Abstract
Medulloblastoma prognosis tends to be poor, despite aggressive therapy, but defining molecular subgroups may identify patients who could benefit from targeted therapies. This study used human gene array and associated clinical data to identify a new molecular subgroup of medulloblastoma characterized by coactivation of the Sonic hedgehog (SHH) and CXCR4 pathways. SHH–CXCR4 tumors were more common in the youngest patients where they were associated with desmoplastic histology. In contrast to tumors activating SHH but not CXCR4, coactivated tumors exhibited greater expression of Math1 and cyclin D1. Treatment with the CXCR4 antagonist AMD3100 inhibited cyclin D1 expression and maximal tumor growth in vivo. Mechanistic investigations revealed that SHH activation stimulated CXCR4 cell surface localization and effector signaling activity, whereas SHH absence caused CXCR4 to assume an intracellular localization. Taken together, our findings define a new medulloblastoma subgroup characterized by a functional interaction between the SHH and CXCR4 pathways, and they provide a rationale to clinically evaluate combined inhibition of SHH and CXCR4 for medulloblastoma treatment.
Introduction
Medulloblastoma is the most common malignant brain tumor of childhood. Despite aggressive multimodal therapy, 5-year event-free survival and overall survival rates remain less than 70% (1). Furthermore, survivors often suffer severe neurologic, cognitive, and endocrine side effects. Thus, there is a need for new and improved therapy. The development of novel therapies will be facilitated by definition of molecular subgroups of disease in which the primary drivers of malignant growth are distinguished and the cell(s) of origin identified.
Multiple prognostically significant histologic subtypes of medulloblastoma are recognized (2, 3). Recent gene expression profiling has also revealed medulloblastoma to comprise distinct molecular subgroups characterized by dysregulations in (i) WNT, (ii) Sonic hedgehog (SHH), (iii) Myc, or (iv) as yet unidentified signaling pathways (4–6). Moreover, genetic analyses and lineage tracings have determined that at least 2 distinct progenitor populations give rise to medulloblastoma, cerebellar granule neuron precursors (GNP), and cells of the lower rhombic lip (7–9).
Medulloblastomas derived from GNPs frequently exhibit constitutive activation of the SHH pathway as a result of mutations in patched (Ptc), suppressor of fused (Sufu; refs. 10–13), or smoothened (Smo; ref. 14). Sonic subgroup tumors can exhibit desmoplastic, classic, or anaplastic large cell histologies. These differing histologies are associated with distinct outcomes with desmoplastic tumors possessing superior survival and anaplastic large cell tumors possessing inferior survival, compared with classic tumors (1). Thus, SHH pathway activation alone does not determine medulloblastoma biology, even within the SHH subgroup tumors.
A role for coacting pathways during tumorigenesis is also indicated by studies of a mouse model of medulloblastoma secondary to heterozygous loss of Ptc. Ptc nullizygous preneoplastic lesions develop in the cerebellum of greater than 50% of these mice. However, only 14% to 20% of these animals develop medulloblastoma, indicating that events in addition to mutational activation of the SHH pathway are necessary for tumorigenesis (15, 16).
A potentially important coacting factor is the chemokine CXCL12. During GNP development CXCL12 and its receptor CXCR4 are required for maximal SHH-induced proliferation (17). Moreover, CXCL12 and CXCR4 are expressed in several brain tumors including medulloblastoma, and the level of CXCR4 expression has prognostic significance (18–20). Inhibition of CXCR4 with the specific antagonists AMD3100 or AMD3465 blocked the intracranial growth of xenografts of the desmoplastic medulloblastoma-derived cell line Daoy, suggesting that CXCR4 activation may also be important for maximal SHH-driven medulloblastoma growth (21).
Herein we report that overexpression of CXCR4 occurs in the SHH subgroup of medulloblastoma. Moreover, in vitro as well as in vivo data indicate that the SHH pathway enhances CXCR4 surface localization and signaling, resulting in a progrowth transcriptional response, maximal tumor growth, and a sensitivity to the antitumor effects of AMD3100. On the basis of these findings, we propose an additional molecular subgroup of medulloblastoma in which the SHH and CXCR4 pathways synergize to promote tumor growth, and for which combined SHH and CXCR4 antagonism may provide a therapeutic benefit.
Materials and Methods
Animals were used in accordance with an established Animal Studies Protocol approved by The Washington University School of Medicine Animal Studies Committee. Chemicals were obtained from Sigma unless otherwise noted.
Cell culture
Primary ND2:SmoA1 medulloblastoma isolates
Tumors were isolated fromSmoA1 mice by surgical dissection. Meninges were removed and the tissue was dissociated with Accutase-enzyme cell detachment medium (eBiosciences) containing 125 units/mL DNase (3× for 15 minutes). Cells were strained through a 70-µm nylon mesh (Falcon) and plated onto poly-d-lysine–coated (20 µg/mL) plates (2 × 106/mL) in Dulbecco's Modified Eagle's Medium/F12 (Life Technologies) containing N2 (Life Technologies), 20 mmol/L KCl, and 36 mmol/L glucose.
Human medulloblastoma (Daoy) cells were obtained from American Tissue Culture Collection and used at low passage (<10) as previously described (21).
GNP cells
Primary cultures of purified GNPs were prepared from postnatal day 6 wild-type C57/BL6 mice as previously described (21).
Growth factor and drug treatments
In vitro treatments
SmoA1 tumor, GNPs, Daoy, 293T, or HOSX4 cells were serum starved (24 hours) and treated with 1 µg/mL CXCL12 (PeproTech). Pretreatments were with/without 1 µg/mL SHH (R&D) or 10 µmol/L cyclopamine [in 2% sodium phosphate/citrate buffer (pH = 3) containing 2-hydropropyl-β-cyclodextrin (HPBCD)] for 12 hours prior to CXCL12 treatment.
AMD3100 treatment of ND2:SmoA1 flank xenografts
Subcutaneous flank implants of 4 × 106 SmoA1+/− medulloblastoma cells in Matrigel were made in nude mice (Jackson). Tumor growth was monitored using serial caliper-based volumetric measures. Mice harboring tumors that exhibited steady growth over a 4-week period and were approximately 10 mm × 10 mm (length × width) underwent subcutaneous insertion of Alzet minipumps filled with AMD3100 (30 µg/mL; n = 7) or PBS (n = 5). Serial tumor volume measurements continued until mice were euthanized 11 days after initiating treatment. Tumors were isolated and fixed in Prefer for immunohistochemistry or lysed in TRIzol for RNA analysis.
cAMP measurement
Intracellular cAMP concentrations in SmoA1 medulloblastoma or Daoy cells pretreated with/without cyclopamine (10 µmol/L, 12 hours) then CXCL12 (1 µg/mL) for varying times (0–20 minutes) were determined as described (22).
Western blot analyses
Whole-cell lysates
SDS-PAGE of cell lysates (40 µg/lane) was carried out as described (21).
Gαi activation assays
Activated Gαi pull-down assays (NewEast Biosciences) followed the manufacturer's protocol. In brief, serum-starved cells (48 hours) were stimulated with CXCL12 (1 µg/mL, 10 minutes). Cell lysates were incubated for 1 hour with an anti-active Gαi antibody and Protein A-agarose beads. Activated Gαi was quantified by Western blot analysis using an anti-Gαi antibody.
Cell surface protein purification
Daoy cells (10 × 106) were pretreated with cyclopamine or vehicle for 12 hours followed by CXCL12 (1 µg/mL, 30 minutes) or vehicle. The Cell Surface Protein Isolation Kit (Pierce) was used as described (23).
