Abstract
We report the discovery and characterization of SM-406 (compound 2), a potent and orally bioavailable Smac mimetic and an antagonist of the inhibitor of apoptosis proteins (IAPs). This compound binds to XIAP, cIAP1 and cIAP2 proteins with Ki values of 66.4 nM, 1.9 nM and 5.1 nM, respectively. Compound 2 effectively antagonizes XIAP BIR3 protein in a cell-free functional assay, induces rapid degradation of cellular cIAP1 protein and inhibits cancer cell growth in various human cancer cell lines. It has good oral bioavailability in mice, rats, non-human primates and dogs, is highly effective in induction of apoptosis in xenograft tumors and is capable of complete inhibition of tumor growth. Compound 2 is currently in Phase I clinical trials for the treatment of human cancer.
Introduction
Apoptosis is a cellular process critical to the normal development and homeostasis of multicellular organisms. Dysfunction of the apoptosis machinery has been linked to many human diseases, including cancer, inflammation and neurological conditions.1–3 Defects in the apoptosis machinery confer on cancer cells resistance to therapeutic agents and are known to compromise current anticancer therapies, leading ultimately to their failure.2–4 Avoidance of apoptosis is a hallmark of human cancer4 and targeting key apoptosis regulators with the goal of overcoming the evasion of apoptosis by tumor cells is an exciting therapeutic strategy for the treatment of human cancer.1
The inhibitor of apoptosis proteins (IAPs) are a class of key apoptosis regulators.5–7 Extensive studies have shown that although their role is not limited to regulation of apoptosis, 9,10 X-linked IAP (XIAP) and cellular IAP 1 and IAP 2 (cIAP1 and cIAP2) are in central positions as inhibitors of death signals that proceed through a number of pathways.5–7 XIAP functions as a potent apoptosis inhibitor by directly binding to and effectively inhibiting three caspases, caspase-3 and -7, and -9.5–8 The third BIR domain (BIR3) of XIAP selectively targets caspase-9,15,16 while the BIR2 domain, together with the linker preceding it, inhibits both caspase-3 and caspase-7.17–19 By inhibiting these three caspases, XIAP plays a central role in the inhibition of apoptosis in both death receptor-mediated and mitochondria-mediated pathways.5–7 cIAP1 and cIAP2 were originally identified through their ability to interact directly with tumor necrosis factor associated factor 2 (TRAF2).20 Through their interactions with TRAF2, cIAP1 and cIAP2 are recruited to TNF receptor 1- and 2-associated complexes where they suppress caspase-8 activation and death-receptor-mediated apoptosis.5 Furthermore, IAPs also influence a multitude of other cellular processes, such as ubiquitin (Ub) dependent signaling events that regulate activation of nuclear factor κB (NFκB), which in turn drive the expression of genes important for inflammation, immunity, cell migration and cell survival.10 IAPs also modulate signaling events that promote the activation of cell motility kinases and metastasis11 and they regulate mitogenic kinase signaling, proliferation and mitosis. Many of these cellular processes are frequently deregulated in cancer and contribute directly or indirectly to disease initiation, tumor maintenance and/or progression.12,13 IAP proteins are therefore considered as attractive new cancer therapeutic targets.12,13,14
Smac/DIABLO (second mitochondria-derived activator of caspases or direct IAP binding protein with low pI) has been identified as an endogenous antagonist of XIAP, cIAP1 and cIAP2.21,22 It nullifies the inhibition by XIAP of caspase-9 by binding to the BIR3 domain in XIAP through its Ala-Val-Pro-Ile (AVPI) tetrapeptide binding motif (1 in Figure 1) and directly competing with a similar tetrapeptide, Ala-Thr-Pro-Phe (ATPF) motif, in caspase-9.15,16,23,24 It has been proposed that Smac protein binds to XIAP BIR2 via its AVPI motif, preventing the binding of XIAP to caspase-3/-7.25 cIAP1 interacts with Smac through its BIR3 domain but not other BIR domains.26 Interestingly, in HeLa cells, expression of Smac protein selectively induces degradation of cIAP1/2 but not XIAP.27
Figure 1.
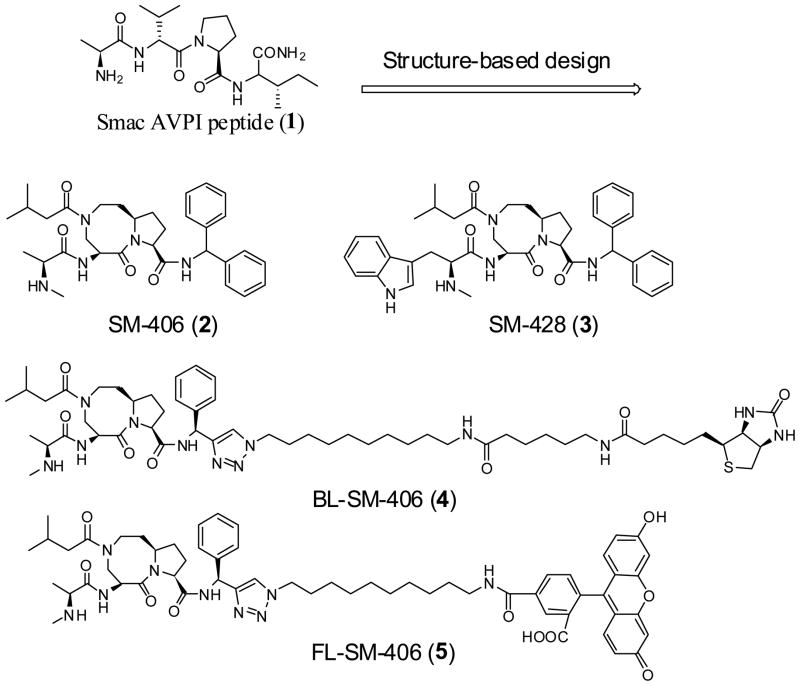
Chemical structures of Smac AVPI peptide, a potent and orally active Smac mimetic 2, its inactive control 3, a biotinylated analogue 4 and a fluorescently tagged analogue 5.
The interaction of Smac with these IAP proteins involves the AVPI tetrapeptide motif in Smac and a well-defined groove in the IAP proteins, and there is considerable interest in the design of non-peptidic, small-molecules as a new class of anticancer drugs that mimic the Smac AVPI peptide and antagonize these IAP proteins.28,29 Employing a structure-based design strategy based upon the crystal structure of Smac in a complex with XIAP BIR3 protein, we have designed and synthesized several classes of conformationally constrained, non-peptide, Smac mimetics.30–38 Our efforts have led to the discovery of SM-406 (2 in Figure 1) as a potent and orally bioavailable Smac mimetic that possesses all the desirable properties as a drug candidate for clinical development, which has subsequently been advanced into Phase I clinical trials. In this paper, we describe the design, synthesis, and in vitro and in vivo evaluations of compound 2 as a potent and orally active Smac mimetic.
Chemistry
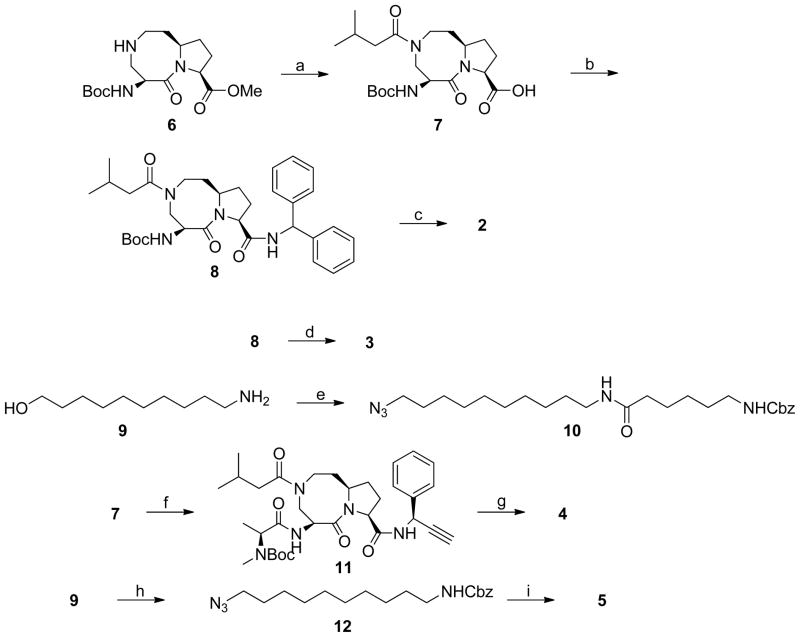
The syntheses of 2, an inactive analogue 3 (SM-428), a biotinylated analogue 4 (BL-SM-406) and a fluorescently tagged compound 5 (FL-SM-406) are outlined in Scheme 1, and their chemical structures are shown in Figure 1.
Scheme 1.
Synthesis of compounds 2–5
Reagents and conditions: (a) i. isovaleryl chloride, N,N-diisopropylethylamine, CH2Cl2; ii. 2 N LiOH, 1,4-dioxane, 82% over two steps; (b) aminodiphenylmethane, EDC, HOBt, N,N-diisopropylethylamine, CH2Cl2, 83%; (c) i. 4 N HCl in 1,4-dioxane, methanol; ii. L-N-Boc-N-methyl-alanine, EDC, HOBt, N,N-diisopropylethylamine, CH2Cl2; iii. 4 N HCl in 1,4-dioxane, methanol, 74% over three steps; (d) i. 4 N HCl in 1,4-dioxane, methanol; ii. L-Boc-Na-methyl-tryptophan, EDC, HOBt, N,N-diisopropylethylamine, CH2Cl2; iii. 4 N HCl in 1,4-dioxane, methanol, 63% over three steps; (e) i. Z-6-aminohexanoic acid, EDC, HOBt, N,N-diisopropylethylamine, CH2Cl2; ii. MsCl, Et3N, CH2Cl2; iii. NaN3, DMF, 110°C, 49% over three steps; (f) i. S-1-phenylprop-2-yn-1-amine, EDC, HOBt, N,N-diisopropylethylamine, CH2Cl2; ii. 4 N HCl in 1,4-dioxane, methanol; iii. L-N-Boc-N-methyl-alanine, EDC, HOBt, N,N-diisopropylethylamine, 69% over three steps; (g) i. 10, CuSO4, sodium L-ascorbate, t-BuOH-H2O 1:1; ii. H2, 10% Pd-C, methanol; iii. (+)-biotin N-hydroxy-succinimide ester, N,N-diisopropylethylamine, CH2Cl2; iv. 4 N HCl in 1,4-dioxane, MeOH, 43% over four steps; (h) i. CbzCl, Et3N, CH2Cl2; ii. MsCl, Et3N, CH2Cl2; iii. NaN3, DMF, 66% over three steps; i. 11, CuSO4, sodium L-ascorbate, t-BuOH-H2O 1:1; ii. H2, 10% Pd-C, methanol; iii. 5-Carboxyfluorescein N-succinimidyl ester, N,N-diisopropylethylamine, CH2Cl2; iv. 4 N HCl in 1,4-dioxane, MeOH, 34% over four steps.
