by the National Institutes of Health (R01-CA136938). AparnaBio has an exclusive option to license histidine-lysine polymer from A.J.M. for uses in gene therapy. A.J.M. also has equity with AparnaBio in which P.V.S. and M.C.W. are employed. The other authors declared no conflict of interest.
Supplementary Material
H3K(+H)4b siRNA polyplexes more effectively buffer the endosomes of MDA-MB-435 cells compared with other HK polyplexes.
Elevated uptake of HK peptide polyplexes in immune (human monocytes, pDC) and MDA-MB-435 cells.
Histidine-rich tails on HK peptides reduced levels of IFN-γ and TNF-α in vivo.
Silencing of luciferase by small and large HK polyplexes in MDA-MB-435-Luc cells.
Uptake of small HK polyplexes is mediated primarily by clathrin endocytosis.
Size of HK polyplexes prepared for in vitro experiments.
REFERENCES
- Stark GR, Kerr IM, Williams BR, Silverman RH., and, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Minks MA, Benvin S, Maroney PA., and, Baglioni C. Synthesis of 2'5'-oligo(A) in extracts of interferon-treated HeLa cells. J Biol Chem. 1979;254:5058–5064. [PubMed] [Google Scholar]
- Robbins M, Judge A., and, MacLachlan I. siRNA and innate immunity. Oligonucleotides. 2009;19:89–102. doi: 10.1089/oli.2009.0180. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS.et al. (2011Structural basis of RNA recognition and activation by innate immune receptor RIG-I Nature 479423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K., and, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kim JY, Choung S, Lee EJ, Kim YJ., and, Choi YC. Immune activation by siRNA/liposome complexes in mice is sequence- independent: lack of a role for Toll-like receptor 3 signaling. Mol Cells. 2007;24:247–254. [PubMed] [Google Scholar]
- Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ.et al. (2008Sequence- and target-independent angiogenesis suppression by siRNA via TLR3 Nature 452591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S.et al. (2005Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7 Nat Med 11263–270. [DOI] [PubMed] [Google Scholar]
- Judge A., and, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W.et al. (2005Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs Nat Biotechnol 231002–1007. [DOI] [PubMed] [Google Scholar]
- Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS.et al. (20085'-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma Nat Med 141256–1263. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Ambegia E, Choi C, Yaworski E, Palmer L.et al. (2008Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation Hum Gene Ther 19991–999. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E., and, MacLachlan I. 2'-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Kedmi R, Ben-Arie N., and, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Mason AJ, Martinez A, Glaubitz C, Danos O, Kichler A., and, Bechinger B. The antibiotic and DNA-transfecting peptide LAH4 selectively associates with, and disorders, anionic lipids in mixed membranes. FASEB J. 2006;20:320–322. doi: 10.1096/fj.05-4293fje. [DOI] [PubMed] [Google Scholar]
- Kichler A, Mason AJ., and, Bechinger B. Cationic amphipathic histidine-rich peptides for gene delivery. Biochim Biophys Acta. 2006;1758:301–307. doi: 10.1016/j.bbamem.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Okuda T, Sugiyama A, Niidome T., and, Aoyagi H. Characters of dendritic poly(L-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomaterials. 2004;25:537–544. doi: 10.1016/s0142-9612(03)00542-8. [DOI] [PubMed] [Google Scholar]
- Midoux P., and, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug Chem. 1999;10:406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- Kichler A, Leborgne C, März J, Danos O., and, Bechinger B. Histidine-rich amphipathic peptide antibiotics promote efficient delivery of DNA into mammalian cells. Proc Natl Acad Sci USA. 2003;100:1564–1568. doi: 10.1073/pnas.0337677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon C, Roufaï MB, Monsigny M., and, Midoux P. Histidylated oligolysines increase the transmembrane passage and the biological activity of antisense oligonucleotides. Nucleic Acids Res. 2000;28:504–512. doi: 10.1093/nar/28.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet K., and, Contreras R. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol Lett. 2005;27:1337–1347. doi: 10.1007/s10529-005-0936-5. [DOI] [PubMed] [Google Scholar]
- Klingen AR., and, Ullmann GM. Negatively charged residues and hydrogen bonds tune the ligand histidine pKa values of Rieske iron-sulfur proteins. Biochemistry. 2004;43:12383–12389. doi: 10.1021/bi0488606. [DOI] [PubMed] [Google Scholar]
- Nicoll AJ., and, Allemann RK. Nucleophilic and general acid catalysis at physiological pH by a designed miniature esterase. Org Biomol Chem. 2004;2:2175–2180. doi: 10.1039/B404730C. [DOI] [PubMed] [Google Scholar]
- Benns JM, Choi JS, Mahato RI, Park JS., and, Kim SW. pH-sensitive cationic polymer gene delivery vehicle: N-Ac-poly(L-histidine)-graft-poly(L-lysine) comb shaped polymer. Bioconjug Chem. 2000;11:637–645. doi: 10.1021/bc0000177. [DOI] [PubMed] [Google Scholar]