The following antibodies were used in Western blot analyses: anti-CXCR4 [Leinco Tech Inc. (1:1,000)], anti-Akt, anti-phospho-Akt (Ser473), anti-ERK, and anti-phospho-ERK (Cell Signaling; 1:1,000); anti-β-actin (Sigma; 1:10,000). Odyssey V3.0 (LI-COR) image acquisition/analysis system was used for detection and quantification of fluorescent bands. Normalization was to Actin or total content of specific proteins as indicated. Data are expressed as percentage of control (mean % SEM). Statistical analysis was carried out by one-way ANOVA. P values are reported in legends.
Measurement of changes in intracellular calcium
Postnatal day 6 GNPs were cultured on poly-d-lysine–coated glass bottom culture dishes (MatTak Corporation) and pretreated with/without SHH (1 ∝/g/mL) for 12 hours. Daoy cells were pretreated with cyclopamine (10 µmol/L) or vehicle for 12 hours. Calcium flux was measured as described (17). Changes in cytosolic-free calcium were determined by monitoring excitation fluorescence using a dual-wavelength excitation source fluorimeter (Fura-2 filter, TILL microscope, TILL photonics GmbH Lochhamer Schlag 19 Germany) in response to sequential excitation at 340 nm and 380 nm. Data are presented as the relative ratio of fluorescence at 340 and 380 nm.
Cell proliferation assay
A total of 1 × 105 SmoA1 tumor cells cultured in poly-d-lysine–coated 96-well flat-bottom plates were treated with AMD3100 (2.5 ng/mL) or CXCL12 (1 µg/mL) as indicated. Proliferation was determined per manufacturer's instructions by the CellTiter 96 AQueous One Cell Proliferation Assay System (Promega) and a μQuant microplate Reader (Bio-Tek Instruments).
Reverse transcription PCR
Complementary DNA and quantitative PCR was carried out with RNA isolated from flank tumors as described (22). Reaction conditions included: 95°C (10 minutes), followed by 40 cycles of 95°C (30 seconds) and various annealing temperatures (1 minute), a final extension at 72°C (1 minute), followed by a melting curve from 55 to 95°C. No-template controls were negative. All reactions were normalized to a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control on the same plate. Fold change was calculated by the formula 2(−ΔΔCt).
Immunohistochemistry
Human medulloblastoma specimens were obtained from the Department of Pathology at Washington University (St. Louis, MO) per an Institutional Review Board–approved protocol for the use of archived pathologic specimens. Staining was accomplished as described (24) with the addition of Tyramide Signal Amplification (Perkin Elmer). Antibodies used and dilutions; CXCR4 clone 12G5 at 1:6,500 (R&D Systems), CXCR7 clone 11G8 at 1:1,000 (kind gift from Mark Penfold, Chemocentryx), CXCL12 at 1:500 (PeproTech), and Mouse IgG 1 µg/mL (Jackson ImmunoResearch).
Human tissue samples, RNA preparation
Primary medulloblastoma (n = 112) and normal cerebella (fetal n = 9, adult n = 5) were obtained in accordance with the Hospital for Sick Children (Toronto, Canada) Research Ethics Board and profiled on Affymetrix GeneChip Human Exon 1.0ST arrays by the Toronto Centre for Applied Genomics (TCAG) as published (4). Molecular subgroups were established as previously described for 103 of 112 samples (4).
CXCR4-high versus -low medulloblastoma class rationale
Samples were divided into CXCR4-high or -low expressers on the basis of their expression patterns relative to average fetal cerebellar levels in a relaxed and stringent analysis. Tumors with more than 1.5-fold (relaxed) and more than 2-fold (stringent) expression levels were classified as CXCR4-high expressers and those with less than −1.5-fold (relaxed) or less than −2-fold (stringent) as CXCR4-low expressers.
Molecular analysis of CXCR4-high versus -low tumors
To detect gene expression changes associated with CXCR4-high versus -low expression, Gene Set Enrichment Analysis (GSEA; ref. 25) was used. As previously discussed (4), a comparative enrichment score for ranked genes in CXCR4-high versus -low expressers was calculated using 1,000 permutations permitting the detection of statistically significant genes and pathways associating with each phenotype.
Results
CXCR4 is overexpressed in the SHH subgroup of medulloblastoma
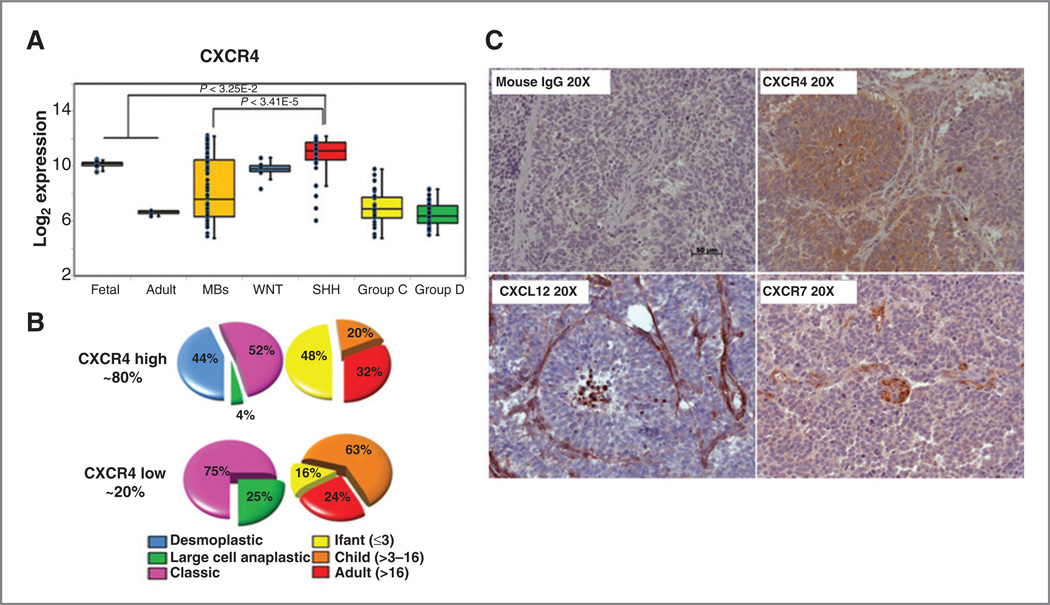
Gene expression profiling has established 4 molecular subgroups of medulloblastoma: WNT, SHH, group C, and group D, with distinct demographics, clinical presentation, transcriptional profiles, genetic abnormalities, and outcome (4). The transcriptional profile of the SHH subgroup resembles an early developmental stage of cerebellar GNPs when their proliferation is driven by SHH (26). During early development, SHH-induced GNP proliferation is modulated by multiple factors including CXCR4, a G-protein–coupled receptor (GPCR), also implicated in medulloblastoma growth (18, 21). We evaluated CXCR4 expression in normal human fetal and adult cerebellum, and across medulloblastoma subtypes. CXCR4 expression was high in the fetal cerebellum and declined significantly with age. A broad range of CXCR4 expression levels were evident in medulloblastoma with Group C and D tumors exhibiting low level, adult cerebellum-like CXCR4 expression, and WNT and SHH subtype tumors exhibiting high level, more fetal cerebellum-like levels of expression (Fig. 1A). While CXCR4 was present in both the WNT and SHH subtype tumors, only the SHH subtype tumors exhibited CXCR4 overexpression relative to the normally high levels found in the fetal cerebellum.