For the synthesis of all these compounds, the common key intermediate compound 6 was prepared using our previously reported method.35 Condensation of 6 with isovaleryl chloride followed by hydrolysis of the methyl ester gave the free acid 7 and condensation of which with diphenylmethylamine yielded the amide 8. Removal of the Boc protecting group from 8, gave an ammonium salt which, upon condensation with L-N-Boc-N-methyl-alanine, followed by removal of the Boc protecting group, gave the designed compound 2. Alternatively, condensation of the ammonium salt with L-Nα-Boc-Nα-methyl-tryptophan and subsequent removal of the Boc protecting group yielded the designed inactive analogue 3.
For the synthesis of biotinylated analogue 4, condensation of compound 9 with N-carbobenzoxy-ε-aminocaproic acid (Z-6-aminohexanoic acid) furnished an amide, mesylation of which, followed by substitution of the mesylate with sodium azide, provided the azide 10. Condensation of 6 with S-1-phenylprop-2-yn-1-amine yielded an amide. Removal of the Boc protecting group from this amide followed by condensation of the resulting ammonium salt with L-N-Boc-N-methyl-alanine afforded amide 11. Cycloaddition of 11 with 10 under the influence of CuSO4 and sodium L-ascorbate furnished a triazole. Removal of the Cbz protecting group in this triazole followed by condensation of the resulted amine with (+)-biotin N-hydroxy-succinimide ester yielded a biotinylated amide, removal of whose Boc protecting group gave the designed biotinylated analogue compound 4.
For the synthesis of fluorescently tagged analogue 5, condensation of compound 9 with benzyl chloroformate furnished a carbamate, mesylation of whose hydroxyl group and substitution of the mesylate with sodium azide provided an azide 12. Cycloaddition of 11 with 12 in the presence of CuSO4 and sodium L-ascorbate furnished a triazole. Removal of the Cbz protecting group in this triazole followed by condensation of the resulted amine with 5-carboxy-fluorescein N-succinimidyl ester yielded an amide. Removal of the Boc protecting group from this amide gave the designed fluorescently tagged analogue compound 5.
Results and Discussion
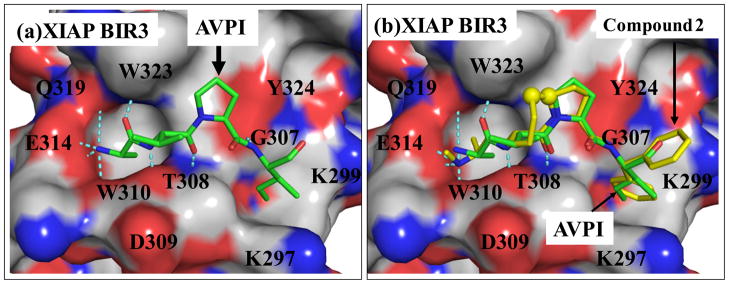
Compound 2 was designed to mimic the interaction of the Smac AVPI peptide with XIAP BIR3 protein. The binding of the Smac AVPI peptide complexed with XIAP BIR3 in the crystal structure23 of Smac protein and the predicted binding model of 2 in a complex with XIAP BIR3 are shown in Figure 2. Compound 2 appears to mimic closely the AVPI peptide in both hydrogen bonding and hydrophobic interactions with XIAP, but has additional hydrophobic contacts with W323 of XIAP. Based upon this model, the terminal methylamino group in 2 inserts into a small, hydrophobic binding pocket associated with W310 in XIAP BIR3. Hence, replacement of the methyl group in 2 with a bulky 3-methyl-1H-indole group disrupts the optimal interaction of 2 with XIAP BIR3, which results in an inactive compound 3.
Figure 2.
Structure-based design of conformationally constrained Smac mimetics. (a). Crystal structure of Smac in a complex with XIAP BIR3. For clarity, only the AVPI peptide in Smac is shown. (b). Superposition of the predicted binding model of compound 2 in a complex with BIR3 on the crystal structure of Smac AVPI peptide complexed with XIAP BIR3. The carbon atoms of AVPI peptide and compound 2 are shown in yellow and green, respectively; the nitrogen and oxygen atoms in both ligands are shown in blue and red, respectively. The protein is represented as its solvent accessible surface and colored by atom types (carbon: gray; nitrogen: blue; oxygen: red). Hydrogen bonds are shown in dashed light blue lines.
To probe the cellular targets for our designed Smac mimetics, we have designed the biotinylated analogue 4. Modeling suggests that the pro-(S) phenyl group in compound 2, without contact with the XIAP protein, is exposed. Accordingly, this phenyl group was replaced by a triazole group. The designed biotinylated analogue 4 was synthesized using a highly efficient “click chemistry” method.39 To accurately determine the binding affinities of our designed Smac mimetics, we have designed and synthesized a fluorescently tagged compound 5 for further optimization of fluorescence-polarization (FP) assays.
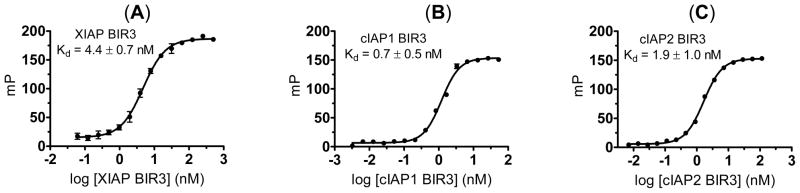
Saturation experiments showed that the fluorescently tagged compound 5 has a Kd = 4.4 nM to XIAP BIR3 protein, 0.7 nM to cIAP1 BIR3 protein and 1.9 nM to cIAP2 BIR3 protein (Figure 3). Furthermore, the dynamic range (ΔmP) for each protein is more than 100, providing a robust signal for the development of competitive FP assays for accurate and quantitative determination of the binding affinities of designed Smac mimetics.
Figure 3.
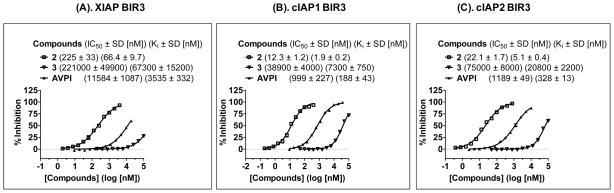
Saturation curves of fluorescently tagged compound 5 to (A). XIAP BIR3 protein; (B). cIAP1 BIR3 protein; and (C). cIAP2 BIR3 protein. The standard deviation for the Kd value to each protein was calculated from three independent experiments.
Using compound 5 as a tracer, a set of competitive FP assays were developed and optimized for XIAP BIR3, cIAP1 BIR3 and cIAP2 BIR proteins. The binding affinities of compounds 2, 3 and the Smac AVPI peptide to these IAP proteins were determined using these assays (Figure 4). Compound 2 binds to XIAP BIR3, cIAP1 BIR3 and cIAP2 BIR3 proteins with a Ki = 66.4 nM, 1.9 nM and 5.1 nM, respectively. The designed inactive control 3 has minimal binding to these IAP proteins at concentrations of 10 μM. Under the same assay conditions, the Smac AVPI peptide binds to XIAP BIR3, cIAP1 BIR3 and cIAP2 BIR3 proteins with Ki = 3.6 μM, 184 nM, and 316 nM, respectively. Hence, compound 2 binds to these three IAP proteins with 50–100 times higher affinities than the Smac AVPI peptide.
Figure 4.
Determination of the binding affinities of compounds 2, 3 and the Smac AVPI peptide to (A). XIAP BIR3; (B). cIAP1 BIR3; and (C). cIAP2 BIR3 using sensitive and quantitative fluorescence-polarization assays. The standard deviation for the IC50 value to each protein was calculated from three independent experiments.
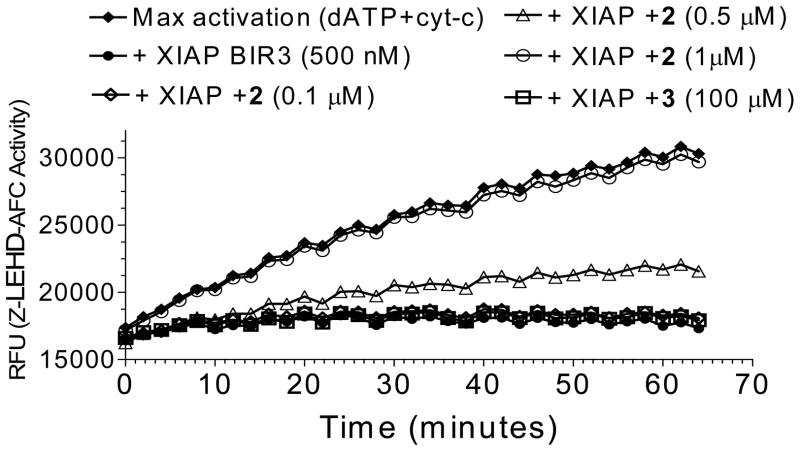
Since XIAP BIR3 binds to and inhibits caspase-9,16 we determined if 2 can effectively antagonize XIAP BIR3 and so promote the activity of caspase-9 (Figure 5). In a cell-free functional assay, addition of cytochrome c and dATP into the MDA-MB-231 cell lysates robustly activates caspase-9, as measured with Z-LEHD-AFC, a substrate specific to caspase-9. Addition of 500 nM of XIAP BIR3 effectively suppresses caspase-9 activity and compound 2 dose-dependently restores it. At a concentration of 1 μM, twice that of the XIAP BIR3 concentration, 2 completely antagonizes XIAP and restores the activity of caspase-9, consistent with its high binding affinity to XIAP BIR3. The inactive control, 3, at concentrations as high as 100 μM fails to antagonize XIAP and to promote caspase-9 activity. 2 is in fact a potent antagonist of XIAP BIR3 protein in this cell-free system.
