Abstract
To test whether the synucleinopathies Parkinson’s disease and multiple system atrophy (MSA) share a common genetic etiology, we performed a candidate single nucleotide polymorphism (SNP) association study of the 384 most associated SNPs in a genome-wide association study of Parkinson’s disease in 413 MSA cases and 3,974 control subjects. The 10 most significant SNPs were then replicated in additional 108 MSA cases and 537 controls. SNPs at the SNCA locus were significantly associated with risk for increased risk for the development of MSA (combined p = 5.5 × 1012; odds ratio 6.2).
Introduction
Multiple system atrophy (MSA) and Parkinson’s disease (PD) are progressive neurodegenerative disorders characterized neuropathologically by deposition of abnormally phosphorylated α-synuclein. In PD, the aggregates are typically found in neurons as Lewy bodies, whereas in MSA, α-synuclein is deposited predominantly in the form of glial cytoplasmic inclusions.1 These observations suggest that PD and MSA share a common pathogenic mechanism.
Although MSA appears to occur sporadically in the community, a number of recent observations have implicated genetic factors in the pathogenesis of the disease. First, neurological signs of parkinsonism are more common in relatives of MSA patients.2,3 Second, affected members within families with SNCA duplication or triplication manifest clinical and pathological features similar to MSA.4–6 Lastly, there are reports of MSA occurring within families, typically with an autosomal recessive inheritance pattern.7,8
We recently completed a genome-wide association study of 1,713 white PD cases and 3,974 white control subjects. Based on this initial cohort, 384 single nucleotide polymorphisms (SNPs) that were most associated with increased risk for development of PD were selected for further testing in an additional cohort of PD cases and control subjects, and we have presented these findings separately.9 To test the hypothesis that MSA and PD share a common genetic causative factor, we tested the same 384 SNPs identified by our PD genome-wide association study in 413 MSA cases and 3,974 healthy control subjects. To confirm our findings, we then replicated the 10 most significant SNPs from this initial screening of MSA cases in an additional cohort of 108 MSA cases and 537 healthy control subjects. Our analysis demonstrated that genetic variants at the SNCA locus coding for α-synuclein were highly significantly associated with increased risk for development of MSA.
Subjects and Methods
Samples
The initial screening cohort consisted of 413 white MSA cases and 3,974 white healthy control subjects. The cases were a mixture of pathologically certain MSA patients (n = 99) and clinically probable or possible cases (n = 314). A total of 283 of 413 MSA cases were included from collaborating centers of the European MSA study group (www.emsa-sg.org). The replication stage was composed of an independent cohort of 108 clinically probable white MSA cases and 537 white healthy control subjects. Diagnosis of patients was based on consensus criteria that Gilman and colleagues10 established. Clinical features and collection sites of cases and control subjects are described in Supplemental Tables 1 and 2. The study was approved by each respective institutional review board, and written informed consent was obtained for each participant.
Genotyping
Genotyping of the 384 SNPs selected for the initial screening stage was performed using custom-made GoldenGate assays on a Veracode platform as per the manufacturer’s instructions (Illumina, San Diego, CA). Raw genotype data were analyzed using Beadstudio software (version 3.1.0; Illumina).
For the replication stage, genotyping was performed by polymerase chain reaction followed by direct sequencing on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) (primer sequences listed in Supplemental Table 3). Genotype information for the control cohort used in the replication stage was extracted from publicly available data of 537 British healthy control subjects who had been previously genotyped on Illumina 610Y SNP chips.
Statistical Analysis
Statistical analysis was performed using PLINK software (v1.04).11 For the screening stage, samples with a call rate less than 90% were excluded from analysis (n = 13 cases and 83 control subjects). SNPs with a minor allele frequency less than 0.01 (n = 3), SNPs with significant departure from Hardy–Weinberg equilibrium (p = 0.001; n = 29), SNPs with a missingness rate greater than 5% (n = 26), or SNPs with inaccurate clustering (n = 2) were excluded from analysis (15 SNPs failed more than one quality-control criterion). Each of the remaining 339 SNPs was then tested for association under allelic, genotypic, dominant, recessive, and trend models, and the lowest p value was calculated for each SNP (pmin). Applying the Bonferroni method to correct for multiple testing, the threshold p value for significance was 2.6 × 10^ −5 (two-sided α of 0.05 divided by [384 SNPs multiplied by 5 models]).