Figure 1.
CXCR4 is overexpressed in the SHH subgroup of medulloblastoma. A, CXCR4 expression in normal cerebella (fetal n = 9, adult n = 5) and medulloblastoma (MB; n = 103) samples profiled on Affymetrix Exon arrays. Statistically significant upregulation of CXCR4 is evident in SHH subgroup tumors relative to normal fetal cerebellar (P < 3.25E-2) and non-SHH medulloblastoma (P < 3.41E-5) levels. B, histologic and age associations for CXCR4-high and -low SHH medulloblastomas. Tumors were classified as high or low on the basis of 2-fold differences compared with the average normal fetal cerebellar levels of CXCR4. C, representative sections from a human desmoplastic medulloblastoma specimen stained for CXCL12, CXCR4, and CXCR7. Brown, positive staining. Mouse IgG controls were negative. Scale bar, 50 µm.
The SHH medulloblastomas could be further categorized into CXCR4-high and -low expressing subgroups (Fig. 1B). The majority of SHH medulloblastomas belonged to the CXCR4-high subgroup (~80%). The level of CXCR4 expression was correlated with other prognostically important features of disease such as histology and age. Desmoplastic tumors were exclusively in the CXCR4-high subgroup. Anaplastic large cell tumors occurred at a rate of only 4% in the CXCR4-high, compared with 25% in the CXCR4-low subgroup. In addition, nearly half of the CXCR4-high tumors occurred in infants, whereas only 13% of CXCR-low tumors occurred in this age group.
While rare mutations in CXCR4 have been described in medulloblastoma (27), CXCR4 activation appears to be ligand dependent. The CXCR4 ligand, CXCL12 was detected in genomic profiling of the SHH subgroup, and as previously described (18, 21), was primarily limited to the endothelium of tumor-associated blood vessels (Fig. 1C). Recently, a second CXCL12 receptor, CXCR7, was identified and shown to be expressed and functional in certain brain tumors such as glioblastoma (28). To determine whether CXCR7 might also mediate CXCL12 effects in the SHH subgroup of medulloblastoma we analyzed genomic data as well as tissue sections for evidence of CXCR7 expression. Little to no CXCR7 mRNA expression was detected in microarray analyses. Immunohistochemical staining of tumor sections from 3 separate human desmoplastic medulloblastoma specimens revealed CXCR7 expression primarily limited to endothelial cells (Fig. 1C). This was in contrast to CXCR4 expression, which as previously described, was evident at high levels throughout the specimens (18, 21). Thus CXCR4 appears to be the sole mediator of CXCL12 effects on medulloblastoma.
SHH pathway activation is required for CXCR4 signaling in medulloblastoma
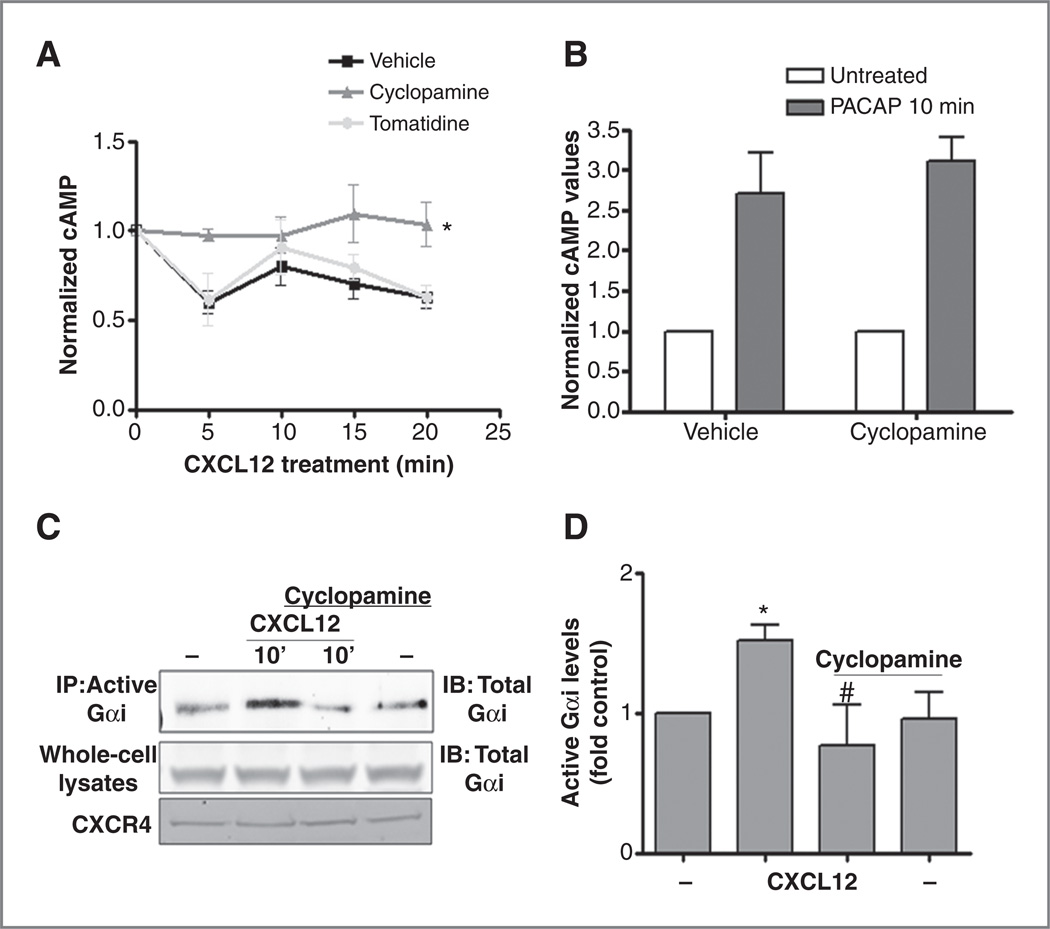
Understanding how the SHH and CXCR4 pathways interact could elucidate new mechanisms of medulloblastoma-genesis and identify novel therapeutic targets. A comparison of CXCR4 activation between postnatal day 6 cerebellar GNPs and Daoy cells revealed that Daoy cells, which possess constitutive activation of the SHH pathway exhibit significantly stronger responses to CXCL12 than GNPs (21). We postulated that among the differences between GNPs and Daoy cells were the degree of SHH pathway activation and that SHH would potentiate CXCR4 function. To test this hypothesis we evaluated whether blockade of SHH signaling in medulloblastoma cells abrogated the effects of CXCL12. For these studies we used primary tumor cells derived from spontaneous medulloblastoma that arise in ND2:SmoA1 mice which possess an activating mutation in smo, constitutive SHH pathway signaling, and a high incidence of medulloblastoma (29). We treated dissociated SmoA1 tumor cells in vitro for 12 hours with the SHH pathway antagonist cyclopamine or vehicle and measured the effects on CXCL12-induced cAMP suppression. CXCL12 stimulation resulted in a significant reduction of intracellular cAMP levels, which was completely blocked by pretreatment with cyclopamine, but not its inactive stereo-isomer, tomatidine (Fig. 2A). The effect of SHH had specificity for CXCR4 as cyclopamine pretreatment had no effect on cAMP elevation in response to PACAP, the ligand for the Gαs-coupled GPCR, PAC1 (Fig. 2B; ref. 30).
Figure 2.
