SUMMARY
Transmembrane semaphorins (Semas) serve evolutionarily conserved guidance roles, and some function as both ligands and receptors. However, the molecular mechanisms underlying the transduction of these signals to the cytoskeleton remain largely unknown. We have identified two direct regulators of Rho family small GTPases, pebble (a Rho guanine nucleotide exchange factor (GEF)) and RhoGAPp190 (a GTPase activating protein (GAP)), that show robust interactions with the cytoplasmic domain of the Drosophila Sema-1a protein. Neuronal pebble and RhoGAPp190 are required to control motor axon defasciculation at specific pathway choice points and also for target recognition during Drosophila neuromuscular development. Sema-1a–mediated motor axon defasciculation is promoted by pebble and inhibited by RhoGAPp190. Genetic analyses show that opposing pebble and RhoGAPp190 functions mediate Sema-1a reverse signaling through the regulation of Rho1 activity. Therefore, pebble and RhoGAPp190 transduce transmembrane semaphorin–mediated guidance cue information that regulates the establishment of neuronal connectivity during Drosophila development.
INTRODUCTION
Normal nervous system function requires the development of elaborate and precise connections among neurons and their targets. Establishing this complex wiring relies on the combined functions of a large and diverse number of axon guidance molecules that coordinate neuronal process pathfinding and target recognition (Dickson, 2002). During development, neurons extend processes that have at their extending tips highly motile structures called growth cones. Receptors expressed on growth cones recognize multiple cues present in the surrounding extracellular environment and manifest their response through the reorganization of neuronal cytoskeletal components, including actin and microtubules (Dent et al., 2011). Though molecular mechanisms that signal cytoskeletal remodeling have been uncovered for certain classes of guidance cue receptors (Bashaw and Klein, 2010; Kolodkin and Tessier-Lavigne, 2011), we are only just beginning to understand how these signaling pathways are integrated in order to allow for discreet steering of neuronal processes; for many guidance cue receptors little is known about the in vivo signaling events they initiate following ligand engagement.
One major class of extracellular guidance cues is the semaphorin protein family, members of which carry out evolutionarily conserved guidance functions during nervous system development through interactions with receptors that include plexins and various other receptors and co-receptors (Mann et al., 2007). A distinctive feature of these proteins is a conserved semaphorin (Sema) domain and a short plexin-semaphorin-integrin (PSI) domain in their extracellular regions; both of these domains are involved in semaphorin homo-multimerization, which is required for the formation of a ligand-receptor signaling complex (Janssen et al., 2010; Liu et al., 2010; Nogi et al., 2010). Both secreted and transmembrane semaphorins function as ligands to mediate a range of repulsive and attractive guidance functions, however, membrane-bound semaphorins can also mediate bidirectional signaling. For example, the transmembrane semaphorin Sema-1a regulates axon-axon repulsion in Drosophila through binding to the plexin A (PlexA) receptor during embryonic development (Winberg et al., 1998; Yu et al., 1998). This canonical “forward signaling” allows semaphorins to act as ligands to activate plexin receptors. More recent work shows that Sema-1a can also participate in “reverse signaling”, reminiscent of the well-characterized signaling events involving ephrin-reverse signaling (Egea and Klein, 2007). Sema-1a reverse signaling in Drosophila can control neuronal process targeting and synapse formation utilizing PlexA, or unknown ligands, to activate its receptor functions (Cafferty et al., 2006; Godenschwege et al., 2002; Komiyama et al., 2007; Yu et al., 2010). Interestingly, the vertebrate class 6 semaphorin Sema6D regulates cardiac morphogenesis through both forward and reverse signaling (Toyofuku et al., 2004). These observations raise questions relating to how forward and reverse transmembrane semaphorin signaling are coordinated during neural development and also, importantly, how the Sema-1a intracellular domain (ICD) transduces Sema-1a reverse signaling.
The Rho family of small GTPases, in combination with their direct regulators (RhoGEFs and RhoGAPs), plays key roles in growth cone steering by mediating localized changes in the actin cytoskeleton (Bashaw and Klein, 2010; Dickson, 2001; Hall and Lalli, 2010; Luo, 2000). Rho GTPases are activated by guanine nucleotide exchange factors (GEFs) that facilitate the exchange of bound GDP with GTP, and they are inactivated by GTPase activating proteins (GAPs) that mediate dephosphorylation of bound GTP to produce GDP. The cycling of Rho GTPases between active and inactive states can, therefore, be regulated by antagonistic relationships between RhoGEFs and RhoGAPs. The activation of Rho GTPases can be regulated spatially through the control of RhoGEF and RhoGAP subcellular localization, and this is influenced by activation of guidance cue receptors that in turn associate directly with GEFs or GAPs (Bashaw and Klein, 2010; Symons and Settleman, 2000). Extracellular cues can also activate signaling pathways that modulate GEF or GAP activity, including phosphorylation or protein-protein interactions that relieve auto-inhibitory intramolecular interactions (Schmidt and Hall, 2002; Shen and Cowan, 2010). Finally, Rho proteins and their regulators have been implicated in mediating repulsive guidance signaling (Derijck et al., 2010; Govek et al., 2005; Hall and Lalli, 2010).
Links between Rho GTPase signaling and Sema–plexin–mediated guidance prompted us to examine interactions between Drosophila RhoGEFs, RhoGAPs, and receptor-type guidance molecules. We identified pebble (Pbl), a RhoGEF for Rho1, and RhoGAPp190 (p190), a RhoGAP for Rho1, as signaling molecules with the potential to function downstream of Sema-1a reverse signaling in neurons. Our genetic analyses suggest that Pbl and p190 play key opposing roles in Sema-1a reverse signaling.
RESULTS
Pebble and RhoGAPp190 Bind to Semaphorin-1a
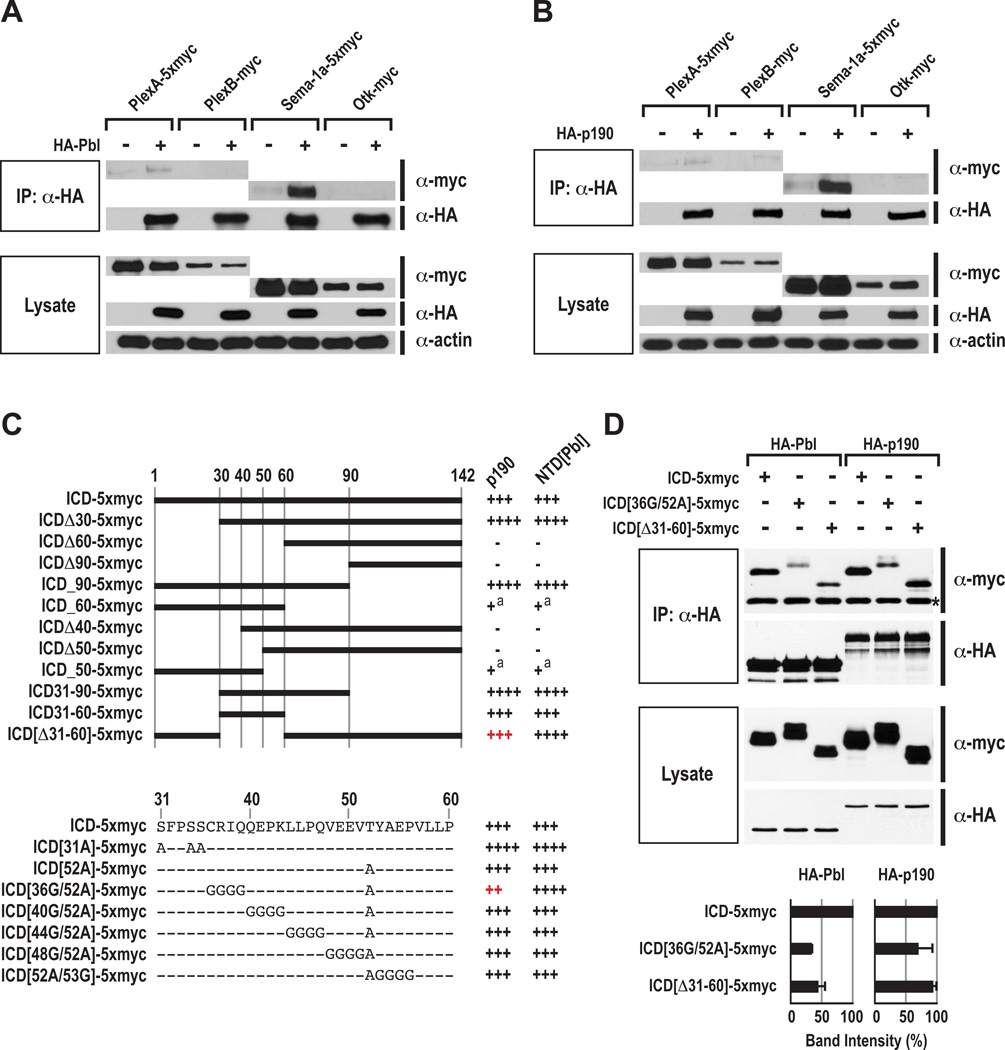
To investigate links between Rho GTPase regulators and semaphorin/plexin–mediated neuronal guidance, we screened several RhoGEF and RhoGAP proteins for their ability to interact with Drosophila PlexA, PlexB, and Sema-1a in Drosophila S2R+ cells in vitro. We found that Pbl weakly interacts with PlexA, while p190 weakly interacts with both PlexA and PlexB (Figures 1A and 1B). However, when we performed these same protein interaction assays using the PlexA ligand Sema-1a, we found that both Pbl and p190 proteins robustly interact with Sema-1a, to a much greater degree than with either PlexA or PlexB (Figures 1A and 1B). These strong interactions are apparently specific since other transmembrane proteins, including Drosophila Off-track (Otk), do not coimmunoprecipitate with either Pbl or p190 when coexpressed in S2R+ cells in these same experiments (Figures 1A and 1B). We also observed in coimmunoprecipitation (Co-IP) experiments that neuronally expressed embryonic HA-Pbl and HA-p190 robustly bind to endogenous Sema-1a in vivo (Figure S1A). These observations suggest that Pbl and p190 participate in intracellular signaling cascades downstream of Sema-1a (Figure 2E).