Figure 5.
Functional antagonism of compound 2 against XIAP BIR3 to restore caspase-9 activity in a cell-free assay. Compound 3 was used as the control compound.
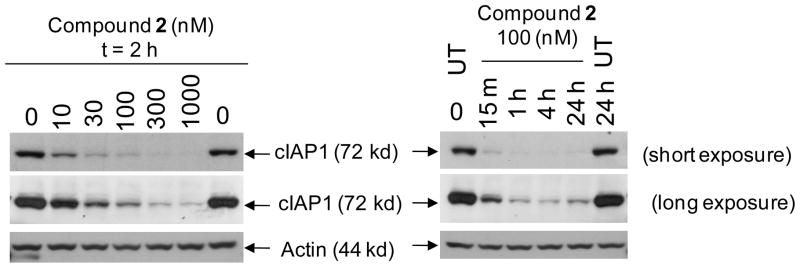
Several recent studies have demonstrated that Smac mimetics induce rapid degradation of cIAP1 and that such degradation is a key early event in apoptosis induction by Smac mimetics as single agents in cancer cells.40–44 Since 2 binds to cIAP1 with a Ki = 1.9 nM, we evaluated its ability to induce cIAP1 degradation in the MDA-MB-231 cancer cells and the associated kinetics, with the results shown in Figure 6. Compound 2 effectively induces cIAP1 degradation at concentrations as low as 10 nM, consistent with its very high binding affinity to cIAP1. Induction of cIAP1 degradation by 2 is also very rapid; treatment of MDA-MB-231 cells by 2 at 100 nM for 15 min causes complete degradation of the cIAP1 protein. Thus, compound 2 is highly potent and effective in induction of rapid cIAP1 degradation.
Figure 6.
Compound 2 induces rapid cIAP-1 degradation in the MDA-MB-231 cancer cell line. Cells were treated with compound 2 as indicated and Western blot analysis was performed using specific antibody against cIAP1 and Actin.
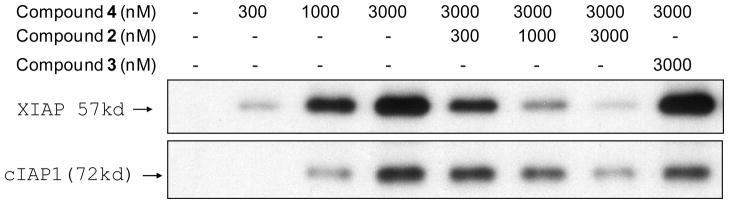
To probe the cellular targets for 2, we have designed and synthesized compound 4, a biotinylated analogue of 2. Compound 4 binds to recombinant XIAP, cIAP1 and cIAP2 BIR3 proteins with Ki = 25.9 nM, <1 nM and 1.7 nM, respectively. We have performed streptavidin-biotin pull-down experiments with 4 and competitive assays to probe the cellular targets of 2 in the MDA-MB-231 cell lysates and the results are given in Figure 7. In the MDA-MB-231 cells which have high levels of XIAP and cIAP1 proteins but very low levels of cIAP2 protein, biotinylated compound 4 dose-dependently pulls down the cellular XIAP and cIAP1 proteins. In the competitive experiment, 2 blocks the interaction of cellular XIAP/cIAP1 with compound 4 in a dose-dependent manner while the inactive control 3 fails to do so. These pull-down experiments show that, consistent with the FP-based binding data using recombinant IAP proteins, both 2 and 4 bind to cellular XIAP and cIAP1 proteins,.
Figure 7.
Biotin-streptavidin pull-down experiments to probe the binding of compounds 2 and 3 to cellular XIAP and cIAP1 in the MDA-MB-231 breast cancer cell lysates using biotinylated compound 4.
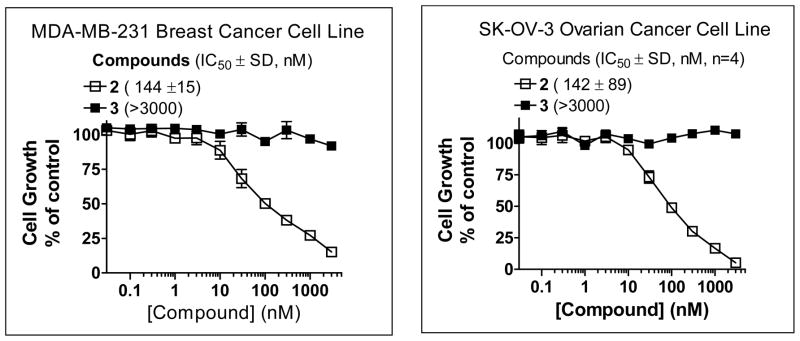
Screening of 2 against more than 100 human cancer cell lines revealed that it is effective in inhibition of cell growth in approximately 15% of these cell lines and its activity is not limited to a single tumor type. For example, in a 4-day WST assay, 2 potently inhibits cell growth in the MDA-MB-231 breast and SK-OV-3 ovarian cancer cell lines with IC50 = 144 nM and 142 nM, respectively, (Figure 8). The inactive control 3 has an IC50 value >10 μM in the same assay, demonstrating the specificity for compound 2.
Figure 8.
Inhibition of cell growth by compounds 2 and 3 in the MDA-MB-231 breast cancer and SK-OV-3 ovarian cancer cell lines, as determined in a water soluble tetrazolium (WST) cell proliferation assay. Four independent experiments were performed for each compound in each cell line.
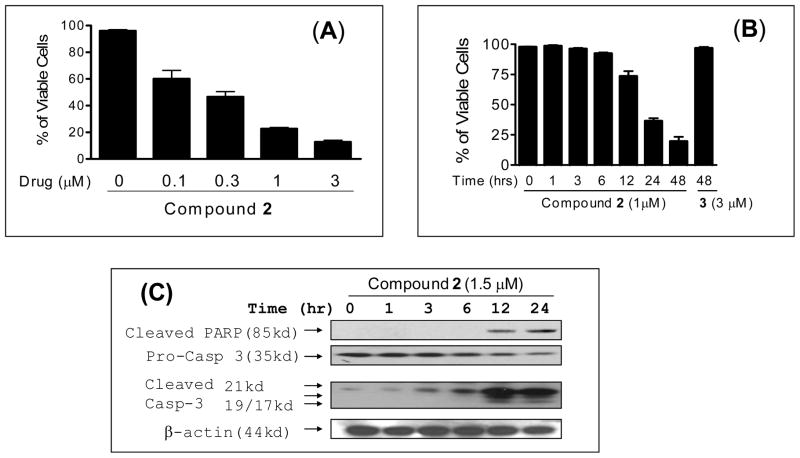
Compound 2 effectively induces cell death in a time- and dose-dependent manner (Figure 9A and 9B). More than 50% of the MDA-MB-231 cancer cells underwent cell death upon treatment with compound 2 at 1 μM for 24 hours. Western blot analysis showed that at 1.5 μM, 2 induces robust activation of caspase-3 and cleavage of PARP in the MDA-MB-231 cancer cells in as little as 12 hr (Figure 9C). Hence, compound 2 is a potent activator of caspases and effectively induces apoptosis in the MDA-MB-231 breast cancer cell line.
Figure 9.
Analysis of induction of cell death and apoptosis by compound 2 in the MDA-MB-231 cell line. MDA-MB-231 cells were treated with compound 2 at (A) different concentrations for 24 hours (B) with 1 μM concentration for different time points. (C) Western blot analysis of caspase processing and cleavage of poly (ADP-ribose) polymerase (PARP). MDA-MB-231 cells were treated with 1.5 μM concentration of compound 2 for 24 hours and cleaved PARP, pro-caspase-3 and cleaved caspase-3 were probed with specific antibodies and actin was used as a loading control.
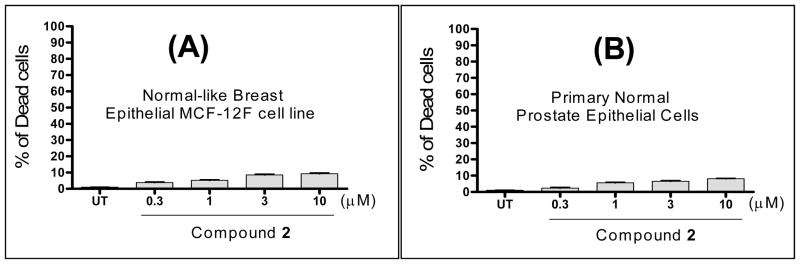
An ideal anticancer drug should be selectively toxic against cancer cells with minimal toxicity to normal cells. We have evaluated the toxicity against normal or normal-like human epithelial cells using the trypan blue cell death assay. Our data, presented in Figure 10, show that 2 has minimal toxicity against these normal or normal-like human epithelial cells at concentrations as high as 10 μM.
Figure 10.
Induction of cell death by compound 2 in (A): normal-like human breast epithelial MCF-12F cells. (B): Primary human normal prostate epithelial cells; Cells were treated with different concentrations of compound 2 for 48 hours and cell viability was determined using the trypan blue exclusion assay.
Compound 2 was evaluated for its pharmacokinetic (PK) properties in mice, rats, non-human primates and dogs. It has an excellent PK profile and good oral bioavailability in each of these four species (Table 1). For example, in non-human primates (NHP), 2 has Cmax = 2.2 μg/ml (~4 μM) in the plasma, T1/2 = 5.4 hours, area-under-the-curve (AUC) = 13.5 μg*h/mL at 5 mg/kg oral dosing, and oral bioavailability of 42%. In dogs, 2 achieves Cmax = 8.5 μg/ml (17 μM), T1/2 = 27 hours, AUC = 198.2 μM*h/mL at 10 mg/kg oral dosing, and an oral bioavailability of 55%.
Table 1.