SHH pathway activation is required for CXCR4 signaling in medulloblastoma. A, CXCL12-induced changes in cAMP levels were measured by ELISA in SmoA1 tumor cells pretreated (12 hours) with cyclopamine (10 µmol/L, gray triangles), its inactive isomer tomatidine (10 µmol/L, lighter gray circles), or vehicle control (HPBCD, black squares) and normalized to t = 0 values (*, P < 0.001 for the difference between the cyclopamine and vehicle/tomatidine conditions by 2-way ANOVA, n = 3). B, cyclopamine pretreatment did not affect PACAP-activated cAMP induction (n = 3). C, CXCL12 (1 µg/mL, 10 minutes) induced significant Gαi activation (GTP loading) without changing total Gαi or CXCR4 levels in whole-cell lysates. Cyclopamine (10 µmol/L) pretreatment blocked CXCL12 effects. D, the mean # SEM (n = 3) for CXCL12 activation of Gαi (*, P < 0.001) and cyclopamine blockade of this effect (#, P < 0.001). IB, immunoblotting; IP, immunoprecipitation.
The earliest stages of CXCR4 signaling involve Gαi activation. Thus, we examined the effect of cyclopamine on CXCR4-Gαi coupling. Intracellular Gαi activation was measured by immunoprecipitation of the GTP-bound (activated) form of Gαi. Acute stimulation of SmoA1 tumor cells with CXCL12 (1µg/mL, 10 minutes) resulted in a significant increase in Gαi activation, which was blocked by cyclopamine pretreatment (Fig. 2C and D). These data indicated that CXCL12-induced cAMP suppression in SmoA1 cells was a direct consequence of CXCL12-induced Gαi activation and that the SHH pathway is required for this action. Together, these data support the hypothesis that mutational activation of SHH pathway sensitizes CXCR4 function.
We used Daoy cells to further explore the molecular basis for SHH effects on CXCR4 signaling. Similar to SmoA1 cells, CXCL12-induced activation of Gαi and its downstream effectors in Daoy cells was dependent upon activation of the SHH pathway (Supplementary Fig. S1A and S1B). CXCL12-induced cAMP suppression (Supplementary Fig. S1C), F-actin polymerization (Supplementary Fig. S1D), calcium mobilization (Supplementary Fig. S1E and S1F), and extracellular signal-regulated kinase (ERK) 1/2 and Akt activation (Supplementary Fig. S1G and S1H) were all blocked by cyclopamine pretreatment. Consistent with Gαi-dependant signaling, CXCL12-induced calcium flux and F-actin polymerization was completely abolished by pertussis toxin (data not shown). Inhibition of CXCR4 function was not the result of changes in total CXCR4 expression which was unaffected by cyclopamine (Supplementary Fig. S1G).
To address the possibility that cyclopamine might have direct off-target effects on CXCR4, we treated HOSX4 cells with CXCL12 in the presence and absence of cyclopamine. HOSX4 cells are an osteosarcoma cell line that overexpresses CXCR4 but does not express components of the SHH pathway such as Ptc and Smo. In these cells, CXCL12-induced ERK1/2 phosphorylation was unaffected by cyclopamine (Supplementary Fig. S2A and S2B). Thus, CXCR4 signaling is not always dependent upon SHH action and cyclopamine has no direct effects on CXCR4.
CXCR4 function is SHH dependent in GNPs
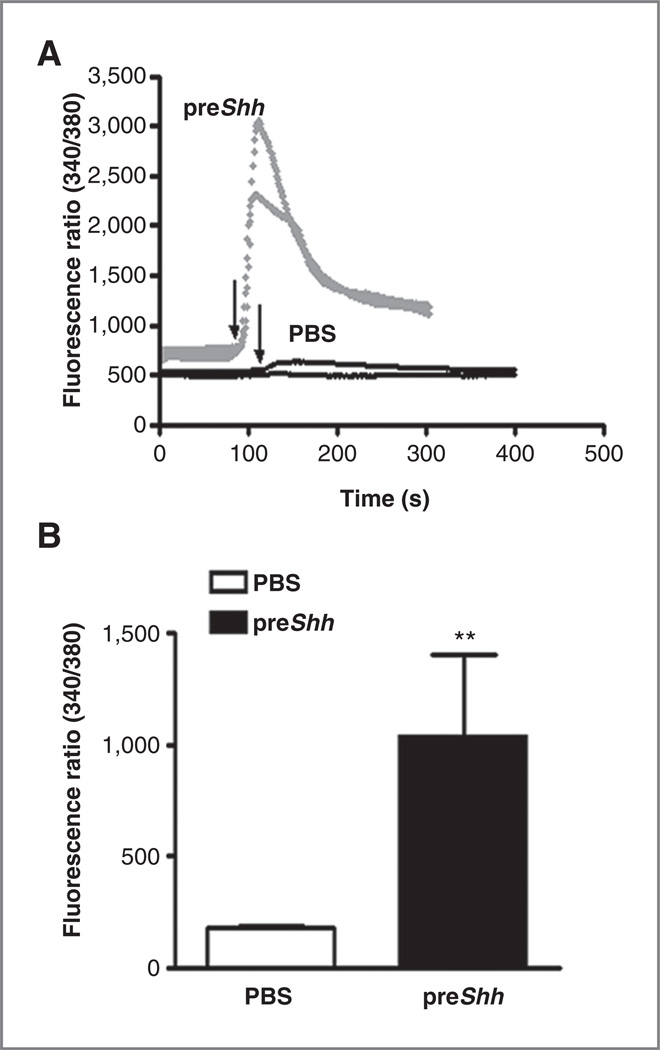
To determine whether regulation of CXCR4 by the SHH pathway might be limited to transformed cells, we cultured normal GNPs in the presence or absence of SHH for 12 hours and measured CXCL12-stimulated calcium flux. In the absence of continuous SHH treatment, SHH pathway activation is downregulated in GNPs by 12 hours postisolation. This is evident in mRNA abundance of the SHH transcriptional target Gli1 (Supplementary Fig. S3A). CXCL12 treatment of GNPs in the absence of SHH had little to no effect on calcium flux (Fig. 3A). In contrast, pretreatment of GNPs with SHH for 12 hours sensitized them to CXCL12 treatment, resulting in substantial CXCL12-induced calcium flux (Fig. 3A and B). Thus, SHH pathway activation can promote CXCR4 pathway responsiveness even in normal, nonneoplastic cells.
Figure 3.
SHH enhances CXCL12-induced CXCR4 signaling in GNP cells. A, CXCL12-induced (1 µg/mL) calcium flux was measured in P6 GNPs in the absence (PBS, black tracings) or presence (preSHH, gray tracings) of 1 µg/mL SHH pretreatment. Arrows indicate the start of CXCL12 treatment. Representative fluorescence ratios (340/380) from 2 individual cells from each treatment group are presented. B, mean and SEM for differences between peak calcium flux ratios without SHH pretreatment (white bar) or with SHH pretreatment (black bar) was determined from 3 separate experiments with 5 groups of cells analyzed per experiment. **, P < 0.001 as determined by 2 tailed t-test.