Figure 1. Pebble and RhoGAPp190 Physically Interact with Semaphorin-1a in S2R+ Cells.
(A) Interactions between Pbl and PlexA, PlexB, Sema-1a, and Otk in Drosophila S2R+ cells. Pbl and p190 were HA-tagged N-terminally, and PlexA, PlexB, Sema-1a and Otk were myc-tagged C-terminally. Pbl was cotransfected with or without PlexA, PlexB, Sema-1a, or Otk. Cell lysates were immunoprecipitated with anti-HA antibody and then blotted with anti-HA or anti-myc antibodies. Input lysates were also blotted with anti-myc, anti-HA, and anti-actin antibodies in the bottom.
(B) Interaction between p190 and PlexA, PlexB, Sema-1a, and Otk in Drosophila S2R+ cells. Co-IP experiments were performed as in Figure 1B.
(C) Schematic diagram of the Sema-1a ICD constructs used for protein interaction assays with p190 and with the N-terminal domain of Pbl (NTD[Pbl]) (right, a summary of their relative binding abilities). The intracellular domain is 142 amino acids long and numbered at the top. In the right column, the symbols “+” or “−” represent the presence or absence of protein interaction, respectively. The number of + symbols is correlated with relative binding levels. Superscript “a” indicates very weak interactions. At the bottom, amino acids and their mutated sequences within ICD residues 31–60 used for protein interaction assays with p190 and NTD[Pbl] are shown.
(D) Binding of wild-type and mutant Sema-1a ICD proteins to full-length Pbl and p190. Asterisk indicates non-specific IgG bands. Relative binding of Pbl and also p190 to Sema-1a ICD derivatives was measured by averaging band intensities from two experiments, as shown at the bottom. In each column, coimmunoprecipitated wild-type ICD-5xmyc was used as a control (equal to 100% in band intensity). Error bars indicate standard deviation.
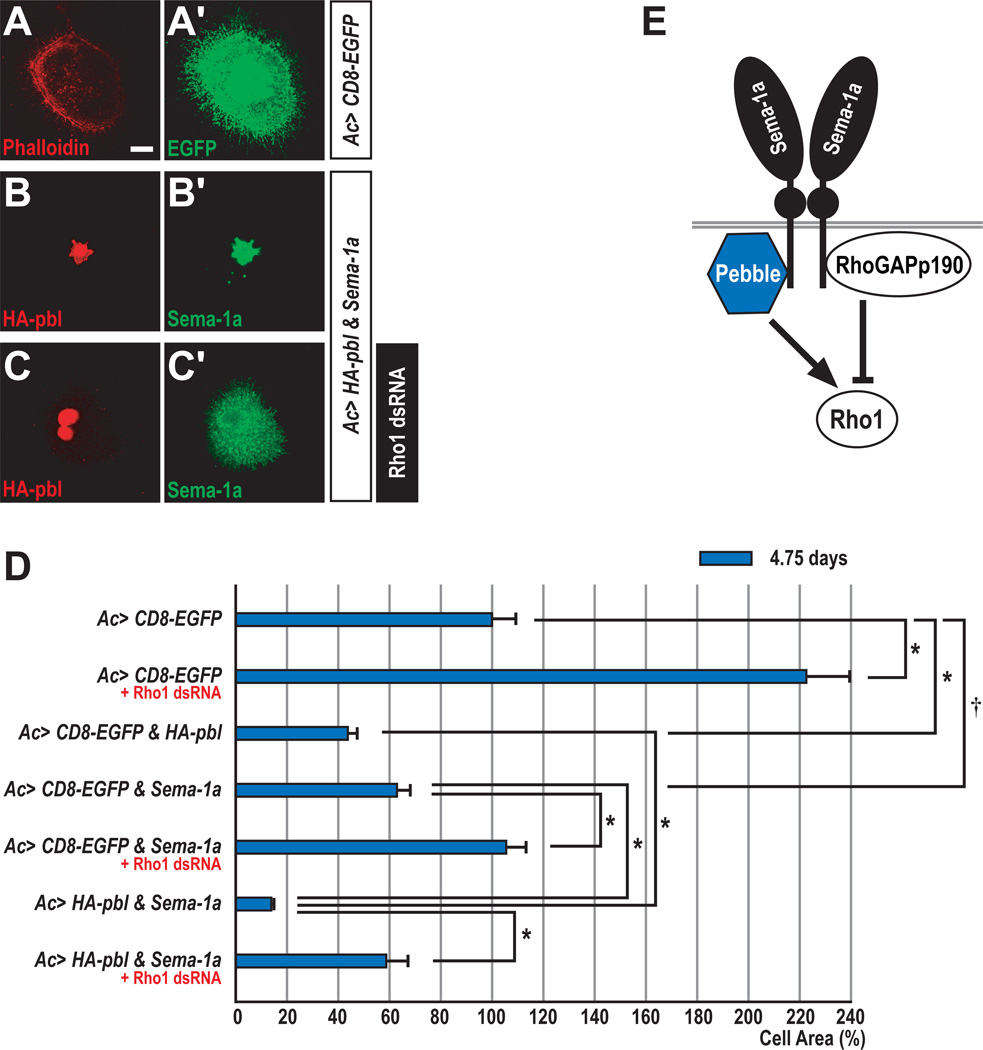
Figure 2. Collaboration between Pebble and Semaphorin-1a in Drosophila Cells.
(A–C) When cultured on concanavalin A-coated coverslips, most ML-DmBG2 cells become flattened. Overall cellular morphology was visualized with anti-GFP or anti-Sema-1a.
(A–A’) ML-DmBG2 cells transfected with membrane-tethered EGFP (CD8-EGFP) were stained with rhodamine-phalloidin (red) and anti-GFP (green).
(B–B’) ML-DmBG2 cells cotransfected with HA-pbl and Sema-1a were stained with anti-HA (red) and anti-Sema-1a (green). Ectopically expressed HA-Pbl protein was predominantly localized to the nucleus.
(C–C’) ML-DmBG2 cells cotransfected with HA-pbl and Sema-1a were treated with Rho1 dsRNA for 4.75 days and then stained with anti-HA (red) and anti-Sema-1a (green). Cells with multiple nuclei were frequently observed following Rho1 dsRNA treatment.
(D) Quantification of Sema-1a, pbl and Rho1 effects on cell size. CD8-EGFP-expressing cells were used as a control (100% cell area). Error bars indicate S.E.M. by t-test (*p<0.0001 and †p=0.001).
(E) Proposed interactions between Pbl and p190 with Sema-1a, highlighting potential antagonistic regulation of the small GTPase Rho1.
Scale bar, 10 µm (A–C).
Mutations in the Sema-1a Intracellular Domain (ICD) Reduce Interactions with Pebble and with RhoGAPp190
To further characterize the specificity of these interactions between Sema-1a and Pbl, we mapped the regions of Pbl responsible for interactions with Sema-1a, revealing that the N-terminal domain (NTD), which encompasses two tandem BRCT (BRCA1 C-terminal) domains, is necessary and sufficient for mediating Sema-1a binding (Figure S1B). Through a systematic deletion and mutagenesis analysis of the Sema-1a ICD, we found that Sema-1a ICD[Δ31–60], in which ICD amino acid residues 31–60 are deleted, and ICD[36G/52A] exhibited differential binding properties to full-length p190 and truncated NTD[Pbl] (Figure 1C). To address whether this difference is due to the absence of the Pbl C-terminal domain (CTD), we next tested the ability of full-length Pbl and p190 to bind to these mutant forms of the Sema-1a ICD; we observed a significant reduction in Pbl binding to both ICD[Δ31–60] and ICD[36G/52A] (Figure 1D). In addition, the latter mutation also reduced the binding of full-length p190 to the Sema-1a ICD, but to a lesser extent than we observed for Pbl (Figure 1D). These mutations appear to result in reduced access by the Sema-1a ICD to the NTD region of overexpressed full-length Pbl since they lead to increased interaction with the NTD alone (Figure 1C). Therefore, multiple mutations in the Sema-1a ICD are required to affect interactions with both Pbl and p190, defining Sema-1a ICD regions likely to participate in the association of these signaling proteins with Sema-1a.
To address whether Pbl and p190 compete for binding to Sema-1a, we performed competition binding assays in S2R+ cells. Increasing relative levels of Pbl decreased the binding of p190 to Sema-1a by ~50%, whereas increasing relative levels of p190 did not alter Pbl binding to Sema-1a (Figure S1C). These competition data are consistent with our finding that Pbl and p190 both bind to the same Sema-1a domain (Figure 1C), and they suggest that the Pbl association with Sema-1a predominates over p190 association with this same Sema-1a ICD region.