For the replication stage, one SNP was excluded because of departure from Hardy–Weinberg equilibrium in control subjects (p = 0.01). The remaining SNPs were tested for association under a recessive model, because this model was the best fit in the screening stage. Based on Bonferroni correction for multiple testing, a p value less than 0.005 was considered significant (two-sided α of 0.05 divided by 10 SNPs tested). The power of this cohort to replicate loci at this significance level with the odds ratios observed in the screening stage is shown in Supplemental Figure 2.
Results
Screening Stage
A total of 384 SNPs were genotyped in a cohort of 413 MSA cases and 3,974 control subjects. After quality-control filters were applied, 339 SNPs were tested for association with disease in a final dataset of 400 cases and 3,891 control subjects under allelic, genotypic, dominant, recessive, and trend models (results of the screening stage are shown in the Table and in Supplemental Figure 1).
Replication Stage
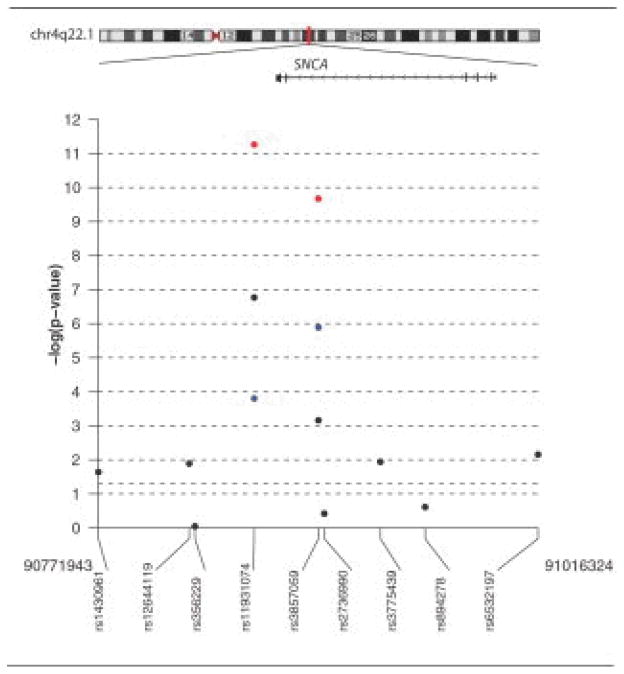
To replicate these findings, we genotyped the 10 most significantly associated SNPs identified in the screening stage in an independent, additional cohort of 108 MSA samples and 537 control samples (see the Table1). Sequence analysis demonstrated a likely genotyping error for rs10515822; reexamination of cluster plots confirmed this error, and this SNP was removed from further analysis. Applying a recessive model, we observed highly significant associations exceeding the Bonferroni threshold for two of these SNPs, namely, rs11931074 (p = 1.6 × 10^ −4) and rs3857059 (p = 1.3 × 10^ −6). When data from the replication stage were combined with data from the screening stage, the p value for rs11931074 was 5.5 × 10^ −12 (odds ratio for homozygous risk allele carriers = 6.2 [95% confidence interval [CI]: 3.4–11.2]), and for rs3857059 was 2.1 × 10^ −10 (odds ratio for homozygous risk allele carriers = 5.9 [95% CI: 3.2–10.9]) (see Supplemental Table 4 for details). These two SNPs are in complete linkage disequilibrium (r2 = 1.0 in the Centre d’Etude du Polymorphisme Humain HapMap population from Utah), and lie in intron 4 of SNCA (rs3857059) and downstream of SNCA (rs11931074) (Fig1). None of the remaining eight SNPs reached significance in the replication stage or in the combined analysis.
Table.