It is possible that SHH effects on GNPs were not the result of direct effects of SHH on CXCR4 but rather a consequence of SHH's mitogenic effects and expansion of a CXCL12-responsive subpopulation of cells. To exclude this possibility, we carried out parallel studies in 293T cells. 293T cells exhibited basal and SHH-induced activation of the SHH pathway as evidenced by both SHH-stimulated and cyclopamine-inhibited Gli1 expression (Supplementary Fig. S3B–S3D). Neither SHH nor cyclopamine had any effect on 293T cell number (Supplementary Fig. S3E). Basal responsiveness of the CXCR4 pathway was also evident as CXCL12 treatment resulted in Gαi activation, cAMP suppression and phosphorylation of ERK1/2 and Akt (Supplementary Fig. S4A–S4F). Consistent with SHH pathway–dependent activation of CXCR4 signaling, each of these CXCL12 effects was blocked by pretreatment with cyclopamine. Therefore, we conclude that the SHH pathway directly modulates CXCR4 signaling rather than merely expanding the relative abundance of a CXCL12-responsive subpopulation.
Together, these in vitro studies clearly indicate that crosstalk between the SHH and CXCR4 pathways, in both normal and neoplastic cells, can be an important regulator of CXCR4 signaling.
SHH pathway regulates cell surface expression of CXCR4
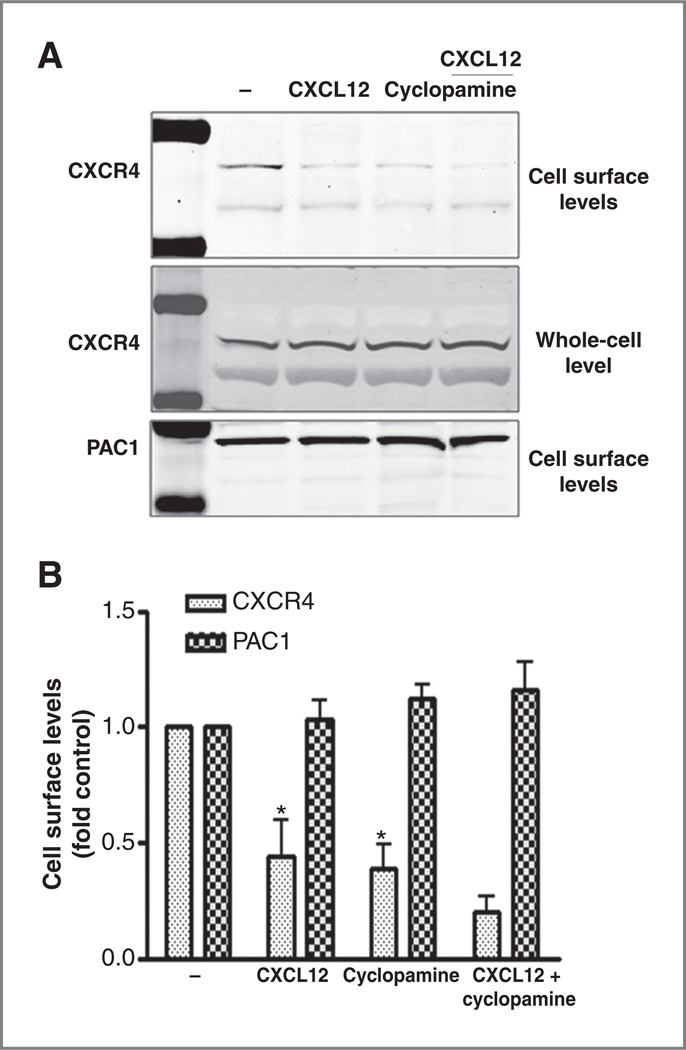
A potential mechanism for regulating CXCR4 coupling to Gαi activation is through control of CXCR4 cell surface localization. To quantitate SHH pathway effects on CXCR4 surface localization we carried out surface biotinylation and surface protein isolation on Daoy cells treated with CXCL12 and cyclopamine. CXCL12 stimulation for 30 minutes led to significant loss of surface CXCR4 through internalization (Fig. 4A and B). Interestingly, exposure to cyclopamine for 12 hours also significantly reduced cell-surface CXCR4, which was further reduced upon subsequent CXCL12 treatment.
Figure 4.
SHH regulates cell surface CXCR4 levels. A, biotinylated plasma membrane proteins from Daoy cells treated with cyclopamine (10 µmol/L, 12 hours) or vehicle, followed by CXCL12 stimulation (1 µg/mL for 30 minutes) were isolated. Surface and total CXCR4 as well as surface PAC1 content were determined by Western blot analysis. B, means ± SEM for cell surface CXCR4 (dark gray bars) and PAC1 receptor (light gray bars) band densities normalized to control (vehicle). CXCL12 and cyclopamine pretreatment significantly lowered CXCR4 levels (*, P < 0.001, n = 3).
Cell surface CXCR4 levels are determined by the balance between receptor internalization and insertion in the plasma membrane. These data suggest that cyclopamine might block the membrane insertion of CXCR4, leading to an overall reduction in surface receptor levels. Neither CXCL12 nor cyclopamine had any effect on the surface levels of PAC1, again suggesting that SHH effects have specificity for CXCR4 rather than for GPCRs in general. Taken together, these data suggest that SHH modulates CXCR4 signaling by regulating the cell surface fraction of CXCR4.
CXCR4 is necessary for maximal growth of SHH-driven medulloblastoma
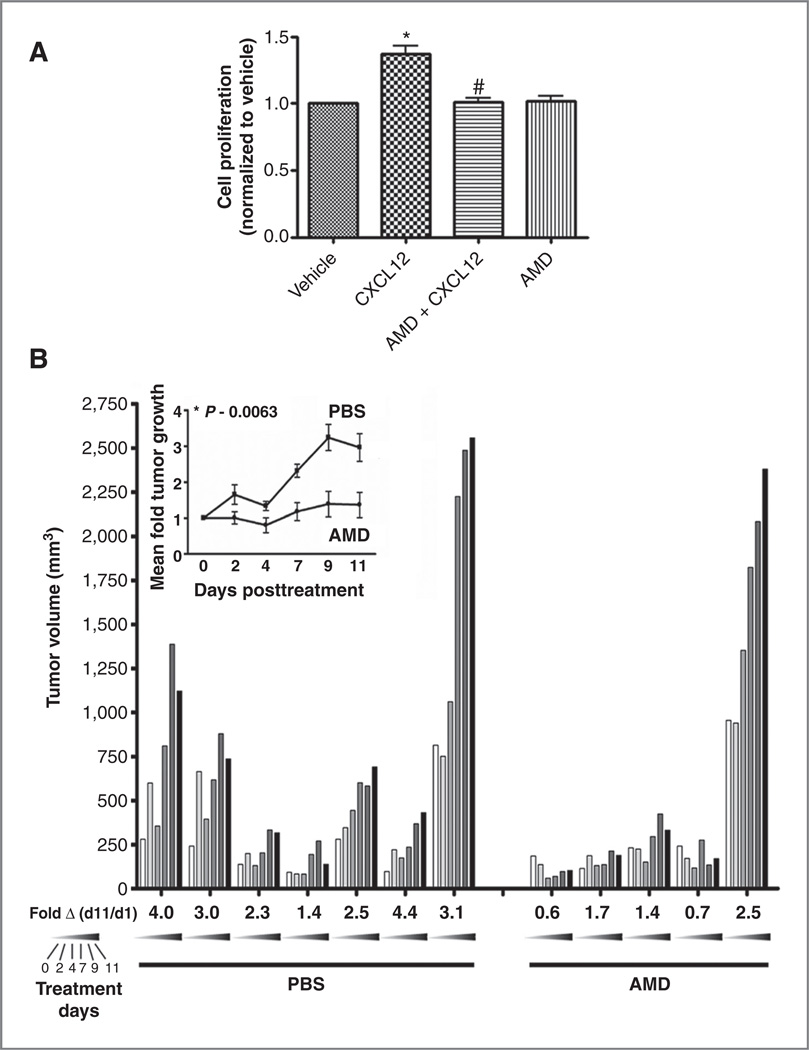
CXCL12 has a positive effect on SHH-induced GNP proliferation (17). To determine whether crosstalk between the SHH and CXCR4 pathways similarly affected the growth of medulloblastoma cells, we measured proliferation of SmoA1 cells in response to CXCL12. Addition of CXCL12 to primary tumor cultures for 12 hours had a small but significant effect on cell proliferation. This effect was completely blocked by the CXCR4 antagonist AMD3100 suggesting that maximal tumor cell proliferation may require both the SHH and CXCR4 pathways (Fig. 5A).