Pharmacokinetic properties of compound 2 in four different species.
| Species | Route | Dose (mg/kg) | AUC (0-t) (hr*μg/mL) | Cmax (μg/mL) | T1/2 (hr) | F% |
|---|---|---|---|---|---|---|
| Mice | i.v. | 10 | 3.8 | 3.0 | ||
| p.o. | 30 | 4.3 | 1.2 | 2.3 | 38 | |
| Rats | i.v. | 5 | 1.1 ± 0.1 | - | 1.4 ± 0.2 | |
| p.o. | 25 | 2.3 ± 0.7 | 0.66 ± 0.27 | 1.8 0.3 | 45 | |
| NHP | i.v. | 2.5 | 17.9 ±11.0 | 6.3 ± 0.4 | ||
| p.o. | 5 | 13.5 ± 7.9 | 2.2 ± 0.6 | 5.4 ± 1.0 | 42 | |
| Dogs | i.v. | 5 | 81.6 ±17.3 | 16.3 ± 3.8 | ||
| p.o. | 10 | 90.3 ± 23.5 | 8.5 ± 2.5 | 27.1 ± 13.7 | 55 |
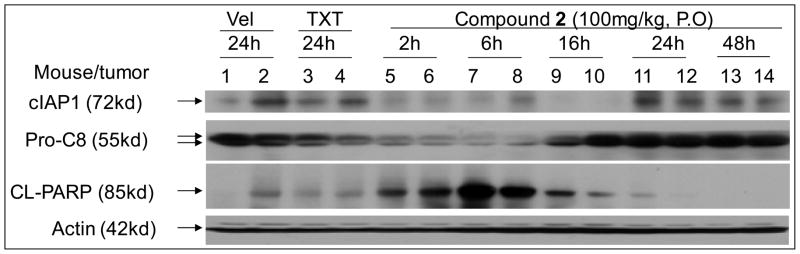
To investigate its mechanism of action in vivo, compound 2 was tested in MDA-MB-231 xenograft tumors in severe combined immune deficiency (SCID) mice. Mice bearing the MDA-MB-231 tumors were given a single oral dose of 2, vehicle control, or intravenous taxotere (TXT). The levels of cIAP1, procaspase-8 and cleaved poly(ADP-ribose) polymerase (PARP) in xenograft tissues were analyzed by Western blotting with the results given in Figure 11. At 100 mg/kg, 2 has effectively induced cIAP1 degradation in tumor tissues at the 2-hr time-point and this effect persists for 16 hr. It also effectively induces processing of procaspase-8 at the 2-hr and 6-hr time-points, as well as cleavage of the apoptosis marker, PARP, at the 2-hr time-point. PARP cleavage reaches a maximum level at the 6-hr time-point and is still evident after 16-hr. In comparison, TXT induces the processing of procaspase-8 but has a minimal effect on cIAP1 degradation and PARP cleavage at the 24-h time-point. Thus 2 is effective in induction of cIAP1 degradation, processing of caspase-8, and PARP cleavage in the MDA-MB-231 xenograft tissues.
Figure 11.
Evaluation of compound 2 on cIAP1, caspase-8 processing and PARP cleavage in the MDA-MB-231 xenograft tissues. Mice bearing the MDA-MB-231 tumors were treated with a single oral dose of compound 2, intravenous taxotere (TXT) or intravenous vehicle. cIAP1, XIAP, pro-caspase-8 (Pro-C8) and cleaved PARP (CL-PARP) were examined by Western blotting.
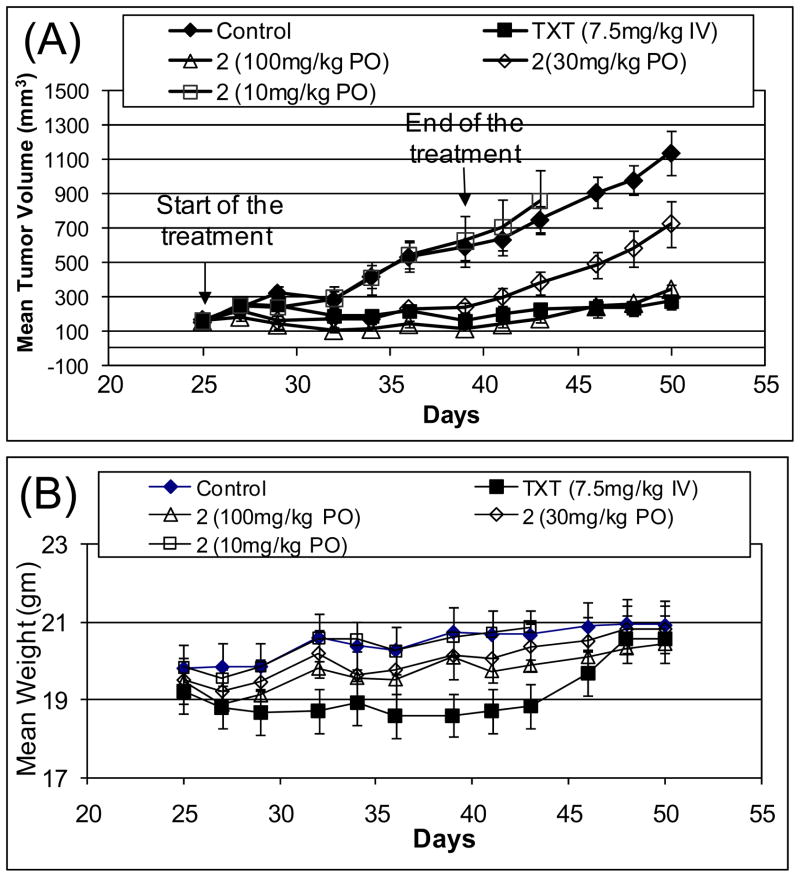
Compound 2 was evaluated for its in vivo antitumor activity as an oral agent in the MDA-MB-231 xenograft model in mice. In our toxicity evaluation, compound 2 was found to be well tolerated in mice at 200 mg/kg, daily for 5 days a week for 2–3 weeks via oral gavage. In this efficacy experiment, each treatment or control group consisted of 8–9 mice, each mouse bearing one tumor. Treatment started on day 25 when the mean tumor size reached 150 mm3. Compound 2 was given via oral gavage daily, 5 days a week for 2 weeks at 10, 30 and 100 mg/kg. Vehicle alone was administered daily, 5 days a week for 2 weeks to a control group of animals. TXT was given intravenously at 7.5 mg/kg, once a week for 2 weeks. The results, shown in Figure 12 indicate that while 2 has no significant antitumor activity at 10 mg/kg, it strongly inhibits tumor growth at 30 and 100 mg/kg and completely inhibits tumor growth during the treatment with 100 mg/kg. The T/C values for 2 at 100 mg/kg on day 36 and 50 are 0.26 and 0.30, respectively, when the control tumors reach 500 and 1100 mm3. Hence in this model, compound 2 is a highly active agent, according to the National Cancer Institute (NCI) criteria (T/C <0.42). The tumor growth inhibition on day 36 at both 30 and 100 mg/kg is statistically highly significant with p values of 0.0083 and 0.0012, respectively, compared to the vehicle control group. The positive control, TXT, also achieves a strong antitumor activity in the MDA-MB-231 xenografts, similar to that of 2 at 100 mg/kg, but it causes significant weight loss in animals during the treatment period (p = 0.01 at day 36), as compared to the vehicle-treated animals. These results indicate that compound 2 is very effective in inhibition of tumor growth in the MDA-MB-231 xenograft model, and has minimal toxicity to animals.
Figure 12.
In vivo antitumor activity of compound 2 in the MDA-MB-231 xenograft model. Taxotere (TXT) was used as the control. (A). Tumor volume. (B). Animal weight.
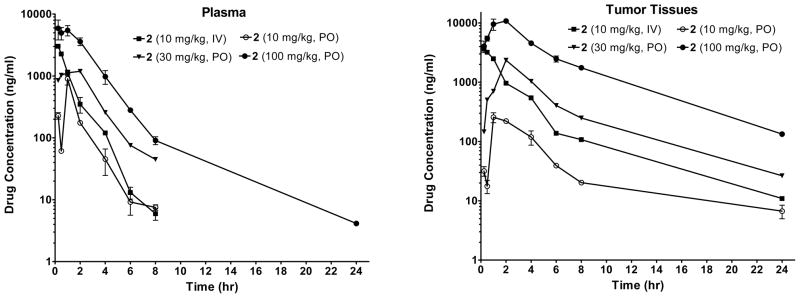
To gain further insights into the PK of compound 2 in both plasma and tumor tissues, we performed an analysis with SCID mice bearing the MDA-MB-231 tumors. SCID mice were given a single dose of compound 2 either intravenously (IV) or via oral gavage (PO) and the concentration of compound 2 was determined in both plasma and tumor tissues. The data are shown in Figure 13 and Table 2. At 10 mg/kg IV, 10 mg/kg PO, 30 mg/kg PO or 100 mg/kg PO dosing, compound 2 achieves Cmax values of 3.0 μg/mL (5.0 μM), 0.9 μg/mL (1.5 μM), 1.2 μg/mL (2.0 μM) and 5.9 (9.9 μM), respectively, and AUC(0-t) values of 3.8, 1.2, 4.3 and 17.4 hr*μg/mL in the plasma. In comparison, at 10 mg/kg IV, 10 mg/kg PO, 30 mg/kg PO or 100 mg/kg PO dosing, compound 2 achieves Cmax values of 3.6 μg/mL (6.0 μM), 0.26 μg/mL (0.5 μM), 2.4 μg/mL (3.9 μM) and 10.9 μg/mL (18.2 μM), respectively, and AUC(0-t) values of 8.3, 1.1, 9.6, and 57.3 hr*μg/mL in the tumor tissues. These data indicate that compound 2 has an excellent tumor tissue penetration. Since compound 2 is effective in inhibition of tumor growth in the MDA-MB-231 xenograft model at 30 mg/kg oral dosing and completely inhibits tumor growth at 100 mg/kg oral dosing, these data support the dose-levels and drug exposures needed for compound 2 to achieve strong antitumor activity in vivo.
Figure 13.
Analysis of pharmacokinetics of compound 2 in plasma and in tumor tissues in SCID mice bearing the MDA-MB-231 xenograft tumors. Compound 2 was dosed via either intravenously (IV) or oral gavage (PO).
Table 2.