Nine Most Significantly Associated Single Nucleotide Polymorphisms
| SNP ID | Chromosome | Gene | Risk allele | Screening stage | Replication stage | Combined | |||
|---|---|---|---|---|---|---|---|---|---|
| P min (test model) | OR (95% CI) [RR vs (RP+PP)] | P recessive | OR (95%CI) [RR vs (RP+PP)] | P recessive | OR (95% CI) [RR vs (RP+PP)] | ||||
| rs11931074 | 4q22.1 | Downstream of SNCA | T | 1.7E–07(recessive)b | 5.4 (2.7–11.1) | 1.6E–04a | 6.6 (2.15–19.93) | 5.5E–12 | 6.2 (3.4–11.2) |
| rs3857059 | 4q22.1 | SNCA | G | 6.9E–04(recessive) | 3.8 (1.7–8.5) | 1.3E–06a | 9.8 (3.20–29.78) | 2.1E–10 | 5.9 (3.2–10.9) |
| rs9480154 | 6q25.1 | Downstream of PPP1R14C | A | 1.6E– 05(recessive)b | 5.0 (2.2–11.2) | 0.99 | 1.0 (0.12–8.81) | 1.3E–04 | 3.9 (1.8–8.2) |
| rs2794256 | 6q22.31 | LOC728727 | T | 1.7E–03(recessive) | 1.7 (1.2–2.5) | 0.17 | 1.6 (0.81–3.19) | 4.0E–04 | 1.7 (1.3–2.4) |
| rs2042079 | 2p24.2 | Intergenic | A | 2.7E–03(recessive) | 1.7 (1.2–2.5) | 0.21 | 1.6 (0.77–3.18) | 8.0E–04 | 1.7 (1.3–2.4) |
| rs13139027 | 4p16.2 | Upstream of MSX1 | A | 2.5E–03(recessive) | 3.9 (1.5–10.1) | 0.53 | 1.5 (0.41–5.63) | 1.8E–03 | 3.2 (1.5–6.9) |
| rs2515501 | 8p23.2 | MCPH1 | T | 6.5E–04(recessive) | 2.4 (1.4–4.1) | 0.45 | 0.6 (0.13–2.52) | 7.0E–03 | 1.9 (1.2–3.2) |
| rs2896159 | 7q31.2 | Intergenic | T | 3.0E–03(recessive) | 0.7 (0.5–1.1) | 0.38 | 1.3 (0.73–2.26) | 0.43 | 1.3 (1.1–1.6) |
| rs2856336 | 12p13.2 | ETV6 | C | 1.6E– 08(recessive)b | 4.6 (2.6–8.3) | 0.12 | —c | 2.4E–05 | 3.1 (1.8–5.5) |
Exceeded Bonferroni significance threshold in the replication stage (i.e., 0.05/10 = 0.005).
Exceeded Bonferroni significance threshold for multiple testing in the screening stage (i.e., 0.05/[384*5] = 2.6E−05).
Unable to calculate odds ratio (OR) because of low allele frequency in cases. SNP = single nucleotide polymorphism; CI = confidence interval; R = risk allele; P = protective allele; HWE = Hardy–Weinberg equilibrium.
Fig. Location of the association signal at the SNCA locus on chromosome 4q22.1.
Association signals are shown for all single nucleotide polymorphisms (SNPs) genotyped in (A) screening-stage samples (black circles), (B) replication-stage samples (blue circles), and (C) for combined screening- and replication-stage samples (red circles). The most associated SNPs, rs11931074 and rs3857059, lie in or near the SNCA gene, and are in complete linkage disequilibrium. The plot were generated using the SNP. plotter package within R version 2.6.1.
Analysis of Pathology-Proved Multiple System Atrophy Cases
To exclude the possibility that PD cases mistakenly clinically diagnosed as MSA might be falsely driving the association with SNCA, we analyzed the SNPs rs11931074 and rs3857059 in pathology-proven MSA cases and healthy control subjects (n = 92 cases and 3,891 control subjects after quality-control filtering). Both SNPs remained significantly associated with increased risk for development of MSA (recessive model p value for rs11931074 = 1.4 × 10^ −11; p value for rs3857059 = 4.9 × 10^ −6; see Supplemental Table 5).