Pebble and Sema-1a Collaborate to Regulate Cell Size Through Rho1 in Drosophila Neuronal Cells In Vitro
In Drosophila S2 cells, RNAi–mediated depletion of Rho1 induces a dramatic increase in cell size (Rogers and Rogers, 2008). Recent analysis of photoreceptor axon guidance in Drosophila shows that Rho1 is involved in Sema-1a reverse signaling (Yu et al., 2010). Therefore, we utilized a Drosophila cell line with neuronal characteristics, called ML-DmBG2-c2 (Ui et al., 1994), to examine links between Sema-1a signaling and Rho1. We depleted endogenous Rho1 in these cells using double-stranded RNA (dsRNA) for 4.75 days and observed a ~2-fold increase in cell size (Figure 2D). In contrast, overexpression of Pbl, which positively regulates Rho1, caused a ~2-fold reduction in cell size. Interestingly, ectopic expression of Sema-1a in these cells also led to a significant reduction in cell size, suggesting a link between Sema-1a signaling and Rho1 activity through the action of Pbl. This idea is supported by the observation that Sema-1a–induced reduction in cell size was suppressed by depleting Rho1 (Figure 2D). To further explore Sema-1a–mediated modulation of Rho1 activity through Pbl, we coexpressed Sema-1a and Pbl and observed a dramatic reduction in cell size (Figures 2B and 2D); these Pbl gain-of-function (GOF)–mediated reductions in cell size were not the result of affecting cytokinesis, a known pbl function (Prokopenko et al., 1999) (data not shown). This synergistic Sema-1a–Pbl reduction in cell size was also significantly attenuated by Rho1 depletion (Figures 2C and 2D). These results suggest that Sema-1a and Pbl collaborate to trigger cell size reduction through the activation of Rho1 in Drosophila neuronal cells in vitro (Figure 2E).
We also asked whether or not an antagonistic relationship exists between Sema-1a and p190 in these cells. As expected, overexpression of p190 resulted in an increase in cell size, and the average size of cells coexpressing both p190 and Sema-1a is significantly reduced as compared to cells expressing p190 alone (Figure S2). Therefore, Sema-1a overexpression is epistatic to p190 overexpression with respect to cell size in vitro.
pebble Regulates Motor Axon Defasciculation and Target Recognition
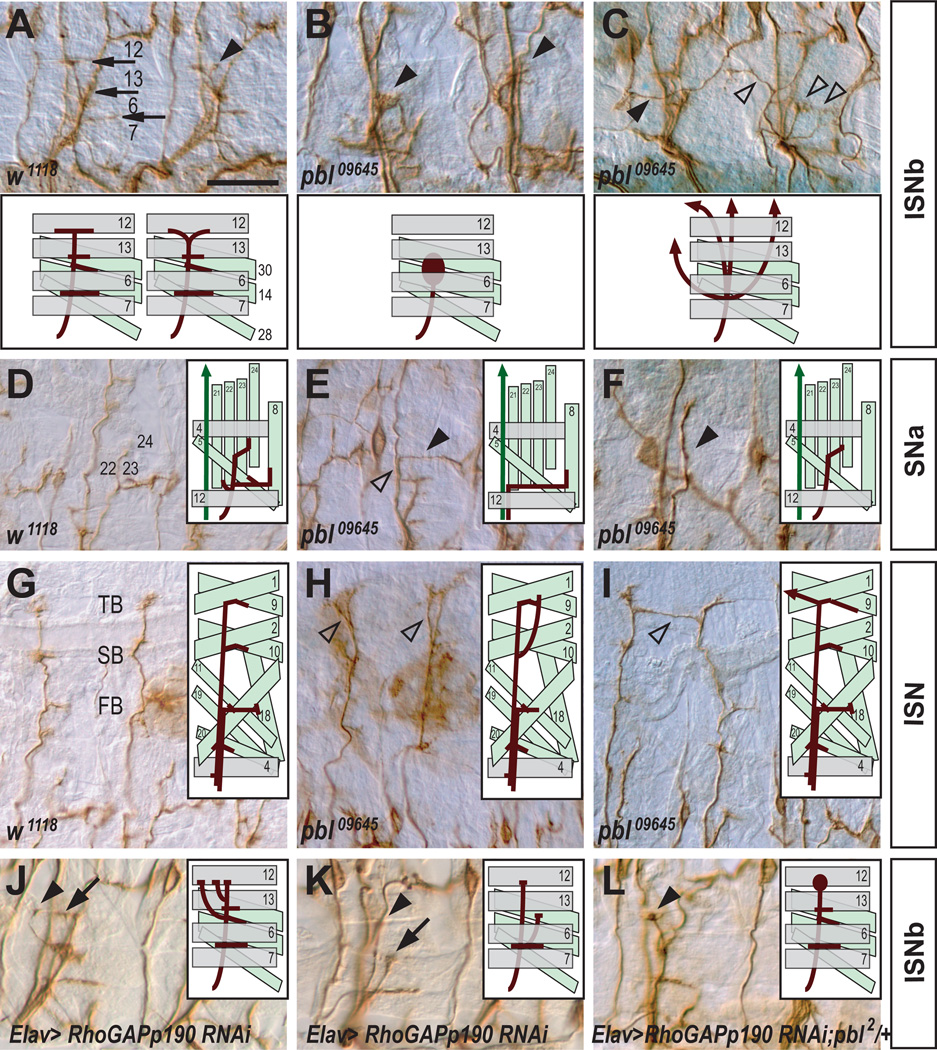
To determine whether pbl plays a role in axon pathfinding, we examined motor axons in hypomorphic pbl alleles, referred to here as pbl09645 and pblKG07669, that have P element insertions in the 5’-untranslated region of pbl (Figure S3A) (Bellen et al., 2004; Prokopenko et al., 2000). Embryos homozygous for these hypomorphic pbl alleles show highly penetrant peripheral nervous system (PNS) axon guidance defects (Figures 3A–3I and 4A). In wild-type embryos, ISNb axons first defasciculate from the ISN near the lateral margins of the CNS and extend to the ventrolateral muscle field (Keshishian et al., 1996). Subsequently, ISNb axons defasciculate from one another and establish presynaptic arborizations between muscles 7 and 6, and at the proximal edges of muscles 13 and 12 (arrows in Figure 3A). ISNb axons in pbl09645 homozygous mutant embryos show highly penetrant guidance defects (98% of mutant hemisegments; Figure 4A). In pbl09645 homozygous mutants, ISNb axons often fail to defasciculate from one another, resulting in a hyperfasciculated phenotype and a failure to reach their muscle targets (Figure 3B). In addition, we frequently observed in pbl mutant embryos that ISNb axons fail to either navigate along their normal trajectories or innervate their normal target muscles, even though these motor axon growth cones do reach the vicinity of their target regions (an apparent target recognition error; Figures 3B and 3C). These fasciculation and target recognition errors are not seen in wild-type embryos (Figure 3A).
Figure 3. pebble and RhoGAPp190 are Required for Motor Axon Defasciculation and Target Recognition.
(A–L) Filleted preparations of late stage 16 embryos stained with anti-FasII (the MAb 1D4) to visualize motor axon projection patterns. Anterior is left and dorsal is up. Schematic diagrams showing motor axon guidance in wild type and indicated mutant backgrounds are presented in each panel. Scale bar, 15 µm (A–C and J–L); 20 µm (D–I).
(A) In wild-type embryos, ISNb axons undergo sequential defasciculation when they arrive between muscles 7 and 6, at the proximal edge of muscle 13, and at muscle 12 to innervate appropriate muscle targets (arrows). Mild premature branching is occasionally observed (arrowhead).
(B) In pbl09645 mutants, ISNb axons fail to defasciculate from one another and the ISNb displays an abnormally thick morphology (closed arrowheads).
(C) ISNb axons in pbl09645 mutants follow aberrant pathways and fasciculate ectopically with adjacent axons (open arrowheads), including the transverse nerve (TN) (closed arrowhead).
(D) In wild-type embryos, SNa axons defasciculate and give rise to two branches. The dorsal branch projects to innervate muscles 22–24 with two characteristic turns, while the lateral branch extends posteriorally and innervates its target muscles.
(E) In pbl09645 mutants, SNa axons follow the ISN trajectory (detour phenotype, open arrowhead) and the dorsal branch of the SNa is often missing (closed arrowhead).
(F) In pbl09645 mutants, lateral SNa branches are missing (closed arrowhead).
(G) In wild-type embryos, ISN axons project to the dorsal-most muscle fields and elaborate three distinct branches: first (FB), second (SB), and third (TB).
(H) In pbl09645 mutants, ISN axons defasciculate prematurely (open arrowheads).
(I) In pbl09645 mutants, ISN axons often cross the segment boundary and ectopically contact the adjacent ISN (open arrowhead).
(J) RNAi–mediated knockdown of p190 causes premature defasciculation (arrow) and probably a defect in muscle target recognition (arrowhead) since two ISNb axons are thought to target the proximal edge of muscle 12.
(K) Premature defasciculation rarely initiates before ISNb axons reach muscle 6 (arrow); an innervation defect is also shown (arrowhead).
(L) RNAi–mediated knockdown of p190 in a heterozygous pbl2 background leads to a characteristic defasciculation defect at the edge of muscle 12 (arrowhead).