Pharmacokinetics of compound 2 in mice in plasma and in tumor tissues. Compound 2 was dosed via either intravenously (IV) or oral gavage (PO).
| Dose(mg/kg) (route) | AUC (0-t) (hr*μg/mL) | Cmax (μg/mL) | Tmax (hr) | T1/2 (hr) | F (%) | |
|---|---|---|---|---|---|---|
| Plasma | 10(IV) | 3.8 | 3.0 | 3.0 | ||
| 10 (PO) | 1.2 | 0.9 | 1 | 2.4 | 31 | |
| 30 (PO) | 4.3 | 1.2 | 2.0 | 2.3 | 38 | |
| 100 (PO) | 17.4 | 5.9 | 0.25 | 5.4 | 46 | |
| Tumor tissues | 10 (IV) | 8.3 | 3.6 | 0.25 | ||
| 10 (PO) | 1.1 | 0.26 | 1.0 | |||
| 30 (PO) | 9.6 | 2.4 | 2.0 | |||
| 100 (PO) | 57.3 | 10.8 | 2.0 |
In summary, we have designed and synthesized compound 2, a non-peptidic Smac mimetic. Our data demonstrate that compound 2 binds to XIAP, cIAP1 and cIAP2 with nanomolar affinities and is a potent antagonist against these IAP proteins. It also achieves an excellent oral bioavailability in rodents and non-rodents and has good aqueous solubility (>50 mg/mL). As a single agent, it effectively inhibits cancer cell growth in a subset of human cancer cell lines and is highly effective in inhibition of tumor growth with oral dosing, while causing no signs of toxicity in mice. Based upon these data, it was selected for further evaluation as a candidate for clinical development. Investigational New Drug (IND) enabling studies under FDA guidelines were conducted. These IND-enabling studies demonstrated that compound 2 has good metabolic stability, a minimal interaction with cytochrome P450s, does not inhibit hERG ion channel at concentrations as high as 30 μM and is well tolerated in dogs (data not shown). Taken together, compound 2 represents an excellent compound for clinical development. Compound 2 (renamed AT-406 by Ascenta Therapeutics) is now being evaluated in Phase I clinical trials as a new therapy for the treatment of solid and hematological human tumors.
Experimental Section
Chemistry
General Methods
1H NMR spectra were measured at 300 MHz. 1H chemical shifts are reported with CHCl3 (7.27 ppm), DHO (4.79 ppm) and CD3OH (3.34 ppm) as internal standards. Final products were purified by HPLC on a C18 reverse phase semi-preparative column with solvent A (0.1% of TFA in water) and solvent B (0.1% of TFA in CH3CN) as eluents and the purity of the final products was checked by analytical HPLC using the C18 reverse phase column to be over >95% purity.
tert-butyl ((5S,8S,10aR)-8-(benzhydrylcarbamoyl)-3-(3-methylbutanoyl)-6-oxodecahydropyrrolo[1,2-a] [1,5]diazocin-5-yl)carbamate (8). Isovaleryl chloride (0.2 mL) and N,N-diisopropylethylamine (0.4 mL) were added to a solution of 6 (340 mg, 1 mmol) in CH2Cl2 (20 mL). The solution was stirred at room temperature for 3 h then concentrated. The residue was purified by chromatography to give an amide. 2N LiOH solution (2.0 mL) was added to a solution of this amide in 1,4-dioxane (3.0 mL). After TLC showed all of the amide had been consumed, the mixture was neutralized with 1N HCl to pH 5 and then extracted with EtOAc (3 × 10 mL). The combined organic layers were dried over Na2SO4 and then concentrated to give the acid 7 (340 mg, 82% over two steps), which was used directly, without purification, in the next step. To a solution of 7 (105 mg, 0.25 mmol) in CH2Cl2 (10 mL) was added aminodiphenylmethane (55 mg, 0.3 mmol), EDC (58 mg, 0.3 mmol), HOBt (45 mg, 0.33 mmol) and N,N-diisopropylethylamine (0.5 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to give 8 (120 mg, 82%). 1H NMR (CDCl3): δ 7.70 (brd, J = 7.0 Hz, 1H), 7.35–7.16 (m, 10H), 6.25 (d, J = 7.5 Hz, 1H), 5.90 (brd, J = 7.0 Hz, 1H), 4.70 (m, 1H), 4.45 (m, 1H), 4.08–3.70 (m, 3H), 2.80–2.50 (m, 3H), 2.42 (m, 1H), 2.30–1.50 (m, 7H), 1.47 (brs, 9H), 1.02 (m, 6H); ESI MS:m/z 577.3 (M + H)+.
(5S,8S,10aR)-N-benzhydryl-5-((S)-2-(methylamino)propanamido)-3-(3-methylbutanoyl)-6-oxodeca-hydropyrrolo[1,2-a][1,5]diazocine-8-carboxamide (2). HCl (4N in 1,4-dioxane, 1 mL) was added to a solution of 8 (58 mg, 0.1 mmol) in MeOH (10 mL). The solution was stirred at room temperature overnight and then concentrated to give an ammonium salt. To a mixture of this salt in CH2Cl2 (10 mL) was added L-N-Boc-N-methyl-alanine (30 mg, 0.15 mmol), EDC (29 mg, 0.15 mmol), HOBt (22 mg, 0.16 mmol) and N,N-diisopropylethylamine (0.3 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to furnish an amide. To a solution of this amide in 5 mL of MeOH was added HCl solution (4N in 1,4-dioxane, 1 mL). The solution was stirred at room temperature overnight and then concentrated to give crude product which was purified by HPLC to give pure compound 2 (salt with TFA, 48 mg, 74% over three steps). The gradient ran from 70% A and 30% B to 50% A and 50% B in 30 min. The purity was determined by reverse analytical HPLC to be over 95%. 1H NMR (CD3OD, 300 M Hz): δ 8.97 (d, J = 7.5 Hz, 1H), 7.38–7.25 (m, 10H), 6.16 (d, J = 7.5 Hz, 1H), 4.80 (m, 1H), 4.58 (m, 1H), 4.25 (m, 1H), 3.96 (m, 3H), 3.47 (m, 1H), 2.72 (s, 3H), 2.49 (d, J = 7.2 Hz, 2H), 2.37–2.32 (m, 1H), 2.15–2.00 (m, 6H), 1.84–1.80 (m, 1H), 1.56 (d, J = 5.4 Hz, 3H), 1.00 (d, J = 6.0 Hz, 6H), ESI-MS m/z 562.3 (M + H)+.
(5S,8S,10aR)-5-((S)-3-(1H-indol-3-yl)-2-(methylamino)propanamido)-N-benzhydryl-3-(3-methylbutanoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocine-8-carboxamide (3). HCl (4N in 1,4-dioxane, 1 mL) was added to a solution of 8 (58 mg, 0.1 mmol) in MeOH (10 mL). The solution was stirred at room temperature overnight and then concentrated to give an ammonium salt. To a mixture of this salt in 10 mL of CH2Cl2 was added L-Nα-Boc-Nα-methyl-tryptophan (48 mg, 0.15 mmol), EDC (30 mg, 0.15 mmol), HOBt (22 mg, 0.16 mmol) and N,N-diisopropylethylamine (0.3 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to furnish an amide. To a solution of this amide in 5 mL of MeOH was added HCl (4N in 1,4-dioxane. 1 mL). The solution was stirred at room temperature overnight and then concentrated to give crude product which was purified by HPLC to give 3 (salt with TFA, 49 mg, 63% over three steps). The gradient ran from 70% A and 30% B to 40% A and 60% B in 30 min. The purity was determined by reverse analytical HPLC to be >95%. 1H NMR (CD3OD, 300 M Hz): δ 8.88 (d, J = 8.4 Hz, 1H), 7.33–7.08 (m,13H), 6.95 (m, 2H), 6.10 (m, 1H), 4.40 (m, 1H), 4.34 (m, 1H), 4.27(m, 1H), 4.23 (m, 1H), 3.97 (m, 2H), 3.83 (m, 2H), 3.44 (m, 1H), 3.10 (m, 2H), 2.73 (2S, 3H), 2.52 (m, 2H), 2.36 (m, 1H), 2.18–1.72 (m, 6H), 1.00 (m, 6H), ESI-MS m/z 677.4 (M + H)+.
tert-butyl methyl((S)-1-(((5S,8S,10aR)-3-(3-methylbutanoyl)-6-oxo-8-(((R)-1-phenylprop-2-yn-1-yl) carbamoyl)decahydropyrrolo[1,2-a][1,5]diazocin-5-yl)amino)-1-oxopropan-2-yl)carbamate (10). S-1-phenylprop-2-yn-1-amine (79 mg, 0.6 mmol), EDC (116 mg, 0.6 mmol), HOBt (80 mg, 0.6 mmol) and N,N-diisopropylethylamine (1 mL) were added to a solution of 7 (205 mg, 0.5 mmol) in CH2Cl2 (15 mL) at 0°C. The mixture was stirred at room temperature for 6 h then concentrated. The residue was purified by chromatography to give an amide. To a solution of this amide in MeOH (15 mL) was added HCl solution (4N in 1,4-dioxane, 2 mL). The solution was stirred at room temperature overnight and then concentrated to give an ammonium salt. To a mixture of this salt in CH2Cl2 (20 mL) was added L-N-Boc-N-methyl-alanine (124 mg, 0.6 mmol), EDC (120 mg, 0.6 mmol), HOBt (83 mg, 0.6 mmol) and N,N-diisopropylethylamine (1 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to give 11 (210 mg, 69% over three steps). 1H NMR (300 MHz, CDCl3, TMS) δ 7.58 (m, 1H), 7.54–7.49 (m, 2H), 7.36–7.20 (m, 4H), 5.95 (dd, J = 8.5, 2.4 Hz, 1H), 4.70 (m, 1H), 4.60–4.45 (m, 2H), 3.95 (m, 1H), 3.92–3.80 (m, 2H), 2.79 (s, 3H), 2.70–2.35 (m, 5H), 2.35–1.75 (m, 7H), 1.50 (brs, 9H), 1.33 (d, J = 7.2 Hz, 3H), 1.05–0.95 (m, 6H); ESI MS m/z 610.3 (M+H)+.