Analysis of Clinical Multiple System Atrophy Subtypes
From available records, 136 patients could be unequivocally assigned to the MSA-P subtype, and 75 patients were MSA-C cases (see Supplemental Table 2 for further details on these cohorts). An analysis in these subgroups could not detect the association between SNCA variants and increased risk for development of MSA (MSA-P: rs11931074, p = 0.194; rs3857059, p = 0.183; MSA-C: rs11931074, p = 0.075; rs3857059, p = 0.069; recessive model using Fisher’s exact test), probably because of lack of power in the relatively small subgroups. However, this result also suggests that the association is not driven just by one MSA sub-phenotype.
Discussion
In this study, we have demonstrated that genetic variants within the SNCA locus are associated with increased risk for development of MSA. These data represent the first genetic variants convincingly identified for patients with MSA. This study is important in that genetic factors play a greater role in the pathogenesis of MSA, which entity primarily suggests thought of as sporadic in occurrence. The veracity of our findings is underscored by the strength of the association that clearly exceeded the conservative Bonferroni threshold for statistical significance, by the successful replication of our findings in an independent cohort, and by the role that SNCA is already known to play in the disease process based on neuropathological findings.4–6
Previous studies (including sequencing of SNCA coding sequence, gene dosage measurements, microsatellite testing, and haplotype studies) have failed to identify significant association of SNCA variants with MSA.12–16 These negative results can be explained by the smaller sample sizes of these studies, and by the fact that none of the SNCA risk variants identified in our study was tested. Our replication of the association between SNCA variants and MSA in an independent patient and control cohort indicates that population stratification was unlikely to be falsely driving the finding. The failure to replicate our findings in MSA-P and MSA-C clinical subgroups was likely due to small sample size and the diagnostic uncertainty inherent to clinical criteria.17 A combination of these factors would negatively impact the power to detect association within these patient subsets, and studies of larger cohorts will be required to dissect the true pathogenic role of SNCA variants within each of these clinical categories. In contrast, analysis in the smaller, but diagnostically accurate, subset of pathology-proven MSA cases clearly demonstrates that SNCA variants are associated with increased risk for disease.
The significant associations with increased risk for MSA were most clearly observed under the recessive model. However, it is possible that the relatively small size of our case–control cohort was powered only to identify individuals carrying two risk alleles, but that an undetected additive risk at these loci exists. Additional studies involving larger patient cohorts are required to determine whether persons with a single copy of the risk allele are at increased risk for development of MSA.
How does genetic variation at the SNCA locus confer an increased risk for development of MSA? Previous sequence analysis of SNCA coding sequence failed to identify pathogenic mutations; thus, direct alteration of the amino acid sequence is considered an unlikely mechanism of disease.12,16 The most plausible explanation, therefore, would be a change in gene expression regulation. This explanation is supported by the observation that duplication or triplication of SNCA leads to glial cytoplasmic inclusion formation in the brains of affected individuals, and that in some subjects, the clinical presentation resembles a MSA phenotype.4–6 A modest alteration in gene expression levels, although pathogenic in a given individual, may have escaped detection in previous SNCA expression studies of small sample size.18–20 The identified risk variants may also alter the splicing pattern of SNCA in a pathogenic manner, or alter SNCA messenger RNA processing, or additional genetic factors may be responsible for the different manners of synuclein accumulation in PD and MSA.
How do the results of our candidate SNP association study in MSA compare with our genome-wide association study in PD? We identified significant association with the SNCA locus in both diseases.9 The odds ratio associated with carrying a single risk allele of the SNCA SNP rs3857059 was 1.3 in both diseases (95% CI in PD: 1.2–1.5; 95% CI in MSA: 1.1–1.6), whereas the odds ratio for homozygous carriers was 3.8 (95% CI: 2.4–5.9) in PD and 5.9 (95% CI: 3.2–10.9) in MSA.