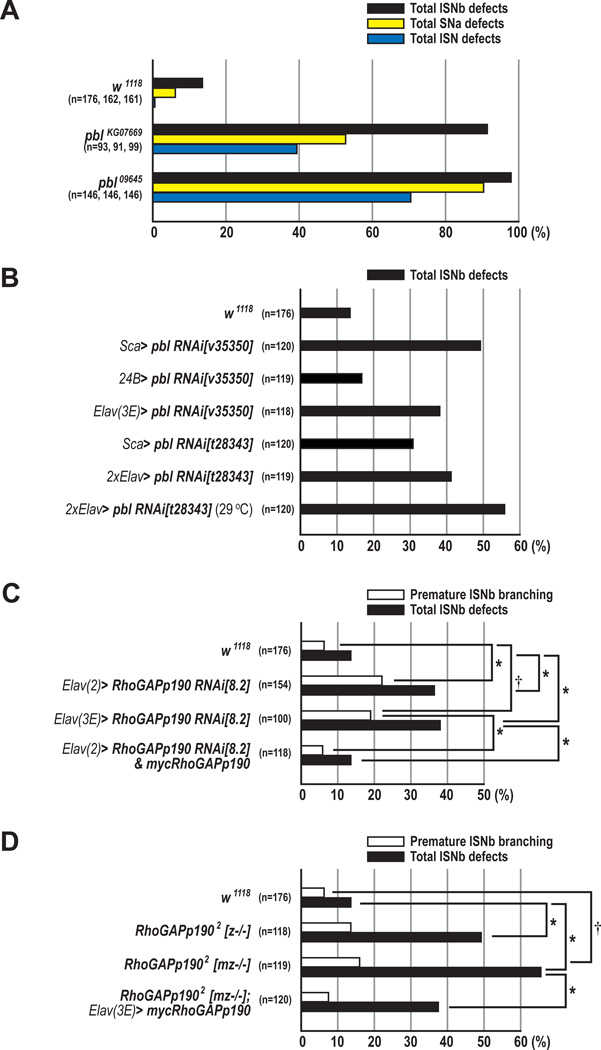
Figure 4. Neuronal pebble and RhoGAPp190 are Required for Motor Axon Guidance.
(A) The percentages of ISNb, SNa, and ISN guidance defects in w1118 (wild type), pblKG07669, and pbl09645 embryos.
(B) Total ISNb defects in embryos in which endogenous pbl is knocked down in either neuroblasts, post-mitotic neurons or all muscle cells using Sca-GAL4, Elav-GAL4, or 24B-GAL4 respectively.
(C) The percentages of total ISNb defects (closed bars) and premature ISNb branching phenotypes (open bars) of the ISNb in w1118, p190 knockdown, and rescue embryos (*p<0.001 and †p<0.01, chi-square test).
(D) The percentages of total ISNb defects (closed bars) and premature ISNb branching phenotypes (open bars) in w1118, p1902, and rescue embryos. z−/− and mz−/− indicate zygotic null, and maternal/zygotic null for p190, respectively (*p<0.001 and †p=0.012, chi-square test).
(A–D) n=the number of abdominal hemisegments scored for each genotype.
Most axons in the segmental nerve a (SNa) pathway also exhibited severe defasciculation defects and/or target recognition failure in pbl09645 homozygous mutant embryos (90% of hemisegments; Figures 3E, 3F and 4A). In wild-type embryos, SNa axons separate from the SN nerve and project to the dorsolateral muscle field (Langraf et al., 1997; Van Vactor et al., 1993). Subsequent defasciculation of SNa axons gives rise to a dorsal and lateral branch. The dorsal branch establishes synaptic arborizations between muscles 21, 22, 23, and 24, while the lateral branch innervates muscle 5 and 8 (Figure 3D). In pbl09645 mutants, the dorsal or lateral SNa branches were often missing (Figures 3E and 3F). These SNa phenotypes are not observed in wild-type embryos (Figure 3D).
Wild-type ISN axons navigate to the dorsal-most muscle field and form three distinctive branches: the first (FB), second (SB), and third branch (TB) (Figure 3G). The ISN axons in pbl09645 homozygous mutant embryos exhibit a failure of correct muscle target recognition. The first or second branches of the mutant ISN motor axons often extend dorsally beyond the correct muscle fields (Figure 3H). Occasionally, mutant ISN axons cross the segment boundary and fasciculate with adjacent ISN axons (Figure 3I). To confirm that these defects in motor axon guidance result from the loss of pbl function, we characterized homozygous mutant embryos from a second P element insertion line called pblKG07669 (Figure S3A). These embryos also displayed highly penetrant axon guidance defects in the PNS, although they were somewhat less severe than those observed in pbl09645 mutants (Figure 4A). Furthermore, we found that embryos transheterozygous for pbl09645 and pblKG07669 showed similar penetrance of these defects but less severity compared to pbl09645 mutants (data not shown). These genetic data show that pbl plays an important role in establishing normal neuromuscular connectivity during embryogenesis.
In addition to motor axon pathfinding defects in the PNS, pbl09645 homozygous mutants showed 2.6% total CNS defects, pblKG07669 homozygous mutants displayed 0.0% total CNS defects, and mutants transheterozygous for pbl09645 and pblKG07669 showed 0.93% total CNS defects (data not shown). These findings indicate that pbl contributes very little to CNS axon guidance.
Neuronal pebble is Required for Motor Axon Pathfinding
To assess whether or not the defects we observe in motor axon pathfinding result from the loss of pbl function in muscles or in neurons, we expressed a HA-pbl transgene in all muscles of pbl09645 mutant embryos using the 24B-GAL4 driver (Luo et al., 1994). These embryos showed motor axon pathfinding defects similar to those seen in pbl09645 mutants (Figures S3F and S3G). In contrast, expression of HA-pbl in neurons using Sca-GAL4, which is expressed in all neuroblasts and their progeny (Klaes et al., 1994), led to partial but significant rescue of pbl LOF phenotypes in both ISNb and SNa, but not ISN, motor neuron pathways (Figures S3E and S3G). For example, in pbl09645 mutant embryos, ISNb axons frequently failed to innervate muscles 6/7 (81.5% of hemisegments; Figure S3G), whereas neuronal expression of HA-pbl in pbl09645 mutant embryos strongly suppressed these innervation defects (34.1% of hemisegments; Figures S3E and S3G). We find that the pblKG07669 allele, which exhibits highly penetrant axon pathfinding defects in the PNS (Figure 4A), does not alter cell fate specification or proliferation of CNS neurons compared to wild-type embryos (Figures S4G and S4H, and data not shown). Furthermore, in pblKG07669 homozygous mutants, the embryonic pattern and morphology of muscles 21–24 and ventrolateral muscles is apparently normal with respect to muscle fiber size, shape, and position (Figures S4B and S4E). These data suggest that neuronal, but not muscle, functions of pbl contribute to correct ISNb and SNa motor axon guidance.
To further address whether or not neuronal Pbl is required for motor axon pathfinding, we used a transgenic RNA interference (RNAi) approach. When the pbl gene was knocked down by overexpression of pbl RNAi[v35350] or pbl RNAi[t28343] (Supplementary Experimental Procedures, and Figure S5) transgenes using Sca-GAL4, projection patterns of ISNb axons showed increased fasciculation and defects in target recognition (49.2% or 30.8% of hemisegments, respectively; Figure 4B). Knockdown of pbl in all muscles using 24B-GAL4 resulted in no significant ISNb pathfinding defects (Figure 4B). To address whether pbl axon guidance and cytokinesis functions are separable, we knocked down pbl gene function using the post-mitotic driver Elav-GAL4. Embryos overexpressing pbl RNAi[v35350] under the control of Elav-GAL4 exhibited ISNb defects in 38% of hemisegments (Figure 4B). A similar phenotype was observed with the t28343 RNAi line under the control of two copies of Elav-GAL4. Since the GAL4/UAS system is temperature-sensitive, we allowed these embryos to develop at 29°C to increase GAL4–mediated expression of pbl RNAi and observed an increase in the penetrance of motor axon pathfinding defects as compared to 25°C (55.8% vs. 41.2%; Figure 4B and Figures S3C–S3D). These data strongly suggest that neuronal Pbl is required post-mitotically for normal motor axon pathfinding.
Neuronal RhoGAPp190 is also Required for Motor Axon Pathfinding
Since we observed that p190, like Pbl, also exhibits a strong physical association with Sema-1a and that two potential p190 enhancer GAL4 lines drive reporter expression in the CNS (Figures S3H–S3J), we examined the role played by p190 in motor axon pathfinding using transgenic RNAi lines (Billuart et al., 2001). Overexpression of the p190 RNAi transgene using Elav-GAL4 resulted in premature defasciculation of ISNb axons prior to reaching muscle 13, and sometimes muscle 6: reflecting either increased defasciculation or a defect in muscle target recognition (~20% of hemisegments; Figures 3J, 3K, 4C, and S6). This premature branching phenotype was rescued to wild-type levels when one copy of a UAS-mycp190 transgene (Billuart et al., 2001) was introduced along with p190 RNAi (5.9% of hemisegments; Figure 4C). Furthermore, when premature branching is observed in wild-type embryos it is qualitatively distinct from what we observe following p190 LOF, often occurring between the ventral and dorsal surfaces of muscle 13 rather than prior to ISNb arrival at muscle 13 (compare arrowhead in Figure 3A to arrows in Figures 3J and 3K). In addition, premature ISNb branching phenotypes qualitatively and quantitatively similar to those we observe in p190 RNAi lines were noted in p1902 maternally and zygotically-derived null alleles, and total ISNb defects were significantly rescued by reintroduction of the neuronal mycp190 transgene (Figure 4D). These results show that neuronal p190 is required post-mitotically for motor axon pathfinding.