(5S,8S,10aR)-5-((S)-2-(methylamino)propanamido)-3-(3-methylbutanoyl)-6-oxo-N-((S)-(1-(10-(6-(5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamido)hexanamido)decyl)-1H-1,2,3-triazol-4-yl)(phenyl)methyl)decahydropyrrolo[1,2-a][1,5]diazocine-8-carboxamide (4). Z-6-aminohexanoic acid (790 mg, 3 mmol), EDC (590 mg, 3 mmol), HOBt (395 mg, 3 mmol) and triethylamine (2 mL) were added to a solution of 10-amino-1-decanol (9, 520 mg, 3 mmol) in CH2Cl2 (15 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to furnish an amide. Methanesulfonyl chloride (0.3 mL) and triethylamine (1 mL) in CH2Cl2 (20 mL) were added to a solution of this amide at 0°C. The solution was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to yield a mesylate. Sodium azide (200 mg) was added to a solution of this mesylate in DMF (10 mL) and the mixture was stirred at 110°C overnight and then partitioned between EtOAc (50 mL) and brine (30 mL). The organic layer was dried over Na2SO4 and then concentrated. The residue was purified by chromatography to provide 10 (650 mg, 49% over three steps). CuSO4 (25 mg, 0.1 mmol) and sodium L-ascorbate (100 mg, 0.5 mmol) in water (5 mL) were added to a solution of 10 (122 mg, 0.2 mmol) and 9 (90 mg, 0.2 mmol) in acetonitrile (3 mL) and tert-butanol (3 mL). The mixture was stirred at room temperature overnight and then partitioned between EtOAc (30 mL) and brine (20 mL). The organic layer was dried over Na2SO4 and then concentrated. The residue was purified by chromatography to give a triazole which, mixed with 10% Pd-C (100 mg) in MeOH (20 mL), was stirred under H2 overnight and then filtered through celite. The filtration was concentrated to yield an amine. To a solution of this amine in CH2Cl2 (20 mL) was added (+)-biotin N-hydroxy-succinimide ester (78 mg, 0.2 mmol) and N,N-diisopropylethylamine (0.5 mL). The solution was stirred at room temperature overnight and then concentrated to give a residue which was purified by chromatography to afford a biotinylated amide. To a solution of this amide in MeOH (10 mL) was added HCl solution (4N in 1,4-dioxane, 2 mL). The mixture was stirred overnight and then concentrated to furnish a crude product which was purified by a semi-preparative HPLC using a reverse phase C18 column to give pure compound 4 (salt with TFA, 98 mg, 43% over four steps). The gradient ran from 70% of A and 30% of B to 40% of A and 60% of B in 30 min. The purity of the compound was determined by an analytical HPLC using a reverse phase C18 column to be over 95%. 1H NMR (D2O): δ 7.70 (brs, 1H), 7.35–7.05 (m, 5H), 6.05 (brs, 1H), 4.70 (m, 1H), 4.50–4.30 (m, 2H), 4.30–4.05 (m, 3H), 4.05–3.60 (m, 2H), 3.60–2.80 (m, 5H), 2.80–2.50 (m, 6H),2.40–1.05 (m, 44H), 0.80 (m, 6H); ESI MS: m/z 1047.6 (M + H)+.
2-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-5-((10-(4-((S)-((5S,8S,10aR)-5-((S)-2-(methylamino)prop-anamido)-3-(3-methylbutanoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocine-8-carboxamido)(phenyl) methyl)-1H-1,2,3-triazol-1-yl)decyl)carbamoyl)benzoic acid (5). Benzyl chloroformate (0.5 mL) and triethylamine (2 mL) were added to a solution of 10-amino-1-decanol (10, 520 mg, 3 mmol) in CH2Cl2 (15 mL) at 0°C. The mixture was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to furnish a carbamate. Methanesulfonyl chloride (0.5 mL) was added dropwise at 0°C to a solution of this carbamate in CH2Cl2 (20 mL) with triethylamine (1 mL). The solution was stirred at room temperature for 6 h and then concentrated. The residue was purified by chromatography to yield a mesylate. Sodium azide (300 mg) was added to a solution of this mesylate in DMF (10 mL). The mixture was stirred at 110°C overnight and then partitioned between EtOAc (50 mL) and brine (30 mL). The organic layer was dried over Na2SO4 and then concentrated. The residue was purified by chromatography to provide 12 (650 mg, 66% over three steps.) A mixture of CuSO4 (25 mg, 0.1 mmol) and sodium L-ascorbate (100 mg, 0.5 mmol) in water (5 mL) was added to a solution of 11 (122 mg, 0.2 mmol) and 12 (66 mg, 0.2 mmol) in acetonitrile (3 mL) and tert-butanol (3 mL). The mixture was stirred at room temperature overnight and then partitioned between EtOAc (30 mL) and brine (20 mL). The organic layer was dried over Na2SO4 and then concentrated. The residue was purified by chromatography to give a triazole. A mixture of this triazole and 100 mg of 10% Pd-C in MeOH (20 mL) was stirred under H2 overnight and then filtered through celite. The filtration was concentrated to yield an amine. To a solution of this amine in CH2Cl2 (20 mL) was added 5-carboxyfluorescein N-succinimidyl ester (95 mg, 0.2 mmol) and N,N-diisopropylethylamine (0.5 mL). The solution was stirred at room temperature overnight and then concentrated. The residue was purified by chromatography to afford a biotinylated amide. To a solution of this amide in MeOH (10 mL) was added HCl (4N in 1,4-dioxane, 2 mL). The mixture was stirred overnight and then concentrated to furnish a crude product which was purified by semi-preparative HPLC using a reverse phase C18 column to give compound 5 (salt with TFA, 72 mg, 34% over four steps). The gradient ran from 70% of A and 30% of B to 40% of A and 60% of B in 30 min. The purity of the compound was determined by analytical HPLC using a reverse phase C18 column to be over 95%. 1H NMR (CD3OD, 300 MHz): δ 8.48 (s, 1H), 8.22 (m, 1H), 7.70 (s, 1H), 7.50–7.20 (m, 5H), 6.75 (brs, 2H), 6.70–6.50 (m, 4H), 6.30 (brs, 1H), 4.70 (m, 1H), 4.65–4.50(m, 2H), 4.40–4.30 (m, 2H), 4.30–4.15 (m, 2H), 4.05–3.80 (m, 3H), 3.50–3.40 (m, 2H), 2.70 (brs, 3H), 2.52–2.01 (m, 7H), 2.01–1.60 (m, 7H), 1.60–1.50 (m, 3H), 1.50–1.20 (m, 11H), 0.95 (m, 6H); ESI MS: m/z 1066.5 (M + H)+.
Computational docking simulation
The crystal structure of XIAP BIR3 in a complex with the Smac protein23 (PDB code:1G73) was used to predict the binding models of XIAP BIR3 bound to compound 2. The binding pose of compound 2 with XIAP BIR3 was predicted with the GOLD program (version 3.1.1).46,47 In the docking simulation, the center of the binding site was set at Thr308 of XIAP BIR3 and the radius of the binding site was defined as 13 Å, sufficient to cover all the binding pockets. For each genetic algorithm (GA) run, a maximum of 200,000 operations were performed on a population of 5 islands of 100 individuals. Operator weights for crossover, mutation and migration were set to 95, 95 and 10 respectively. The docking simulation was terminated after 20 runs for compound 2. GoldScore, implemented in Gold, was used as the fitness function to evaluate the docked conformations. The 20 conformations ranked highest by each fitness function were saved for analysis of the predicted docking modes. The docking protocol was validated using a series of Smac mimetics.45 The predicted binding pose of compound 2 shown in Figure 2 is the highest ranked conformation from the docking simulations.
Fluorescence polarization based assays for XIAP, cIAP1 and cIAP2 BIR3 proteins
Sensitive and quantitative fluorescence polarization (FP)-based assays were used to determine the binding affinities of our designed Smac mimetics to XIAP BIR3, cIAP1 BIR3, and cIAP2 BIR3 proteins.
Human XIAP BIR3-only (residues 241–356) was cloned into a pET28 vector (Novagen) containing an N-terminal 6xHis tag. Protein was produced in E. coli BL21(DE3) cells grown at 37°C in 2xYT containing kanamycin to an OD600 of 0.6. Protein expression was induced by 0.4 mM IPTG at 27°C for 4 hours. Cells were lysed by sonication in buffer containing Tris pH 7.5 (50 mM), NaCl (200 mM), ZnAc (50 μM), 0.1% βME and Leupectin/Aprotin protease inhibitors. Protein was purified from the soluble fraction using Ni-NTA resin (QIAGEN) followed by gel filtration on a Superdex 75 column in Tris pH 7.5 (20 mM), NaCl (200 mM), ZnAc (50 μM), and dithiothretal (DTT, 1 mM). After purification, DTT was added to a final concentration of 10 mM. Human cIAP1 BIR3-only (residues 253–363) and cIAP2 BIR3-only (residues 238–349) were cloned into pHis-TEV vector, produced and purified using the same method as for the XIAP BIR3 protein.
The fluorescently tagged compound 5 was employed to develop a set of new FP assays for determination of the binding affinities of our designed Smac mimetics to XIAP, cIAP-1, and cIAP-2 BIR3 proteins. The Kd value of compound 5 to each IAP protein was determined by titration experiments using a fixed concentration of compound 5 and different concentrations of the protein up to full saturation. Fluorescence polarization values were measured using an Infinite M-1000 plate reader (Tecan U.S., Research Triangle Park, NC) in Microfluor 2 96-well, black, round-bottom plates (Thermo Scientific). To each well, compound 5 (2nM, 1nM, and 1nM for experiments with XIAP BIR3, cIAP-1 BIR3, and cIAP-2 BIR3, respectively) and different concentrations of the protein were added to a final volume of 125 μl in the assay buffer (100 mM potassium phosphate, pH 7.5, 100 μg/ml bovine γglobulin, 0.02% sodium azide, Invitrogen, with 4% DMSO). Plates were mixed and incubated at room temperature for 2–3 hours with gentle shaking. The polarization values in millipolarization units (mP) were measured at an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Equilibrium dissociation constants (Kd) were then calculated by fitting the sigmoidal dose-dependent FP increases as a function of protein concentrations using Graphpad Prism 5.0 software (Graphpad Software, San Diego, CA).
In competitive binding experiments for XIAP3 BIR3, a tested compound was incubated with 30 nM of XIAP BIR3 protein and 5 nM of compound 5 in the assay buffer (100 mM potassium phosphate, pH 7.5; 100 μg/ml bovine gamma globulin; 0.02 % sodium azide, Invitrogen). In competitive binding experiments for cIAP1 BIR3 protein, 10 nM of the protein and 2 nM of compound 5 were used. In competitive binding experiments for cIAP2 BIR3, 25 nM of the protein and 2 nM of compound 5 were used.