In summary, our study has conclusively demonstrated that genetic variants in SNCA play a role in the pathogenesis of MSA, and that these genetic factors overlap with those found in PD. These data support the general notion that variability at the gene that encodes the major pathologically deposited species is a risk factor in neurological diseases involving protein deposition21 but highlights that often large sample sizes are required to see such an effect. Additional genetic loci undoubtedly remain to be identified in the pathogenesis of this fatal neurodegenerative disease.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute on Aging, the National Institute on Neurological Disorders and Stroke and the Institute of Environmental Health Services, Department of Health and Human Services; project numbers Z01-AG000957-06 (S.W.S., J.R.G., J.S.S., J.B., J.D., B.J.T., S.A., R.R.Z., A.B.S.) and Z01-ES101986 (H.C.). We gratefully acknowledge support by the Medical Research Council (MRC) (H.H. G108/638 Clinician Scientist Fellowship; N.W., J.A.H.), the Michael J. Fox Foundation (H.H., C.P.R., J.A.H.), the Reta Lila Weston Institute for Neurological Studies (J.A.H., T.R., A.L., J.H.), the Sarah Matheson Trust for Multiple System Atrophy (H.H., T.R., J.H.), the Alzheimer Research Trust (J.A.H.), the National Organization for Rare Disorders (NORD) (H.H.), Ataxia UK (H.H.) and the Progressive Supranuclear Palsy (Europe) Association (J.A.H., T.R., J.H., A.L.). This work was performed under the auspices of the European MSA Study Group (EMSA-SG; http://www.emsa-sg.org/), which is supported by an EU project (QLK6-CT2000-00661). The KORA research platform (KORA: Cooperative Research in the Region of Augsburg; www.gsf.de/KORA) was initiated and financed by the GSF-National Research Centre for Environment and Health, which is funded by the German Federal Ministry of Education, Science, Research, and Technology and by the State of Bavaria (T.I.); furthermore, the study was funded by the German National Genome Network (NGFNplus; German Ministry for Education and Research, #01GS08134) (T.G., C.S., M.S.) and the Helmholtz association (alliance for mental health in an ageing society) (T.G., C.S., M.S.). DNA samples contributed by the Parkinson Institute-Istituti Clinici di Perfezionamento were from the “Human genetic bank of patients affected by PD and parkinsonisms” (http://www.parkinson.it/dnabank.html), which is supported by the Italian Telethon (n. GTB07001).
We thank the participants of this study. We gratefully acknowledge the Queen Square Brain Bank and the New York Brain Bank at Columbia University for providing tissue samples. Human tissue was also obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland and the Human Brain and Spinal Fluid Resource Center, Veterans Affairs West Los Angeles Healthcare Center, which is sponsored by the NIH (National Institute of Neurological Disorders and Stroke/National Institute of Mental Health), the National Multiple Sclerosis Society, and the Department of Veteran Affairs.