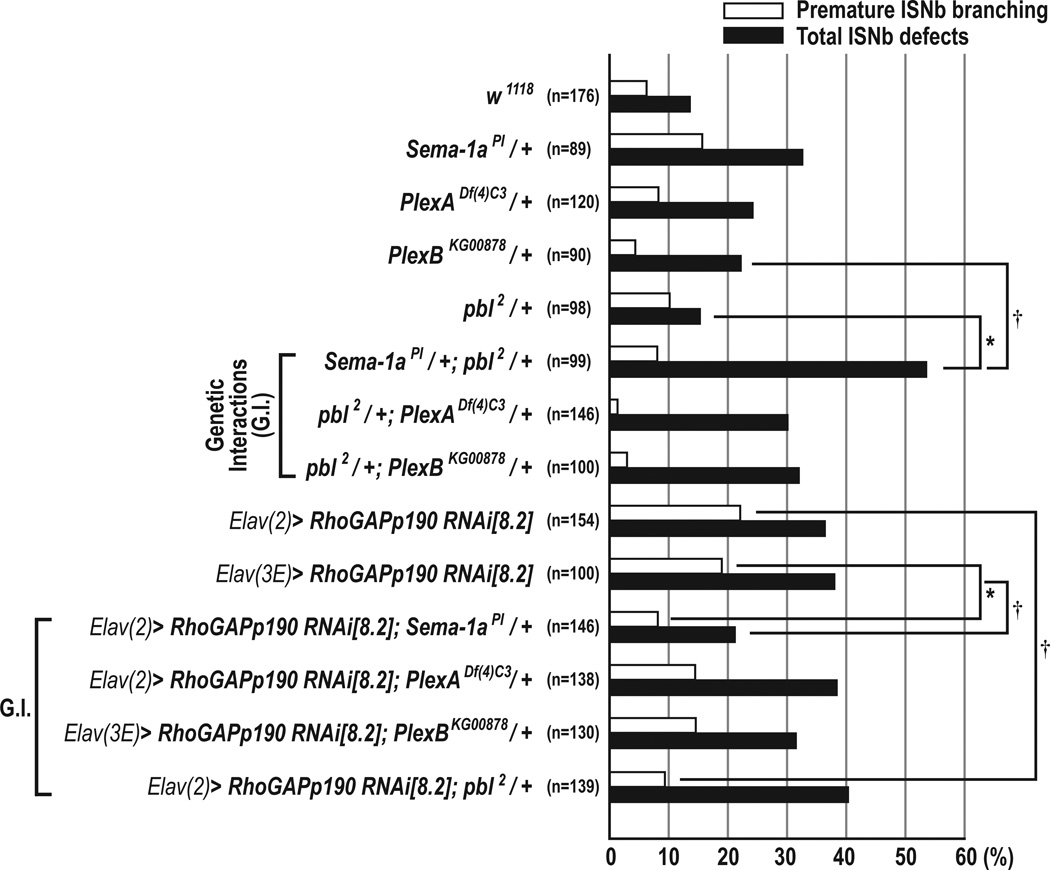
Semaphorin-1a–mediated Axon Guidance is Regulated Positively by pebble and Negatively by RhoGAPp190
To test whether pbl plays a role in Sema-1a–mediated motor axon guidance, we investigated genetic interactions between pbl and Sema-1a, PlexA, and PlexB. When either a PlexA or PlexB null allele was introduced into pbl2 heterozygotes, total ISNb and premature branching defects were not significantly affected. However, when a single Sema-1a mutant allele was introduced into the pbl heterozygous background, we observed a significant increase in the frequency of total ISNb defects (53.5%) compared to embryos heterozygous for either Sema-1a or pbl, though premature branching was apparently unaffected (Figure 5). Similar patterns of genetic interactions were also observed with pbl3, and pbl5 null alleles (Figure S6). The amino acid Val531, which is changed to aspartate in the pbl5 mutant (V531D), is located within the Pbl DH domain and is known to be required for nucleotide exchange activity in the DH domains of other GEFs (Liu et al., 1998; Prokopenko et al., 1999). These results suggest that Sema-1a and Pbl act together in the same signaling pathway to promote motor axon guidance through the regulation of Pbl GEF activity and, since similar genetic interactions are not observed with PlexA, that Pbl functions as an intracellular mediator downstream of Sema-1a rather than PlexA. However, heterozygosity for pblKG07669 significantly enhances the Sema-1a null phenotype (Figure S6), indicating that additional Pbl guidance functions cooperate with Sema-1a signaling.
Figure 5. Genetic Interactions among Sema-1a, pebble, and RhoGAPp190 Mutants.
Premature ISNb branching phenotypes (open bars) and total ISNb defects (closed bars) in embryos with indicated genotypes (*p<0.001 and †p<0.01, chi-square test). Total ISNb defects in embryos transheterozygous for Sema-1a and pbl are greater than those observed in either Sema-1a or pbl heterozygous embryos. Removing one copy of Sema-1a significantly suppresses p190 knockdown–mediated premature ISNb branching phenotypes and also total ISNb defects. Removing one copy of pbl also leads to significant suppression of p190 knockdown–mediated premature ISNb branching phenotypes. n=the number of abdominal hemisegments scored for each genotype.
Since p190 physically associates with Sema-1a, we next asked whether or not p190 is involved in Sema-1a–mediated motor axon guidance. First, we observed that the p190 RNAi phenotype was suppressed by loss of a single copy of Sema-1a: from 22.1% to 8.2% for premature ISNb branching, and from 36.4% to 21.2% for total ISNb defects (Figure 5). In contrast, PlexA or PlexB null alleles did not affect the p190 RNAi phenotype. These suppressive genetic interactions between p190 and Sema-1a were also observed using a different p190 RNAi line and the p1902 null allele (Figure S6). Therefore, p190 functions to antagonize Sema-1a signaling, but not PlexA or PlexB signaling. This is consistent with physical association between p190 and Sema-1a being stronger than with either PlexA or PlexB (Figure 1B). Taken together, these genetic interaction data suggest that Pbl and p190 exert opposing control over Sema-1a–mediated motor axon.
Next, we examined whether pbl and p190 genetically interact. When a single pbl2 mutant allele was introduced to p190 RNAi knockdown embryos, the premature branching phenotype was significantly reduced, from 22.1% to 9.4% (Figure 5). In these embryos, we also observed a significant increase in motor axon defasciculation defects (excluding premature branching phenotypes) at the last ISNb choice point (Figures 3L and 5). To test whether increasing Pbl levels affects premature ISNb branching phenotypes, we overexpressed HA-pbl alone or with coexpression of p190 RNAi in neurons. Increasing Pbl leads to a significant increase in premature branching phenotypes, but does not affect the p190 RNAi phenotype (Figrue S7D). These data suggest that premature ISNb branching is largely controlled by antagonistic functions of p190 and pbl.
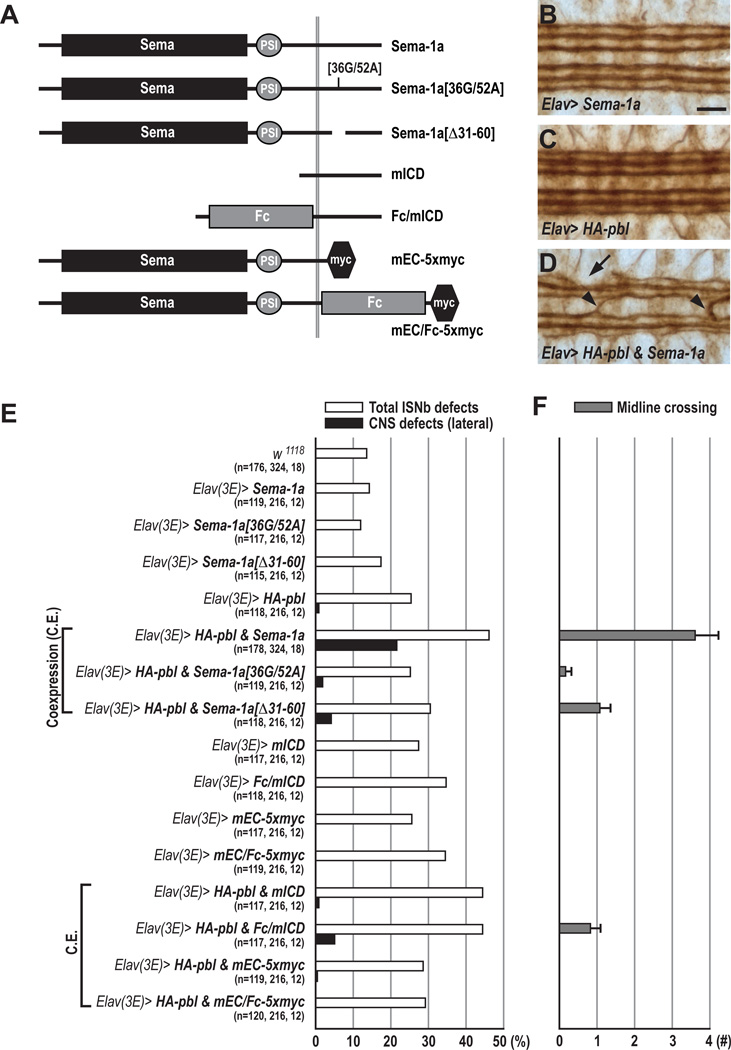
The Sema-1a ICD is Required for Pebble–mediated PNS and CNS Axon Guidance
We find that Sema-1a and Pbl collaborate to induce cell contraction in vitro (Figure 2). To test whether they also collaborate in vivo, the two ICD mutations 36G/52A and Δ31–60, which show a 33% and 44% reduction in binding to Pbl, respectively (Figure 1D), were introduced into the wild-type Sema-1a protein. We then overexpressed HA-pbl and modified Sema-1a transgenes using the post-mitotic driver Elav-GAL4. When both Sema-1a and pbl transgenes were coexpressed in neurons, ISNb defects increased from 25.4% to 46.1%; interestingly, CNS defects in the lateral-most FasII+ longitudinal connective increased ~20 fold, from 0.9% to 21.6%, when compared to overexpression of HA-pbl alone (Figures 6B–6E). In embryos expressing both Sema-1a and Pbl in post-mitotic neurons, we also observed a dramatic increase in ectopic CNS midline crossing: from 0.0 to 3.6 crossings per embryo (Figures 6D and 6F). These synergistic effects were not observed in embryos coexpressing HA-Pbl and PlexA, suggesting they result from specific signaling interactions between Pbl and Sema-1a (Figure 7B). A truncated form of Sema-1a (mEC-5xmyc, Figure 6A), which lacks the entire ICD, did not exhibit any synergistic interactions with HA-Pbl (Figure 6E), commensurate with our observations that the ICD binds to Pbl (Figure 1D). Next, we examined two Sema-1a mutant transgenes harboring the mutations 36G/52A and Δ31–60. When these altered Sema-1a proteins were coexpressed with HA-pbl, total ISNb and CNS defects were not significantly increased above HA-pbl overexpression alone (Figures 6E and 6F). Coexpression of Sema-1a[Δ31–60] with HA-pbl did cause a modest increase in lateral CNS defects (4.2%) and midline crossing phenotypes (1.1 per animal); these defects are far less robust than those observed with coexpression of wild-type Sema-1a and Pbl, and they are consistent with our observation that the Pbl NTD is able to bind in vitro to ICD[Δ31–60] (Figures 6E, 6F, and 1C). In addition, the synergistic Sema-1a–Pbl–mediated increase in premature ISNb branching phenotypes in vivo, and also the reduction in cell size in vitro, was significantly attenuated when either ICD Sema-1a mutant was coexpressed with HA-Pbl (Figures S7D and S2). These data show that pbl and Sema-1a can collaborate in these GOF paradigms to affect axon guidance in vivo and cell size in vitro, and that this likely occurs through interactions between Pbl and the Sema-1a ICD.