For each competitive binding experiment, polarization values were measured after 2–3 hours incubation using an Infinite M-1000 plate reader. The IC50 value, the inhibitor concentration at which 50% of the bound tracer was displaced, was determined from the plot using nonlinear least-squares analysis. Curve fitting was performed using the PRISM software (GraphPad Software, Inc., San Diego, CA). A Ki value for each compound was calculated based upon the IC50 value using a previously reported algorithm and the associated computer program.48
Cell-free functional assay against XIAP BIR3 protein
MDA-MB-231 cell lysates were prepared by solubilizing cells in ice cold buffer containing KCl (50 mM), EGTA (5 mM), MgCl2 (2 mM) DTT (1 mM), 0.2% CHAPS and HEPES, (50 mM, pH 7.5), containing cocktail protease inhibitors, incubating on ice for 10 minutes, then freezing in liquid nitrogen. Cytochrome c and dATP were added to the cell lysates, which were then incubated at 30°C in a water bath for 60 minutes to activate caspase-9. Addition of recombinant XIAP BIR3 protein dose-dependently suppressed the activity of caspase-9. Different concentrations of a tested Smac mimetic (1 nM – 100 μM) were added to determine the restoration of the activity of these caspases.
For determination of caspase-9 activity, the caspase-9 substrate (Z-LEHD-AFC (25 μM) (BioVision Inc.) was added. Fluorescence detection of substrate cleavage by caspase-9 for the substrate was carried out on an ULTRA READER using an excitation wavelength of 400 nm and an emission wavelength of 505 nm. The reaction was monitored for 1–2 hours.
Biotin-streptavidin pull-down experiment
MDA-MB-231 cells were lysed in NETN buffer (50 mM Tris (pH 8), 1 mM EDTA, 150 mM NaCl, 0.5% NP-40). Lysates were pre-cleared with streptavidin-agarose beads to remove non-specifically adsorbed proteins. Pre-cleared lysates were incubated with biotinylated compound 4 for 1 hr, followed by incubation with new streptavidin-agarose beads for 2 hr to pull-down proteins bound to compound 4. Beads were washed with NETN buffer, proteins eluted by boiling in SDS-PAGE sample buffer, and analyzed by Western blotting using specific antibodies. DMSO alone was used as a control for non-specific pull-down. In the competition experiments, pre-cleared cell lysates were incubated with different concentrations of compound 2 for 30 min, followed by incubation with biotinylated 4 and streptavidin-agarose beads.
Cell growth inhibition and cell-death assays
The MDA-MB-231 breast cancer and SK-OV-3 ovarian cancer cell lines were purchased from the American Type Culture Collection (ATCC). Cells were seeded in 96-well flat bottom cell culture plates at a density of 3–4×103 cells/well with compounds and incubated for 4 days. The rate of cell growth inhibition after treatment with different concentrations of the inhibitors was determined by assaying with (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium mono-sodium salt (WST-8; Dojindo Molecular Technologies Inc., Gaithersburg, Maryland). WST-8 was added to each well to a final concentration of 10%, and then the plates were incubated at 37°C for 2–3 h. The absorbance of the samples was measured at 450 nm using a TECAN ULTRA Reader. Concentration of the compounds that inhibited cell growth by 50% (IC50) was calculated by comparing absorbance in the untreated cells and the cells treated with the compounds.
A trypan blue exclusion assay was used for determination of cell viability. Cells were treated with compounds at different concentrations and time-points. Cell viability was determined using the trypan blue exclusion assay. Blue cells or morphologically unhealthy cells were scored as dead cells. At least 100 cells from each treatment, performed in triplicate, were counted.
Western blot analysis
Cells and xenograft tumor tissues were lysed using radioimmunoprecipitation assay (RIPA) lysis buffer (PBS containing 1% NP-40, 0.5% Na-deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) supplemented with 1 μM phenylmethylsulfonyl fluoride, and 1 protease inhibitor cocktail tablet per 10 ml on ice for 20 min, and lysates were then cleared by centrifugation prior to protein concentration determination using the Bio-Rad protein assay kit according to the manufacturer’s instructions. Proteins were electrophoresed onto 18% SDS-PAGE gels (Invitrogen, Grand Island, NY) and transferred to PVDF membranes. Following blocking in 5% milk, membranes were incubated with a specific primary antibody, washed, and incubated with horseradish peroxidase–linked secondary antibody (Amersham, Piscataway, NJ). The signals were visualized with the Chemiluminescent HRP antibody detection reagent (Denville Scientific, Metuchen, NJ). When indicated, the blots were stripped and reprobed with a different antibody.
The following primary antibodies were used in the study: anti-cleaved-caspase 8, anti-XIAP, anti-PARP (Cell Signaling Technology, Beverly, MA), anti-cIAP1 (from R&D), anti-cIAP2 (R&D), anti-caspases-3, -9 and anti-procaspase-8 (Stressgen Biotechnologies, Victoria, Canada) for Western blot analysis.
In Vivo pharmacodynamic (PD) studies
For in vivo PD studies, the MDA-MB-231 xenograft tumor model was employed. To develop xenograft tumors, 5 × 106 MDA-MB-231 cancer cells with matrigel were injected subcutaneously on the dorsal side of the severe combined immunodeficient mice (SCID mice from Charles River), one tumor per mouse. Mice bearing MDA-MB-231 xenograft tumors were administered with a single dose of compound 2 in its HCl salt form at 100 mg/kg via oral gavage, Taxotere at 7.5 mg/kg intravenously or vehicle control. Tumor tissues were harvested at indicated time points. Tumor tissues were analyzed using Western blotting to examine levels of cIAP1 and XIAP, caspase-8 processing and PARP cleavage in tumor tissues. All animal experiments were performed under the guidelines of the University of Michigan Committee for Use and Care of Animals.
In Vivo Pharmacokinetic studies in plasma and MDA-MB-231 tumor tissues in SCID mice
To develop xenograft tumors, 5 × 106 MDA-MB-231 cancer cells with matrigel were injected subcutaneously on the dorsal side of the severe combined immunodeficient mice (SCID mice from Charles River), two tumors (left and right sides) per mouse. Mice bearing MDA-MB-231 xenograft tumors were administered with a single dose of compound 2 in its HCl salt form at 100 mg/kg via oral gavage. Blood and tumor samples were collected from each mouse by terminal cardiac puncture at 0.25, 0.5, 1, 2, 4, 6, 8, 24 h post-dose. Samples were taken from three mice at each time point. Blood samples were collected into potassium heparin treated tubes and centrifuged at 2000g and 4°C for 10 min. Plasma was collected and stored at −80°C prior to analysis. Isolated tumor tissues were immediately frozen and ground with a mortar and pestle in liquid nitrogen, then stored at −80°C until analysis.
The quantitative LC/MS/MS analyses were conducted using an Agilent 1200 HPLC system coupled to an API 3200 mass spectrometer (Applied Biosystems, MDS Sciex Toronto, Canada) equipped with an API electrospray ionization (ESI) source. Mobile phase A and B were water and methanol, and both contained 5 mM ammonium acetate and 0.1% formic acid (v/v). The flow gradient was initially 90:10 v/v of A:B for 2 min, linearly ramped to 0:100 over 4 min, held at 0:100 for 2 min, and then returned to 90:10 over 0.1 min. This condition was held for a further 5 min prior to the injection of another sample. The mass spectrometer was operated in ESI positive ion mode and detection of the ions was performed in the multiple reaction monitoring (MRM) mode, monitoring the transition of m/z 562.3 precursor ion [M+H]+ to the m/z 167.1 product ion for compound 2 and m/z 610.2 precursor ion [M+H]+ to the m/z 167.1 product ion for IS using an analogue of compound 2 (SM-408). The ion spray voltage was set at 5500 V. Ionization temperature was set as 700°C. The instrument parameters, curtain gas, Gas 1 and Gas 2 (auxillary gas), were set at 20, 60 and 60, respectively. Compounds parameters, declustering potential (DP), collision energy (CE), entrance potential (EP), collision entrance energy (CEP) and collision exit potential (CXP) were 61, 45, 5, 26, 4 V and 61, 45, 9, 27.3, 4 V for compound 2 and the IS, respectively. Data acquisition and quantitation were performed using analyst software version 1.4.2 (Applied Biosystems, MDS Sciex Toronto, Canada).
Compound 2 was dissolved in 50% acetonitrile and the IS solution was prepared by dissolving the IS in acetonitrile to yield a final concentration of 100 ng/ml. Plasma samples were prepared by mixing 50 μl plasma with 10 μl of 50% acetonitrile and 140 μl IS solution. The solution was vortexed for 1 min at high speed and centrifuged at 13,000 rpm for 10 min to precipitate protein. The clear supernatants were transferred to vial inserts for LC/MS/MS analysis. Ten calibration standard solutions for constructing standard curve were prepared by mixing 10 μl of compound 2 at 2.5, 5, 25, 50, 250, 500, 2500, 5000, 10000, 25000 ng/ml with 50 μl blank plasma and 140 μl IS solution. To prepare tumor samples, about 60–80 mg of ground tumor powder was weighed and mixed with PBS to obtain final concentrations (w/v) of 200 mg/ml. Samples were homogenized using a tissue homogenizer (Tissuemiser Homogenizer, Fisher Scientific) for 20 s. Then 10 μl of 50% acetonitrile and 90 μl IS solution was added and vortexed for 1 min at high speed. The tubes were centrifuged at 13,000 rpm for 10 min to precipitate protein. The clear supernatants were transferred to vial inserts for analysis. Calibration standard solutions for constructing standard curve were prepared by mixing 10 μl of compound 2 at 1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000, 5000 ng/ml with 20 μl of blank tumor homogenate and 90 μl of IS solution. Plasma and tumor concentration-time data were analyzed using non-compartmental pharmacokinetic methods with the WinNolin software package (Pharsight Corporation, CA, USA). The PK experiments were performed under the guidelines of the University of Michigan Committee for Use and Care of Animals.