- Hatefi A, Megeed Z., and, Ghandehari H. Recombinant polymer-protein fusion: a promising approach towards efficient and targeted gene delivery. J Gene Med. 2006;8:468–476. doi: 10.1002/jgm.872. [DOI] [PubMed] [Google Scholar]
- Leng Q, Scaria P, Lu P, Woodle MC., and, Mixson AJ. Systemic delivery of HK Raf-1 siRNA polyplexes inhibits MDA-MB-435 xenografts. Cancer Gene Ther. 2008;15:485–495. doi: 10.1038/cgt.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D.et al. (2005A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids Nucleic Acids Res 33e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Q., and, Mixson AJ. Small interfering RNA targeting Raf-1 inhibits tumor growth in vitro and in vivo. Cancer Gene Ther. 2005;12:682–690. doi: 10.1038/sj.cgt.7700831. [DOI] [PubMed] [Google Scholar]
- Chen QR, Zhang L, Luther PW., and, Mixson AJ. Optimal transfection with the HK polymer depends on its degree of branching and the pH of endocytic vesicles. Nucleic Acids Res. 2002;30:1338–1345. doi: 10.1093/nar/30.6.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ST, Leng Q, Scaria P, Woodle M., and, Mixson AJ. Selective modification of HK peptides enhances siRNA silencing of tumor targets in vivo. Cancer Gene Ther. 2011;18:707–716. doi: 10.1038/cgt.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J Mol Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Pal A, Ahmad A, Khan S, Sakabe I, Zhang C, Kasid UN.et al. (2005Systemic delivery of RafsiRNA using cationic cardiolipin liposomes silences Raf-1 expression and inhibits tumor growth in xenograft model of human prostate cancer Int J Oncol 261087–1091. [PubMed] [Google Scholar]
- Grayson AC, Doody AM., and, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R., and, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–1433. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Türeci ö.et al. (2011Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation Gene Ther 18702–708. [DOI] [PubMed] [Google Scholar]
- Sarkar K, Kruhlak MJ, Erlandsen SL., and, Shaw S. Selective inhibition by rottlerin of macropinocytosis in monocyte-derived dendritic cells. Immunology. 2005;116:513–524. doi: 10.1111/j.1365-2567.2005.02253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Seong J, An JH., and, Oh HJ. Enhancement of tumor radioresponse by wortmannin in C3H/HeJ hepatocarcinoma. J Radiat Res. 2007;48:187–195. doi: 10.1269/jrr.06077. [DOI] [PubMed] [Google Scholar]
- Ohno I, Eibl G, Odinokova I, Edderkaoui M, Damoiseaux RD, Yazbec M.et al. (2010Rottlerin stimulates apoptosis in pancreatic cancer cells through interactions with proteins of the Bcl-2 family Am J Physiol Gastrointest Liver Physiol 298G63–G73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Anantharam V, Kanthasamy A., and, Kanthasamy AG. Neuroprotective effect of protein kinase C delta inhibitor rottlerin in cell culture and animal models of Parkinson's disease. J Pharmacol Exp Ther. 2007;322:913–922. doi: 10.1124/jpet.107.124669. [DOI] [PubMed] [Google Scholar]
- Hansmann L, Groeger S, von Wulffen W, Bein G., and, Hackstein H. Human monocytes represent a competitive source of interferon-alpha in peripheral blood. Clin Immunol. 2008;127:252–264. doi: 10.1016/j.clim.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E., and, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Wolff JA., and, Rozema DB. Breaking the bonds: non-viral vectors become chemically dynamic. Mol Ther. 2008;16:8–15. doi: 10.1038/sj.mt.6300326. [DOI] [PubMed] [Google Scholar]
- Zhang X, Dong X, Sawyer GJ, Collins L., and, Fabre JW. Regional hydrodynamic gene delivery to the rat liver with physiological volumes of DNA solution. J Gene Med. 2004;6:693–703. doi: 10.1002/jgm.595. [DOI] [PubMed] [Google Scholar]
- Chou ST, Leng QX, Scaria P, Woodle MC, Mixson AJ. Modified HK siRNA polyplexes with improved pharmacokinetics enhance tumor inhibition. Mol Ther. 20:S8-S8. [Google Scholar]
- Chen HC, Sun B, Tran KK., and, Shen H. Effects of particle size on toll-like receptor 9-mediated cytokine profiles. Biomaterials. 2011;32:1731–1737. doi: 10.1016/j.biomaterials.2010.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesoon-Wood LA, Kim WH, Kleinman HK, Weintraub BD., and, Mixson AJ. Systemic gene therapy with p53 reduces growth and metastases of a malignant human breast cancer in nude mice. Hum Gene Ther. 1995;6:395–405. doi: 10.1089/hum.1995.6.4-395. [DOI] [PubMed] [Google Scholar]
- Xu M, Kumar D, Srinivas S, Detolla LJ, Yu SF, Stass SA.et al. (1997Parenteral gene therapy with p53 inhibits human breast tumors in vivo through a bystander mechanism without evidence of toxicity Hum Gene Ther 8177–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
H3K(+H)4b siRNA polyplexes more effectively buffer the endosomes of MDA-MB-435 cells compared with other HK polyplexes.
Elevated uptake of HK peptide polyplexes in immune (human monocytes, pDC) and MDA-MB-435 cells.
Histidine-rich tails on HK peptides reduced levels of IFN-γ and TNF-α in vivo.
Silencing of luciferase by small and large HK polyplexes in MDA-MB-435-Luc cells.
Uptake of small HK polyplexes is mediated primarily by clathrin endocytosis.
Size of HK polyplexes prepared for in vitro experiments.