Figure 5.
CXCR4 activation is necessary for maximal growth of SHH-driven medulloblastomas. A, proliferation of SmoA1 tumor cells was measured as described in Materials and Methods after treatment with media alone (vehicle), CXCL12 (1 µg/µL), AMD3100 (2.5 ng/mL), or a combination of the 2. Mean absorbances ± SEM normalized to vehicle are shown (n = 3). CXCL12 induced modest but significant proliferation compared with vehicle (%, P < 0.001); this was blocked by AMD (#, P < 0.001). B, AMD3100 blocked SmoA1 tumor growth in mice. Presented are tumor volume measurements for each mouse in the control (PBS) and AMD3100-treated (AMD) groups obtained over the 11-day treatment period. Each bar represents the volume measurement for a single mouse on a given day. Each day (0, 2, 4, 7, 9, and 11) is represented by a shade between white to black as indicated. Treatment was initiated on day 0, 4 weeks after tumor implantation. Fold growth for day 11 compared with day 1 is indicated beneath each mouse and mean fold tumor growth over the entire treatment period for PBS (n = 7, dark box) and AMD3100-treated (n = 5, dark circle) animals is shown in the inset. AMD treatment significantly reduced tumor volume (%, P < 0.05, by 2-way ANOVA).
To test whether CXCR4 enhanced medulloblastoma growth in vivo, we examined the effect of AMD3100 treatment on the growth of SmoA1 xenografts. Primary SmoA1 tumor cells were isolated and injected into the flanks of nude mice. Tumor volume was monitored over a 4-week period and animals that exhibited steady tumor growth underwent implantation of subcutaneous osmotic infusion pumps filled with either PBS or AMD3100. Control animals (PBS group) all exhibited an increase in tumor volume over the 2-week experimental period. AMD3100 treatment led to significant inhibition of tumor growth indicating that CXCR4 potentiates SHH-driven medulloblastoma growth in vivo (Fig. 5B).
To further examine the importance of CXCR4 to medulloblastoma growth, we treated Ptc+/−, math1GFP mice with AMD3100. Medulloblastoma develops in about 15% of these mice with a peak incidence between 16 and 24 weeks (10, 15, 16, 31, 32). Six tumors developed in the PBS (n = 56) and 8 in the AMD3100 (n = 57) group. While not statistically different, the median time to tumor presentation was delayed to 14.9 weeks in the AMD3100 group compared with 10.6 weeks in the PBS group (Supplementary Fig. S5B).
Coactivation of SHH and CXCR4 pathways results in a unique expression profile in medulloblastoma
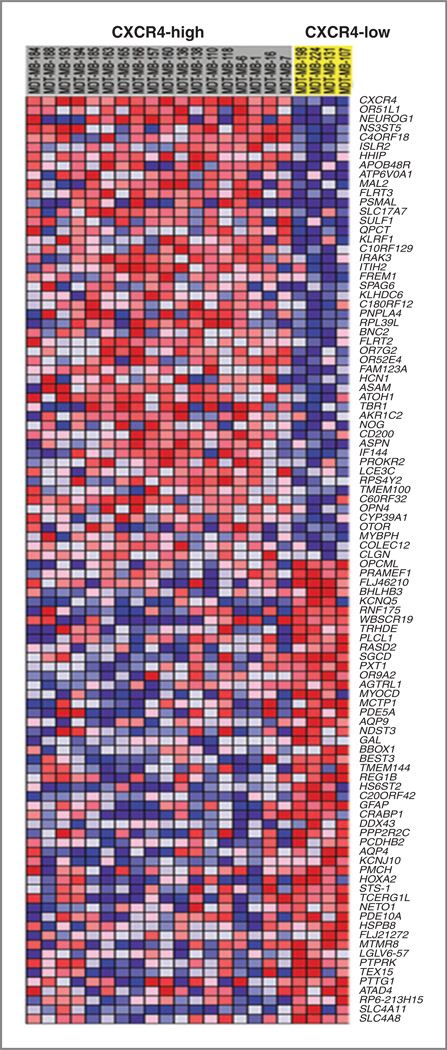
The correlation between CXCR4 expression and histologic subtype of medulloblastoma, together with the impact of CXCR4 activation on model medulloblastoma growth, suggested that coactivation of the SHH and CXCR4 pathways might result in a unique progrowth phenotype. We determined that CXCR4-high and -low medulloblastoma possessed unique transcriptional profiles and could be distinguished by gene array analysis (Fig. 6). Prominent among those genes with differential expression was Atoh1 (Math1), a HLH transcription factor known to regulate GNP proliferation and be required for medulloblastomagenesis. In addition, multiple SHH target genes (Hhip, Neurog1) were more highly expressed in CXCR4-high tumors. To distinguish between genes regulated by CXCR4 and genes merely coregulated along with CXCR4, we isolated SmoA1 tumor cells from flank tumors treated with AMD3100 or PBS and measured mRNA abundance by quantitative PCR (Supplementary Table S1). Four genes appeared to be regulated by CXCR4 (Table 1). Remarkably, CXCR4 significantly suppressed the expression of PPP2R2C (~60%), the regulatory subunit of the phosphatase PP2A and elevated the expression of cyclin D1 (~20%). These changes in gene expression could enhance SHH-driven growth by promoting cell-cycle progression (cyclin D1) and blocking differentiation via PP2A-mediated dephosphorylation of S6 kinase (PPP2R2C; ref. 33). Together, these data suggest a model in which functional interaction between the SHH and CXCR4 pathways results in a unique progrowth phenotype in medulloblastoma.
Figure 6.
CXCR4-high versus -low SHH tumors are molecularly distinct Gene Set Enrichment Analysis (GSEA) heatmap of the most differentially expressed genes in CXCR4-high versus -low SHH tumors identified by phenotypic comparison using signal-to-noise metrics.
Table 1.
Genes that are significantly altered after CXCR4 inhibition of SmoA1 flank tumors: CXCR4-regulated genes
| Sample | mRNA |
Ct (primer) – Ct (GAPDH) |
Fold change [2(−ΔΔCt)] |
P (t test) |
|---|---|---|---|---|
| PBS | Cyclin D1 | 4.43 | 1.00 | |
| AMD | Cyclin D1 | 4.76 | 0.79 | 0.04 |
| PBS | PPP2R2C | 4.17 | 1.00 | |
| AMD | PPP2R2C | 3.52 | 1.58 | <0.01 |
| PBS | PLCβ4 | 7.67 | 1.00 | |
| AMD | PLCβ4 | 6.58 | 2.13 | 0.04 |
| PBS | PLCL1 | 8.15 | 1.00 | |
| AMD | PLCL1 | 7.01 | 2.21 | 0.02 |
Discussion
Medulloblastoma, the most common malignant pediatric brain tumor, arises from multiple cerebellar cell lineages and can be driven by several genetic abnormalities. Recent gene expression profiling and immunohistochemical studies have elucidated 4 distinct molecular subgroups with unique expression profiles (3, 4, 6). Three subgroups are characterized by abnormal activation of the Wnt, SHH, or Myc pathways. The fourth subgroup includes tumors whose molecular foundations are not yet specified.