Figure 6. Pebble and RhoGAPp190 Mediate Semaphorin-1a Reverse Signaling.
(A) Summary of Sema-1a constructs used to make transgenic lines for rescue and gain-of-function studies. Sema, Semaphorin domain; PSI, Plexin-Semaphorin-Integrin domain; Fc, Fragment Crystallizable region of human immunoglobulin G.
(B–D) Filleted preparations of late stage 16 embryos stained with anti-FasII to visualize CNS longitudinal connective projection patterns. Anterior is to the left.
(B) Neuronal overexpression of Sema-1a in a wild-type background does not affect the three major FasII+ longitudinal fascicles.
(C) A majority of embryos overexpressing HA-pbl in a wild-type background exhibits normal CNS longitudinal fascicle projection patterns.
(D) Coexpression of Sema-1a and HA-pbl in most post-mitotic neurons causes occasional disruptions in the outermost FasII+ fascicles (arrow) and frequent midline crossing defects (arrowheads).
(E) Total ISNb defects (open bars) and CNS lateral connective disruptions (closed bars) in embryos of different GOF genotypes. n=the number of hemisegments exhibiting total ISNb defects, lateral CNS defects, and the number of animals scored for midline crossing (Figure 5F).
(F) Quantification of the number of midline crossings per embryo (grey bars). Genotypes are same as in E.
Scale bar, 15 µm (B–D).
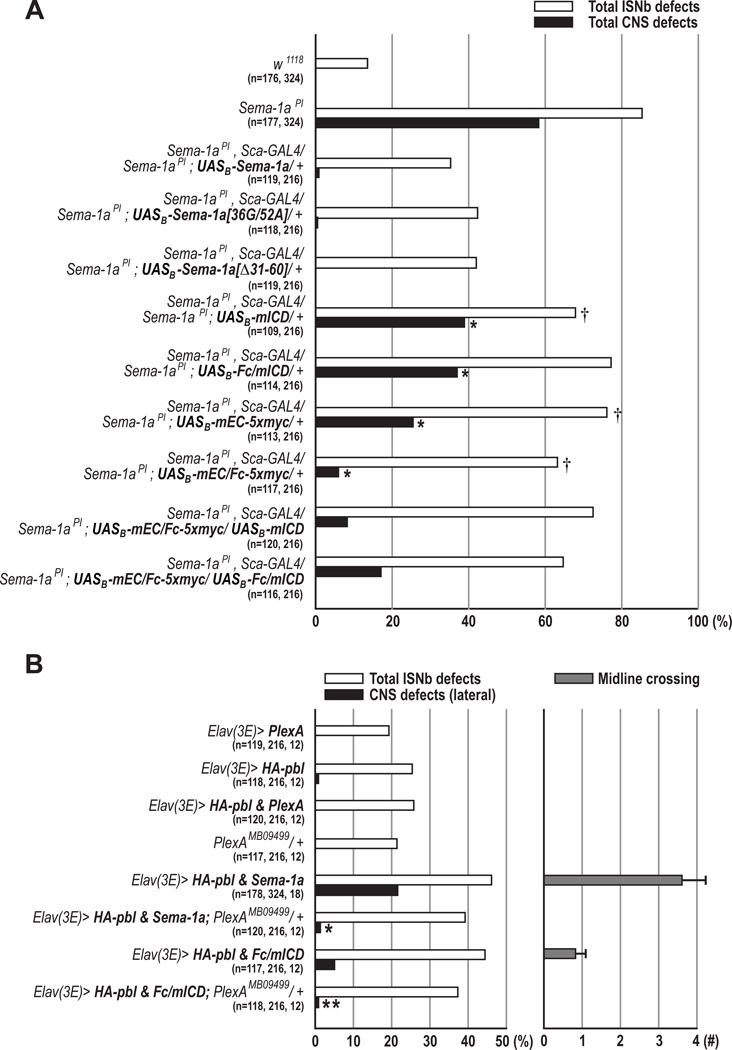
Figure 7. Both Forward and Reverse Sema-1a Signaling is Required for Correct Motor Axon Guidance in the PNS.
(A) Rescue of Sema-1a axon guidance phenotypes with various modified Sema-1a proteins. Embryos with indicated genotypes were scored for the frequency of total ISNb defects (open bars) and total CNS defects (closed bars) (*p<0.0001 and †p=0.05, chi-square test).
(B) Genetic interactions among PlexA, Sema-1a, and pbl. Total ISNb branching phenotypes (open bars), lateral longitudinal CNS defects (closed bars), and midline crossing phenotypes (grey bars) were scored in embryos of the indicated genotypes (*p=0.0001 and **p=0.0245; chi-square test).
(A–B) n=the number of hemisegments scored for total ISNb defects, lateral CNS defects, and the number of animals scored for midline crossing.
Semaphorin-1a Bidirectional Signaling is Required for Embryonic Motor Axon Pathfinding
The Sema-1aPI LOF allele (Yu et al., 1998) has the capacity to impair both forward and reverse signaling. However, it is not clear whether Sema-1a bidirectional signaling is required for PNS and/or CNS axon guidance in embryonic development. Therefore, we made a series of constructs that express truncated and chimeric Sema-1a proteins and then assessed these Sema-1a transgenes for their ability to rescue PNS and CNS guidance defects in homozygous Sema-1a mutants (Figure 6A). Sema-1a homozygous mutants show dramatically increased guidance defects in the ISNb and most lateral FasII+ CNS longitudinal axon pathways (Figures 7A, S3B, and S8C). Interestingly, neuronal expression of the mEC/Fc-5xmyc chimeric receptor, which lacks the entire ICD and thus reverse signaling activity, caused a dramatic reduction in CNS defects compared to the Sema-1a null phenotype (6.0% vs. 58.5%), while the mEC-5xmyc truncated protein led to an intermediate reduction (25.5%; Figure 7A). However, significant but modest rescue of CNS defects was observed in embryos carrying either mICD or Fc/mICD mutant Sema-1a proteins, which lack forward signaling activity (Figure 7A). These results demonstrate that multimerization of the Sema-1a extracellular domain is largely sufficient to mediate Sema-1a functions in CNS axon guidance. Furthermore, we found that neuronal expression of either of these signaling mutant transgenes partially rescued Sema-1a null ISNb pathway phenotypes (Figure 7A). In particular, neuronal expression of mEC/Fc-5xmyc, which should allow forward but not reverse signaling, resulted in only modest rescue of ISNb defects but significant rescue of CNS defects, suggesting an essential role for Sema-1a–mediated reverse signaling in peripheral axon guidance. Next, we performed additional rescue experiments in order to examine whether the introduction of both forward (mEC/Fc-5xmyc) and reverse (mICD or Fc/mICD) signaling mutant transgenes together can further rescue Sema-1a ISNb phenotypes. However, simultaneous expression of these transgenes did not lead to additional resuce of Sema-1a null PNS phenotypes, as compared to neuronal expression of either single transgene (Figure 7A). Given that Sema-1a functions as a ligand for PlexA in the PNS (forward signaling; Winberg et al., 1998), these complementation analyses strongly suggest that both the extracellular and intracellular Sema-1a domains, and therefore the coordinated action of bidirectional signaling, are necessary for Sema-1a–mediated motor axon guidance.
Pebble Mediates Semaphorin-1a Reverse Signaling
Since overexpression of wild-type Sema-1a synergistically enhances pbl misexpression phenotypes in both the PNS and CNS (Figures 6B–6F), we reasoned that if this synergistic enhancement occurs through the potentiation of reverse signaling, the Sema-1a intracellular domain alone should recapitulate the synergistic enhancement we see when wild-type Sema-1a is coexpressed with HA-pbl. We found in our GOF analysis that expression of mICD or Fc/mICD did indeed increase pbl GOF ISNb phenotypes to a similar level as wild-type Sema-1a, but produced only mild defects in CNS patterning (Figures 6E and 6F). However, overexpression of the Sema-1a extracellular domain (mEC-5xmyc or mEC/Fc-5xmyc) did not affect pbl GOF phenotypes in either ISNb or CNS axon guidance. These in vivo GOF data strongly suggest that Pbl mediates Sema-1a reverse signaling.