In Vivo antitumor efficacy study
SCID mice (8–10 per group) bearing MDA-MB-231 xenograft tumors were treated with different doses of compound 2, or 7.5 mg/kg of Taxotere or vehicle control daily, 5 days a week for 2 weeks. Tumor sizes and animal weights were measured 3 times a week during the treatment and twice a week after the treatment. Data are presented as mean tumor volumes ± SEM. Statistical analyses were performed by two-way ANOVA and unpaired two-tailed t test, using Prism (version 4.0, GraphPad, La Jolla, CA). P < 0.05 was considered statistically significant. The efficacy experiment was performed under the guidelines of the University of Michigan Committee for Use and Care of Animals.
Pharmacokinetics of compound 2 in rats, dogs and non-human primates
Pharmacokinetic (PK) studies in male Sprague Dawley rats, beagle dogs and cynomolgus monkeys (non-human primates) were performed by the Division of Pharmacokinetics and Metabolism, Shanghai Medicilon Inc, Shanghai 201203, P. R. China.
Compound 2 in its hydrochloride salt form was used in pharmacokinetic (PK) evaluations and was dissolved in saline to yield final concentration at 25 mg/mL (pH≈7). The solution was administered to animals on preparation. The concentration of compound 2 in dosing solution was confirmed by HPLC.
For PK studies in rats, dogs and monkeys, animals were randomly assigned to the treatment groups and were carotid cannulated before the PK studies. The LC system comprised an Agilent (Agilent Technologies Inc. USA) liquid chromatograph equipped with an isocratic pump (1100 series), an autosampler (1100 series) and a degasser (1100 series). Mass spectrometric analysis was performed using an API3000 (triple-quadruple) instrument from AB Inc (Canada) with an ESI interface. The data acquisition and control system were created using Analyst 1.4 software from ABI Inc. The concentrations in plasma below the limit of quantitation (LOQ = 5 ng/mL) were designated as zero. The pharmacokinetic data analysis was performed using noncompartmental analysis. Oral bioavailability was calculated as F(%)=(Dose(oral)×AUC(0-∞)(oral))/(Dose (iv)×AUC(0-∞) (iv))*100%.
Acknowledgments
We are grateful for the financial support from the Breast Cancer Research Foundation, the National Cancer Institute Grant R01CA109025, Ascenta Therapeutics, and University of Michigan Cancer Center Core grant from the National Cancer Institute P30CA046592. The authors thank Dr. G.W.A. Milne for his critical reading of the manuscript and many useful suggestions and Ms. Karen Kreutzer for her excellent secretary assistance.
References
- 1.Nicholson DW. From bench to clinic with apoptosis-based therapeutic agents. Nature. 2000;407:810–816. doi: 10.1038/35037747. [DOI] [PubMed] [Google Scholar]
- 2.Ponder BA. Cancer genetics. Nature. 2001;411:336–341. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 3.Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Salvesen GS, Duckett CS. Apoptosis: IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- 6.Deveraux QL, Reed JC. IAP family proteins — suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 7.Holcik M, Gibson H, Korneluk RG. XIAP: Apoptotic brake and promising therapeutic target. Apoptosis. 2001;6:253–261. doi: 10.1023/a:1011379307472. [DOI] [PubMed] [Google Scholar]
- 8.Shiozaki EN, Shi Y. Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem Sci. 2004;29:486–494. doi: 10.1016/j.tibs.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasula SM, Ashwell JD. IAPs: what’s in a name? Mol Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–74. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 11.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri D. C IAP Regulation of Metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 13.Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 14.Vucic D, Fairbrother WJ. The inhibitor of apoptosis proteins as therapeutic targets in cancer. Clin Cancer Res. 2007;13:5995–6000. doi: 10.1158/1078-0432.CCR-07-0729. [DOI] [PubMed] [Google Scholar]
- 15.Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, Lee RA, Robbins PD, Fernandes-Alnemri T, Shi Y, Alnemri ES. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 16.Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 17.Chai J, Shiozaki E, Srinivasula SM, Wu Q, Dataa P, Alnemri ES, Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- 19.Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 20.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 21.Du C, Fang M, Li Y, Wang X. Smac, a Mitochondrial Protein that Promotes Cytochrome c– Dependent Caspase Activation by Eliminating IAP Inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 22.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL. Identification of DIABLO, a Mammalian Protein that Promotes Apoptosis by Binding to and Antagonizing IAP Proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 23.Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y. Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature. 2000;406:855–862. doi: 10.1038/35022514. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 25.Huang Y, Rich RL, Myszka DG, Wu H. Requirement of both the second and third BIR domains for the relief of X-linked inhibitor of apoptosis protein (XIAP)-mediated caspase inhibition by Smac. J Biol Chem. 2003;278:49517–49522. doi: 10.1074/jbc.M310061200. [DOI] [PubMed] [Google Scholar]
- 26.Samuel T, Welsh K, Lober T, Togo SH, Zapata JM, Reed JC. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumor necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J Biol Chem. 2006;281:1080–1090. doi: 10.1074/jbc.M509381200. [DOI] [PubMed] [Google Scholar]
- 27.Yang QH, Du C. Smac/DIABLO selectively reduces the levels of c-IAP1 and c-IAP2 but not that of XIAP and livin in HeLa cells. J Biol Chem. 2004;279:16963–16970. doi: 10.1074/jbc.M401253200. [DOI] [PubMed] [Google Scholar]
- 28.For recent reviews see: Wang S. Design of Small-Molecule Smac Mimetics as IAP Antagonists. Curr Top Microbiol Immunol. 2010 Nov 12; doi: 10.1007/82_2010_111. [Epub ahead of print]Mannhold R, Fulda S, Carosati E. IAP antagonists: promising candidates for cancer therapy. Drug Discov Today. 2010;15:210–219. doi: 10.1016/j.drudis.2010.01.003.Sun H, Nikolovska-Coleska Z, Yang CY, Qian D, Lu J, Qiu S, Bai L, Peng Y, Cai Q, Wang S. Design of small-molecule peptidic nonpeptidic Smac mimetics. Acc Chem Res. 2008;41:1264–1277. doi: 10.1021/ar8000553.
- 29.http://clinicaltrials.gov/
- 30.Sun H, Nikolovska-Coleska Z, Yang CY, Xu L, Liu M, Tomita Y, Pan H, Yoshioka Y, Krajewski K, Roller PP, Wang S. Structure-Based Design of Potent, Conformationally Constrained Smac Mimetics. J Am Chem Soc. 2004;126:16686–16697. doi: 10.1021/ja047438+. [DOI] [PubMed] [Google Scholar]
- 31.Sun H, Nikolovska-Coleska Z, Yang CY, Xu L, Tomita Y, Krajewski K, Roller PP, Wang S. Structure-Based Design, Synthesis, and Evaluation of Conformationally Constrained Mimetics of the Second Mitochondria-Derived Activator of Caspase That Target the X-Linked Inhibitor of Apoptosis Protein/Caspase-9 Interaction Site. J Med Chem. 2004;47:4147–4150. doi: 10.1021/jm0499108. [DOI] [PubMed] [Google Scholar]
- 32.Sun H, Nikolovska-Coleska Z, Lu J, Qiu S, Yang CY, Gao W, Meagher J, Stuckey J, Wang S. Design, Synthesis, and Evaluation of a Potent, Cell-Permeable, Conformationally Constrained Second Mitochondria Derived Activator of Caspase (Smac) Mimetic. J Med Chem. 2006;49:7916–7920. doi: 10.1021/jm061108d. [DOI] [PubMed] [Google Scholar]
- 33.Sun H, Nikolovska-Coleska Z, Lu J, Meagher JL, Yang CY, Qiu S, Tomita Y, Ueda Y, Jiang S, Krajewski K, Roller PP, Stuckey JA, Wang S. Design, Synthesis, and Characterization of a Potent, Nonpeptide, Cell-Permeable, Bivalent Smac Mimetic That Concurrently Targets Both the BIR2 and BIR3 Domains in XIAP. J Am Chem Soc. 2007;129:15279–15294. doi: 10.1021/ja074725f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun H, Stuckey JA, Nikolovska-Coleska Z, Qin D, Meagher JL, Qiu S, Lu J, Yang CY, Saito NG, Wang S. Structure-Based Design, Synthesis, Evaluation and Crystallographic Studies of Conformationally Constrained Smac Mimetics as Inhibitors of the X-linked Inhibitor of Apoptosis Protein (XIAP) J Med Chem. 2008;51:7169–7180. doi: 10.1021/jm8006849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y, Sun H, Nikolovska-Coleska Z, Qiu S, Yang CY, Lu J, Cai Q, Yi H, Wang S. Design, Synthesis and Evaluation of Potent and Orally Bioavailable Diazabicyclic Smac Mimetics. J Med Chem. 2008;51:8158–8162. doi: 10.1021/jm801254r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang B, Nikolovska-Coleska Z, Zhang Y, Bai L, Qiu S, Yang CY, Sun H, Wang S, Yikang Wu Y. J Med Chem. 2008;51:7352–7355. doi: 10.1021/jm801146d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun W, Nikolovska-Coleska Z, Qin D, Sun H, Yang CY, Bai L, Qiu S, Ma D, Wang S. Design, Synthesis and Evaluation of Potent, Non-Peptidic Smac Mimetics. J Med Chem. 2009;52:593–596. doi: 10.1021/jm801101z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun H, Lu J, Liu L, Yi H, Qiu S, Yang CY, Deschamps JR, Wang S. Nonpeptidic and Potent Small-Molecule Inhibitors of cIAP-1/2 and XIAP Proteins. J Med Chem. 2010;53:6361–6367. doi: 10.1021/jm100487z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 41.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJ, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Lu J, Bai L, Sun H, Nikolovska-Coleska Z, McEachern D, Qiu S, Miller RS, Yi H, Shangary S, Sun Y, Meagher JL, Stuckey JA, Wang S. SM-164: A novel, bivalent Smac mimetic induces apoptosis and tumor regression by concurrent removal of the blockade of cIAP1/2 and XIAP. Cancer Research. 2008;68:9384–9393. doi: 10.1158/0008-5472.CAN-08-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang CY, Sun H, Chen J, Nikolovska-Coleska Z, Wang S. Importance of ligand reorganization free energy in protein-ligand binding-affinity prediction. J Am Chem Soc. 2009;131:13709–13721. doi: 10.1021/ja9039373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones G, Willett P, Glen RC, Leach AR, Talylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 47.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 48.Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, Roller PP, Krajewski K, Saito NG, Stuckey JA, Wang S. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]