Clinically, Wnt tumors carry the best prognosis and those medulloblastoma with MYC alterations, the worst (34). SHH tumors exhibit an intermediate outcome (1). Identifying the pathways altered in each of these tumor subgroups is necessary for the development of novel and molecularly targeted therapies and in addition, promises to improve outcome through molecular stratification of patients for clinical trials. In this regard, identification of a medulloblastoma as belonging to the SHH subgroup may not provide complete characterization. SHH pathway activation alone does not determine medulloblastoma biology. SHH medulloblastoma can include low-risk desmoplastic tumors or more aggressive anaplastic disease. Hence, secondary pathways, acting in concert with SHH must also determine biology within the SHH subtype. In the current study, we show that the level of CXCR4 expression further divides the SHH group of medulloblastoma into relevant subsets. We identified 2 medulloblastoma populations with either high or low CXCR4 expression. These 2 subsets of SHH-driven medulloblastoma possess distinct molecular, epidemiologic, and histologic profiles. CXCR4-high tumors are predominantly either desmoplastic or classic histology and occur most frequently in the youngest patients. In contrast, CXCR4-low tumors more commonly occur in older patients and exhibit classic or large cell anaplastic histology. Thus, CXCR4 expression may distinguish populations of SHH medulloblastoma with different prognoses.
The impact of CXCR4 in the SHH subgroup of medulloblastomas appears to be a consequence of unique molecular interactions that occur between these pathways. Our data suggest that SHH regulates CXCR4 activity at one of the earliest stages of its signaling. While SHH did not affect overall CXCR4 levels, prolonged inhibition of the SHH pathway reduced the cell surface fraction of CXCR4 and abrogated cellular responses to CXCL12. Cell surface localization of CXCR4, like other GPCRs, is regulated through desensitization, involving receptor phosphorylation and arrestin-mediated internalization, followed by either degradation or recycling to the cell surface (35–37). SHH could potentially affect any of these processes. Recent studies have highlighted the complexity of such heterologous regulation of GPCR internalization/ recycling. Yu and colleagues showed that ligand-induced activation of Neurokinin 1 receptors (NK1R) produced a nonreciprocal inhibition of ligand-induced mu-opioid receptor endocytosis as a consequence of NK1R-dependent sequestration of arrestins on endosome membranes (38). In addition, nerve growth factor (NGF) has been shown to modulate the recycling of delta-opioid receptors to the surface membrane of central synaptic terminals (39).
We previously described alterations in CXCR4 signaling as a consequence of reduced desensitization in astrocytes null for the tumor suppressor neurofibromin (Nf1; ref. 22). Loss of Nf1 resulted in hyperactivation of the RAS-MAP kinase pathway and resultant ERK-dependent inhibition of GRK2, a kinase involved in CXCR4 desensitization. GRK2 inhibition was correlated with reduced phosphorylation of CXCR4 and sustained CXCL12-induced suppression of cAMP. Interestingly, GRK2 and β-arrestin 2 also directly interact with Smo and regulate SHH-signaling (40, 41). Thus, activation of the SHH pathway might modulate CXCR4 signaling through regulation of the desensitization machinery, including GRK2 and β-arrestin 2.
The biologic significance of interactions between the SHH and CXCR4 pathways may lie in the unique transcriptional profile of the CXCR4-high tumors. PPP2R2C is the regulatory subunit of the phosphatase PP2A, which enhances SHH-driven proliferation by stabilizing N-myc (42). In addition, PP2A blocks differentiation within the granule lineage by inactivating S6 kinase (33). SHH directly induces expression of the catalytic subunit of PP2A and here we show that CXCR4 suppresses expression of the regulatory subunit of PP2A. Both effects could increase PP2A activity and promote proliferation.
Cyclin D1 is a regulator of cyclin-dependent kinases such as CDK4/CDK6, which are critical for G1 to S transition and cell-cycle progression. Induction of cyclin D1 expression is essential for SHH-induced proliferation (43, 44). CXCR4 enhanced cyclin D1 expression in the SHH-medulloblastomas and this is likely to be a critical component of CXCR4's positive effects on tumor growth. Furthermore, CXCR4's effect on cyclin D1 expression suggests that high CXCR4 expression and/or activation may feed back and augment SHH signaling.
Significantly, CXCR4 activation appeared to be necessary for maximal proliferation of SHH-driven tumors. In SmoA1 tumor models, the CXCR4 antagonist AMD3100 significantly inhibited tumor growth. These data indicate that dual inhibition of SHH and the CXCR4 pathways may provide additional benefit in treating those SHH subtype medulloblastomas that also express CXCR4. A recent clinical report described a single patient with medulloblastoma whose tumor was initially sensitive to SHH antagonist therapy, but who subsequently experienced an aggressive recurrence because of mutation in Smo (45–47). The ability of AMD3100 to inhibit the growth of SmoA1-driven tumors suggests that perhaps CXCR4 antagonism might address this mechanism of resistance.
The interplay between the SHH and CXCR4 pathways could function in normal development and cancer biology in any circumstances where the elements of both pathways are coexpressed. In addition to medulloblastoma, components of the CXCR4 and SHH pathways are expressed in primary specimens of breast, pancreatic, and prostate cancers as well as in gliomas and rhabdomyosarcomas (24, 48–53). It will be important to determine whether coexpression of these pathways provides additional prognostic value and whether combined antagonism of these pathways may have added benefit in the treatment of these cancers. Defining how SHH and CXCR4 modulate each other's signaling is likely to open up new avenues of inquiry in each of these fields and new approaches to treating any cancer in which both pathways are operative.