To address further Sema-1a receptor function and its regulation by Pbl, we performed additional GOF experiments utilizing apterous-GAL4 (apGAL4), which drives expression of GAL4 in only three neurons per hemisegment; these neurons extend axons longitudinally and do not cross the CNS midline (O’Keefe et al., 1998), and Sema-1a signaling mutants (Figure S7A). Coexpression of Sema-1a and HA-Pbl in the apterous neurons frequently leads to a failure of fasciculation among these apterous+ axons and results in a “midline crossing” phenotype (Figure S7B). Similar fasciculation defects in the apterous+ neurons are also found in embryos coexpressing Fc/mICD and HA-Pbl (Figure S7C). However, the axon fasciculation defects observed in embryos coexpressing mEC/Fc-5xmyc (Sema-1a ecto-domain) and HA-Pbl are not significantly different from those observed in apGAL4/+ animals (Figure S7C). These results provide additional support for Sema-1a functioning as a receptor that signals via Pbl.
To complement these genetic interaction data, we performed similar GOF studies in Drosophila neuronal cells in vitro. Consistent with our in vivo observations, we found that Pbl collaborates with Fc/mICD, but not with mEC/Fc-5xmyc, to induce cell size reduction (Figure S2). Taken together with our protein-protein interaction and genetic complementation data (Figures 1 and 7A), these GOF studies strongly suggest that Pbl mediates Sema-1a reverse signaling through direct interaction with the intracellular domain of Sema-1a.
To determine whether PlexA functions as a ligand for Sema-1a–mediated reverse signaling, one copy of a PlexA loss-of-function mutation (PlexAMB09499; Figure S8; (Bellen et al., 2011)) was introduced into embryos coexpressing Sema-1a and HA-pbl. Interestingly, the CNS defects observed following coexpression of both Sema-1a and HA-pbl transgenes were strongly suppressed in a PlexA heterozygous background (Figure 7B). Similarly, removing one copy of PlexA led to a significant suppression of Fc/mICD and HA-pbl gain-of-function CNS defects, indicating that this PlexA–mediated suppression is not dependent upon the Sema-1a extracellular domain (Figure 7B). However, we did not observe a significant suppression of ISNb defects in embryos that coexpress either Sema-1a or Fc/mICD along with HA-pbl in a PlexA heterozygous background, consistent with previous observations that PlexA functions as a Sema-1a receptor to mediate repulsive signaling during neuromuscular development (Figure 7B; Winberg et al., 1998). These results suggest that PlexA functions in parallel with Sema-1a reverse signaling in the CNS, and further, that PlexA does not function as a major ligand for Sema-1a in both the PNS and the CNS (Figure 8).
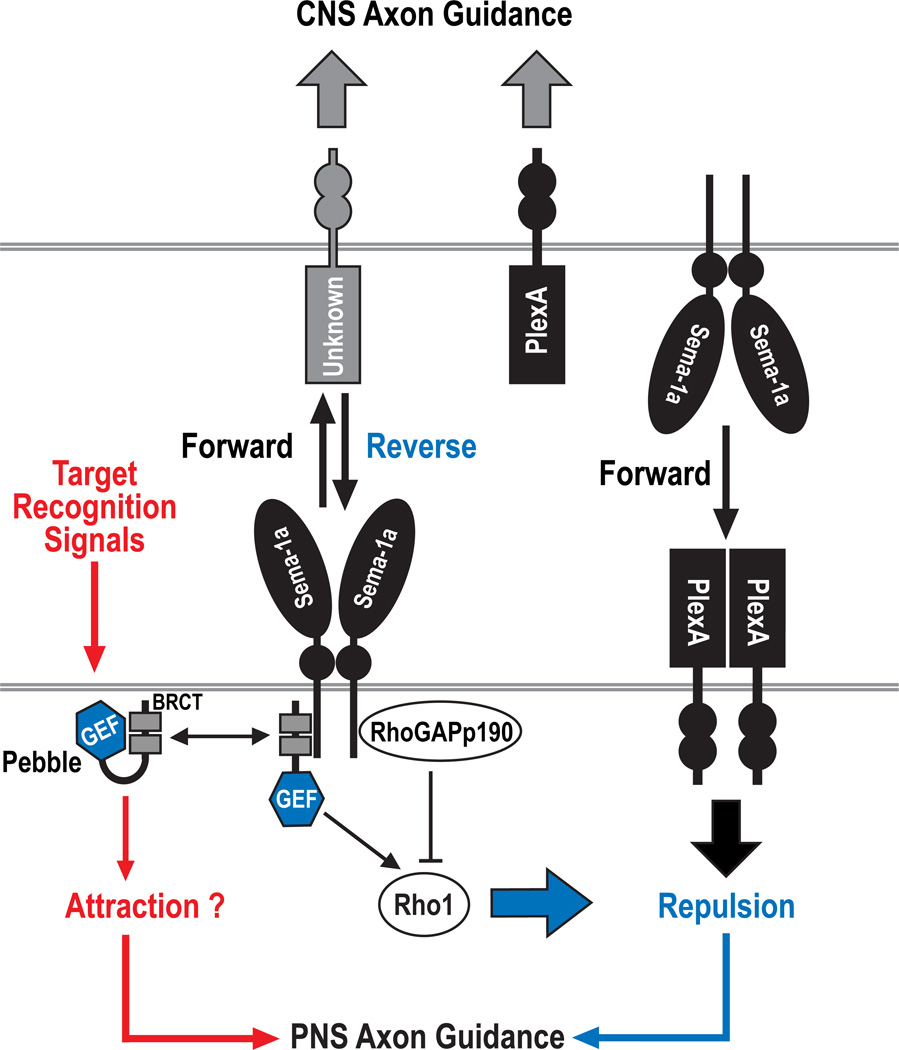
Figure 8. The Control of Semaphorin-1a Reverse Signaling by Pebble and RhoGAPp190.
Repulsive guidance signals specified by Sema-1a bidirectional signaling play an important role in sculpting Drosophila neuromuscular connectivity. In our model, Sema-1a forward signaling largely contributes to CNS axon guidance, whereas both forward and reverse signaling are required to induce axon-axon repulsion at specific pathway choice points in the PNS. In the PNS, two direct but opposing regulators of Rho family small GTPases, pbl and p190, are required for attractive target recognition processes that guide motor axons before reaching, or after leaving, guidance choice points. Furthermore, these proteins mediate Sema-1a reverse signaling through the control of Rho1 activity at guidance choice points and likely participate in the recognition of these choice points by axonal growth cones, an essential prerequisite for selective axon-axon defasciculation mediated by repulsive signaling pathways. We propose that Pbl links choice-point recognition signaling and Sema-1a reverse signaling, and further, that Sema-1a reverse signaling collaborates in parallel with receptor functions of PlexA to induce selective axon-axon repulsion. In addition, Pbl not only competes, but also collaborates, with p190 to regulate Sema-1a reverse signaling in a context dependent manner. Sema-1a/PlexA–mediated repulsive signaling and target recognition signaling are likely integrated and coordinated by pbl and p190, both spatially and temporally, so as to establish precise connections between motoneurons and their muscle targets.
DISCUSSION
We describe here a novel signaling pathway whereby the transmembrane guidance cue Sema-1a regulates axon-axon interactions, providing insight into links between guidance cue recognition and subsequent intracellular signaling events. We find that the RhoGTPase regulators Pbl and p190 physically associate with the cytoplasmic domain of Sema-1a, and our genetic analyses support a role for these signaling molecules in mediating Sema-1a reverse signaling during embryonic axon pathfinding in Drosophila.
Function and Regulation of pebble and RhoGAPp190 in Motor Axon Pathfinding
We find that the pbl09645 hypomorphic allele shows highly penetrant defects in target recognition by ISNb axons (88.4% of hemisegments) in addition to an increased fasciculation phenotype (54.1% of hemisegments). The function of Pbl in target recognition might be directly associated with Sema-1a–mediated defasciculation, since 41.1% of the hemisegments in pbl09645 homozygous mutants exhibit both target recognition errors and severely increased fasciculation. In general, repulsive signaling can be selectively activated to mediate axon-axon defasciculation at choice points, whereas attractive target recognition cues most likely guide axons before reaching, or after leaving, guidance choice points (Kolodkin and Tessier-Lavigne, 2010). Furthermore, the recognition of choice points by axonal growth cones is an essential prerequisite for selective axon-axon defasciculation mediated by repulsive signaling pathways.
Here, we propose that Pbl links choice-point recognition signaling and Sema-1a/PlexA repulsive signaling (Figure 8). In this scenario, Pbl is primed by choice-point recognition signals, and primed Pbl is subsequently activated through direct interaction with the Sema-1a signaling complex. The priming event might be related to the accessibility of the Sema-1a ICD to the BRCT domains of Pbl. In support of this idea, we observed that two mutant forms of Sema-1a (Sema-1a[36G/52A] and Sema-1a[Δ31–60]) that exhibit strong reductions in binding to full-length Pbl and also reductions in synergistic genetic interactions with HA-Pbl in GOF studies can fully rescue the Sema-1a mutant phenotype in complementation tests (Figure 7A). However, these mutations introduced into Sema-1a ICD do not affect the ability of this modified Sema-1a ICD to bind to truncated Pbl NTD proteins that lack the C-terminal domain which, based on the work of others (Kim et al., 2005; Saito et al., 2004), is known to mediate auto-inhibitory intramolecular interactions with BRCT domains. Therefore, endogenous Pbl may undergo a conformational change to relieve auto-inhibitory interactions and/or increase membrane targeting as a result of choice-point recognition, and this could increase the accessibility and binding of the Pbl BRCT domains to Sema-1a. This is in line with previous observations showing, with respect to GEF regulation, that protein-protein interactions and post-translational modifications can result in the relief of auto-inhibitory intramolecular interactions, GEF relocalization, or down-regulation of GEF activity (Schmidt and Hall, 2002).