References
- 1.Galvin JE, Lee VM, Trojanowski JQ. Synucleinopathies: clinical and pathological implications. Arch Neurol. 2001;58:186–190. doi: 10.1001/archneur.58.2.186. [DOI] [PubMed] [Google Scholar]
- 2.Nee LE, Gomez MR, Dambrosia J, et al. Environmental occupational risk factors and familial associations in multiple system atrophy: a preliminary investigation. Clin Auton Res. 1991;1:9–13. doi: 10.1007/BF01826052. [DOI] [PubMed] [Google Scholar]
- 3.Wenning GK, Wagner S, Daniel S, Quinn NP. Multiple system atrophy: sporadic or familial? Lancet. 1993;342:681. doi: 10.1016/0140-6736(93)91789-o. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs J, Nilsson C, Kachergus J, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 5.Gwinn-Hardy K, Mehta ND, Farrer M, et al. Distinctive neuropathology revealed by alpha-synuclein antibodies in hereditary parkinsonism and dementia linked to chromosome 4p. Acta Neuropathol. 2000;99:663–672. doi: 10.1007/s004010051177. [DOI] [PubMed] [Google Scholar]
- 6.Singleton AB, Farrer M, Johnson J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 7.Hara K, Momose Y, Tokiguchi S, et al. Multiplex families with multiple system atrophy. Arch Neurol. 2007;64:545–551. doi: 10.1001/archneur.64.4.545. [DOI] [PubMed] [Google Scholar]
- 8.Wullner U, Abele M, Schmitz-Huebsch T, et al. Probable multiple system atrophy in a German family. J Neurol Neurosurg Psychiatry. 2004;75:924–925. doi: 10.1136/jnnp.2003.025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scholz S, Schulte C PD Genetics Consortium. Genome-Wide Association Study in Parkinson’s Disease: identification of strong associations with SNCA and MAPT [abstract program number 90]; Presented at the annual meeting of The American Society of Human Genetics; November 13, 2008; Philadelphia, Pennsylvania. [Google Scholar]
- 10.Gilman S, Low P, Quinn N, et al. Consensus statement on the diagnosis of multiple system atrophy. American Autonomic Society and American Academy of Neurology. Clin Auton Res. 1998;8:359–362. doi: 10.1007/BF02309628. [DOI] [PubMed] [Google Scholar]
- 11.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen R, Forno L, DiMonte D, et al. Mutation screening in the α-synuclein gene in MSA. Parkinsonism Relat Disord. 1999;5:S28. [Google Scholar]
- 13.Lincoln SJ, Ross OA, Milkovic NM, et al. Quantitative PCR-based screening of alpha-synuclein multiplication in multiple system atrophy. Parkinsonism Relat Disord. 2007;13:340–342. doi: 10.1016/j.parkreldis.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris HR, Vaughan JR, Datta SR, et al. Multiple system atrophy/progressive supranuclear palsy: alpha-synuclein, synphilin, tau, and APOE. Neurology. 2000;55:1918–1920. doi: 10.1212/wnl.55.12.1918. [DOI] [PubMed] [Google Scholar]
- 15.Ozawa T, Healy DG, Abou-Sleiman PM, et al. The alpha synuclein gene in multiple system atrophy. J Neurol Neurosurg Psychiatry. 2006;77:464–467. doi: 10.1136/jnnp.2005.073528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozawa T, Takano H, Onodera O, et al. No mutation in the entire coding region of the alpha-synuclein gene in pathologically confirmed cases of multiple system atrophy. Neurosci Lett. 1999;270:110–112. doi: 10.1016/s0304-3940(99)00475-9. [DOI] [PubMed] [Google Scholar]
- 17.Litvan I, Goetz CG, Jankovic J, et al. What is the accuracy of the clinical diagnosis of multiple system atrophy? A clinicopathologic study. Arch Neurol. 1997;54:937–944. doi: 10.1001/archneur.1997.00550200007003. [DOI] [PubMed] [Google Scholar]
- 18.Langerveld AJ, Mihalko D, DeLong C, Walburn J, Ide CF. Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov Disord. 2007;22:766–77. doi: 10.1002/mds.21259. [DOI] [PubMed] [Google Scholar]
- 19.Ozawa T, Okuizumi K, Ikeuchi T, et al. Analysis of the expression level of alpha-synuclein mRNA using postmortem brain samples from pathologically confirmed cases of multiple system atrophy. Acta Neuropathol. 2001;102:188–190. doi: 10.1007/s004010100367. [DOI] [PubMed] [Google Scholar]
- 20.Vogt IR, Lees AJ, Evert BO, et al. Transcriptional changes in multiple system atrophy and Parkinson’s disease putamen. Exp Neurol. 2006;199:465–478. doi: 10.1016/j.expneurol.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Singleton A, Myers A, Hardy J. The law of mass action applied to neurodegenerative disease: a hypothesis concerning the etiology and pathogenesis of complex diseases. Hum Mol Genet. 2004;13:123–126. doi: 10.1093/hmg/ddh093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.