Supplementary Material
Acknowledgments
The authors thank Nicole Warrington for technical assistance and critical reading of the manuscript. Calcium flux measurements were made through the Regional Center for Biodefense and Emerging Infectious Diseases Research Live Cell Microscopy Core Facility at the Washington University School of Medicine
Grant Support
This work was supported by RO1NS052323-04S1 (RW-Reya) and RO1CA118389 (J.B. Rubin). M.D. Taylor holds a CIHR Clinician-Scientist Phase II Award and is supported by grants from the NIH (R01CA148699), and the Pediatric Brain Tumor foundation.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 4.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 6.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, et al. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form SHH-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corcoran RB, Scott MP. A mouse model for medulloblastoma and basal cell nevus syndrome. J Neuro Oncol. 2001;53:307–318. doi: 10.1023/a:1012260318979. [DOI] [PubMed] [Google Scholar]
- 11.Ellison D. Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol Appl Neurobiol. 2002;28:257–282. doi: 10.1046/j.1365-2990.2002.00419.x. [DOI] [PubMed] [Google Scholar]
- 12.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 13.Taylor MD, Liu L, Raffel C, Hui CC, Mainprize TG, Zhang X, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 14.Reifenberger J, Wolter M, Weber RG, Megahed M, Ruzicka T, Lichter P, et al. Missense mutations in SMOH in sporadic basal cell carcinomas of the skin and primitive neuroectodermal tumors of the central nervous system. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- 15.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 16.Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, et al. Loss of patched and disruption of granule cell development in a preneoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 17.Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, et al. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128:1971–1981. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 18.Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, et al. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc Natl Acad Sci U S A. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bian XW, Yang SX, Chen JH, Ping YF, Zhou XD, Wang QL, et al. Preferential expression of chemokine receptorCXCR4 by highly malignant human gliomas and its association with poor patient survival. Neurosurgery. 2007;61:570–578. doi: 10.1227/01.NEU.0000290905.53685.A2. discussion 578–9. [DOI] [PubMed] [Google Scholar]
- 20.Calatozzolo C, Maderna E, Pollo B, Gelati M, Marras C, Silvani A, et al. Prognostic value of CXCL12 expression in 40 low-grade oligodendrogliomas and oligoastrocytomas. Cancer Biol Ther. 2006;5:827–832. doi: 10.4161/cbt.5.7.2838. [DOI] [PubMed] [Google Scholar]
- 21.Yang L, Jackson E, Woerner BM, Perry A, Piwnica-Worms D, Rubin JB. Blocking CXCR4-mediated cyclic AMP suppression inhibits brain tumor growth in vivo. Cancer Res. 2007;67:651–658. doi: 10.1158/0008-5472.CAN-06-2762. [DOI] [PubMed] [Google Scholar]
- 22.Warrington NM, Woerner BM, Daginakatte GC, Dasgupta B, Perry A, Gutmann DH, et al. Spatiotemporal differences in CXCL12 expression and cyclic AMP underlie the unique pattern of optic glioma growth in neurofibromatosis type 1. Cancer Res. 2007;67:8588–8595. doi: 10.1158/0008-5472.CAN-06-2220. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta R, Burbassi S, Shimizu S, Cappello S, Vallee RB, Rubin JB, et al. Morphine increases brain levels of ferritin heavy chain leading to inhibition ofCXCR4-mediated survival signaling in neurons. J Neurosci. 2009;29:2534–2544. doi: 10.1523/JNEUROSCI.5865-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woerner BM, Warrington NM, Kung AL, Perry A, Rubin JB. Widespread CXCR4 activation in astrocytomas revealed by phospho-CXCR4-specific antibodies. Cancer Res. 2005;65:11392–11399. doi: 10.1158/0008-5472.CAN-05-0847. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Miller HL, Jensen P, Hernan R, Connelly M, Wetmore C, et al. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- 27.Schuller U, Koch A, Hartmann W, Garre ML, Goodyer CG, Cama A, et al. Subtype-specific expression and genetic alterations of the chemokinereceptor gene CXCR4 in medulloblastomas. Int J Cancer. 2005;117:82–89. doi: 10.1002/ijc.21116. [DOI] [PubMed] [Google Scholar]
- 28.Hattermann K, Held-Feindt J, Lucius R, Muerkoster SS, Penfold ME, Schall TJ, et al. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70:3299–3308. doi: 10.1158/0008-5472.CAN-09-3642. [DOI] [PubMed] [Google Scholar]
- 29.Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of Sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- 30.Nicot A, Lelievre V, Tam J, Waschek JA, DiCicco-Bloom E. Pituitary adenylate cyclase-activating polypeptide and Sonic hedgehog interact to control cerebellar granule precursor cell proliferation. J Neurosci. 2002;22:9244–9254. doi: 10.1523/JNEUROSCI.22-21-09244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JY, Koralnik IJ, LeFave M, Segal RA, Pfister LA, Pomeroy SL. Medulloblastomas and primitive neuroectodermal tumors rarely contain polyomavirus DNA sequences. Neuro Oncol. 2002;4:165–170. doi: 10.1093/neuonc/4.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wetmore C, Eberhart DE, Curran T. The normal patched allele is expressed in medulloblastomas from mice with heterozygous germ-line mutation of patched. Cancer Res. 2000;60:2239–2246. [PubMed] [Google Scholar]
- 33.Mainwaring LA, Kenney AM. Divergent functions for eIF4E and S6 kinase by Sonic hedgehog mitogenic signaling in the developing cerebellum. Oncogene. 2011;30:1784–1797. doi: 10.1038/onc.2010.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busillo JM, Armando S, Sengupta R, Meucci O, Bouvier M, Benovic JL. Site-specific phosphorylation of CXCR4 is dynamically regulated by multiple kinases and results in differential modulation of CXCR4 signaling. J Biol Chem. 2010;285:7805–7817. doi: 10.1074/jbc.M109.091173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefkowitz RJ. G protein-coupled receptors. III. New roles for receptor kinases and beta-arrestins in receptor signaling and desensitization. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 37.Freedman NJ, Lefkowitz RJ. Desensitization of G protein-coupled receptors. Recent Prog Horm Res. 1996;51:319–351. discussion 352–3. [PubMed] [Google Scholar]
- 38.Yu YJ, Arttamangkul S, Evans CJ, Williams JT, von Zastrow M. Neurokinin 1 receptors regulate morphine-induced endocytosis and desensitization of mu-opioid receptors in CNS neurons. J Neurosci. 2009;29:222–233. doi: 10.1523/JNEUROSCI.4315-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bie B, Zhang Z, Cai YQ, Zhu W, Zhang Y, Dai J, et al. Nerve growth factor-regulated emergence of functional delta-opioid receptors. J Neurosci. 2010;30:5617–5628. doi: 10.1523/JNEUROSCI.5296-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, Beachy PA, et al. Activity-dependent internalization of smoothened mediated by beta-arrestin-2 and GRK2. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 41.Atkinson PJ, Dellovade T, Albers D, Von Schack D, Saraf K, Needle E, et al. Sonic hedgehog signaling in astrocytes is dependent on p38 mitogen-activated protein kinase and G-protein receptor kinase 2. J Neurochem. 2009;108:1539–1549. doi: 10.1111/j.1471-4159.2009.05900.x. [DOI] [PubMed] [Google Scholar]
- 42.Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The Cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9:327–338. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by Sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 45.Romer J, Curran T. Targeting medulloblastoma: small-molecule inhibitors of the Sonic hedgehog pathway as potential cancer therapeutics. Cancer Res. 2005;65:4975–4978. doi: 10.1158/0008-5472.CAN-05-0481. [DOI] [PubMed] [Google Scholar]
- 46.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holm NT, Abreo F, Johnson LW, Li BD, Chu QD. Elevated chemokine receptor CXCR4 expression in primary tumors following neoadjuvant chemotherapy predicts poor outcomes for patients with locally advanced breast cancer (LABC) Breast Cancer Res Treat. 2009;113:293–299. doi: 10.1007/s10549-008-9921-8. [DOI] [PubMed] [Google Scholar]
- 49.Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64:6071–6074. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 50.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azoulay S, Terry S, Chimingqi M, Sirab N, Faucon H, Gil Diez de Medina S, et al. Comparative expression of Hedgehog ligands at different stages of prostate carcinoma progression. J Pathol. 2008;216:460–470. doi: 10.1002/path.2427. [DOI] [PubMed] [Google Scholar]
- 52.Ehtesham M, Sarangi A, Valadez JG, Chanthaphaychith S, Becher MW, Abel TW, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26:5752–5761. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 53.Diomedi-Camassei F, McDowell HP, De Ioris MA, Uccini S, Altavista P, Raschella G, et al. Clinical significance of CXC chemokine receptor-4 and c-Met in childhood rhabdomyosarcoma. Clin Cancer Res. 2008;14:4119–4127. doi: 10.1158/1078-0432.CCR-07-4446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.