The mammalian p190 protein is required for axon guidance and fasciculation, and its function is regulated by phosphorylation events downstream of cell adhesion molecules (Brouns et al., 2001). Similarly, in fly mushroom body neurons, p190 and cell adhesion/signaling molecules including integrins control axon branch stability (Billuart et al., 2001). We find here that p190 knockdown leads to analogous defects in motor axon pathfinding and fasciculation, further supporting an evolutionarily conserved role for this GAP during neural development. Based upon the strong genetic interactions we observe between p190 and Sema-1a, and also the increased defasciculation phenotypes in p190 knockdown embryos, we propose that p190 negatively regulates Sema-1a repulsive signaling. In addition, the antagonistic genetic interactions we observe between p190 and pbl suggest that they compete to control Sema-1a reverse signaling. This competition could serve to rapidly amplify or inhibit Sema-1a–mediated signaling. Interestingly, we also observed synergistic interactions between p190 and pbl, suggesting employment of a cyclic mode of action for Rho GTPase activation and inactivation in axon guidance (Luo, 2000). These distinct and cooperative functions may contribute to selective activation of Sema-1a repulsive signaling at different choice points. Taken together, our results support a model whereby Pbl and p190 together act to integrate target recognition and repulsive signaling resulting from reverse Sema-1a signal transduction events (Figure 8).
Semaphorin-1a Forward and Reverse Signaling
Sema-1a was initially identified as an axonal repellent that functions as a ligand for PlexA (Yu et al., 1998; Winberg et al., 1998). This Sema-1a ligand function is strongly supported by genetic analyses that define roles for Sema-1a–PlexA forward signaling in PNS motor axon pathfinding (Winberg et al., 1998; this present study). However, differences in Sema-1a and PlexA null mutant phenotypes, and also the lack of genetic interactions between these mutants with respect to CNS defects, suggest that Sema-1a plays a unique role independent of PlexA in CNS axon guidance (Figures S8B–S8E). Here, we provide cellular and genetic evidence that Sema-1a forward signaling is largely responsible for Sema-1a–mediated CNS axon guidance, whereas both forward and reverse signaling are required for Sema-1a–mediated PNS motor axon pathfinding. In addition, Sema-1a reverse signaling is dependent upon opposing Pbl and p190 functions (Figure 8).
Sema-1a is highly expressed on embryonic motor and CNS axons and plays an important role in both CNS and PNS axon guidance (Yu et al., 1998). The neuronal requirement for Sema-1a in thse guidance events fits well with our finding that the Sema-1a receptor function required for PNS axon guidance is controlled by neuronal Pbl and p190. Our genetic interaction analyses, however, suggest that PlexA does not function as a major Sema-1a ligand in both the embryonic PNS and the CNS, consistent with previous observations in the olfactory system (Sweeny et al., 2011), but, rather, cooperates with Sema-1a reverse signaling to mediate repulsion (Figure 8). Given that plexins harbor a GAP activity directed toward Ras GTPases (Oinuma et al., 2004; Yang and Terman, 2012), Sema-1a reverse signaling and the receptor function of PlexA likely converge on Rho and Ras GTPases, respectively, and these two small GTPases likely collaborate to control axonal defasciculation. Recent work suggests that Rap, not Ras, GTPases are the target of the PlexA GAP domain (Wang et al., 2012), and future work will determine which GTPases in vivo are regulated by plexin GAP activity.
Rho1 GTPases in Motor Axon Guidance
The Rho family of small GTPases, which includes Rho, Rac, and Cdc42, controls growth cone behavior through the regulation of actin dynamics (Hall and Lalli, 2010). In Drosophila neuromuscular development, overexpression of a dominant-negative (DN) Cdc42 causes motor neuron growth cone arrest. However, expression of DN Rac1 frequently results in parallel bypass phenotypes, indicative of defects in target recognition and axonal defasciculation (Kaufmann et al. 1998). Here, we observe that Rho1 plays a critical role in Sema-1a–mediated motor axon repulsion, and that its activity is modulated by opposing Pbl and p190 functions specified by Sema-1a–mediated repulsive, and most likely attractive, signals (Figure S6). We also observed highly penetrant motor axon pathfinding defects in Sema-1a, pbl, and p190 loss-of-function mutants (85%, 98%, and 66% of mutant hemisegments respectively). Furthermore, our finding that Sema-1a and Pbl collaborate to reduce cell size in cultured cells, and that this synergistic effect is reversed by inhibition of Rho1 activity, support an essential role for Rho1 in repulsive guidance at specific choice points. Previous studies show that pbl null alleles including pbl2, pbl3, and pbl5, which we find here show strong genetic interactions with Sema-1a, strongly suppress the Rho1-induced rough eye phenotypes in Drosophila, and that Pbl interacts with Rho1, but not with Rac1 or Cdc42, in yeast two-hybrid assays (Prokopenko et al., 1999). Taken together, these observations strongly suggest that Sema-1a regulates the GEF activity of Pbl directed toward the Rho1 GTPase. Phenotypic analysis of pbl and p190 mutants demonstrates an additional role for Rho1 in regulating axon target recognition. In summary, our results suggest that Sema-1a–mediated reverse signaling pathways converge on Rho1 to control motor axon target recognition and gudiance. It will be important to determine whether other transmembrane guidance cues best known as ligands, including vertebrate transmembrane semaphorins, also mediate receptor functions through direct modulation of Rho GTPase activities.
EXPERIMENTAL PROCEDURES
Drosophila Strains and Phenotypic Characterization
We used the w1118 strain as a wild-type control. The following flies were obtained from the Bloomington Stock Center: pbl2, pbl3, pbl5, pblKG07669, pbl09645, UAS-pbl RNAi[t28343], UASp190 RNAi[8.2], UAS-p190 RNAi[5.2], UAS-mycp190, e16E-GAL4, and Rho172F. UAS-pbl RNAi[v35349] and UAS-pbl RNAi[v35350] were obtained from the Vienna Drosophila RNAi Center. All other strains were described previously: Sema-1aPI, and UAS-Sema-1a (Yu et al., 1998); PlexADf(4)C3 (Winberg et al., 1998); PlexBKG00878 (Ayoob et al., 2006); Elav(2)-GAL4, Elav(3E)-GAL4, and 24B-GAL4 (Luo et al., 1994); Sca-GAL4 (Klaes et al., 1994); GMR37C03GAL4 and GMR37D12GAL4 (Pfeiffer et. al., 2008). p1902 was obtained from J. Cho. Phenotypic analyses of axon guidance defects were performed as described previously (Yu et al., 1998).
Expression Constructs and Mutagenesis
PlexB, Sema-1a and Otk (LP17455) open reading frames (ORF) from cDNAs or EST clones were myc-tagged C-terminally and subcloned into pUAST (Brand and Perrimon, 1993). The UAS-PlexA-5xmyc was described previously (Wu et al., 2011). Pbl (SD01796), NTD[Pbl], CTD[Pbl] and p190 (RE10888) were HA-tagged N-terminally and similarly subcloned into pUAST. The UAS-CD8-EGFP (pUAST-DEST16) was obtained from the Drosophila Genomics Resource Center (DGRC). Serially deleted or point mutation constructs of Sema-1a ICD were generated using pUAST as represented in Figure 1C. All constructs for transgenic flies shown in Figure 6A were generated by polymerase chain reaction (PCR) and/or restriction enzyme-based strategies and inserted into a customized version of pUAST (pUAST-attB), which allows site-specific integration into predetermined landing sites (Bischof et al., 2007). To minimize position effects, all transgenic flies were generated using the same landing site (Strain 9750, BestGene Inc.). Integration and orientation were confirmed by a PCR-based assay with attP-F and attB-R primers (Venken et al., 2006).
Cell Contraction Assay in ML-DmBG2-c2 Cells
ML-DmBG2-c2 cells were maintained according to standard procedures (available at http://www.flyrnai.org/DRSC-PRC.html). Immunofluorescence microscopy and RNAi experiments were performed as described previously (Rogers and Rogers, 2008) but with a few modifiections. Cells were fixed with 3.7% paraformaldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, pH 7.0, 10 mM EGTA, 4 mM MgSO4) for 10 min at room temperature. To knock down endogenous Rho1, 10 µg of dsRNA directed against Rho1 was first added to each well 30 min after transfection. After 2.75 days, another 10 µg of dsRNA was added to each well. By quantitative immunoblotting, we verified that Rho1 dsRNA reduced endogenous protein levels by ~70% as compared to control cells (data not shown). More than 28 single, isolated, cells for each transfection experiment were analyzed for cell area using ImageJ. Primary antibodies used in this experiment were as follows: HA (3F10, Roche), myc (9E10, Sigma), GFP (rabbit & 3E6, Invitrogen) and Sema-1a (Yu et al., 1998) antibodies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Liqun Luo and Zhuhao Wu for comments on the manuscript, and Kolodkin laboratory members for helpful discussions throughout this work; Joong Cho, the Bloomington Drosophila Stock Center, and Vienna Drosophila RNAi Center for strains; and the Drosophila Genomics Resource Center for clones and vectors. This work was supported by NIH R01 NS35165 (A.L.K.). A.L.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental data include eight figures, Supplementary Experimental Procedures, and Supplementary References.
REFERENCES
- Ayoob JC, Terman JR, Kolodkin AL. Drosophila Plexin B is a Sema-2a receptor required for axon guidance. Development. 2006;133:2125–2135. doi: 10.1242/dev.02380. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Klein R. Signaling from axon guidance receptors. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a001941. a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, et al. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics. 2011;188:731–743. doi: 10.1534/genetics.111.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brouns MR, Matheson SF, Settleman J. p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat. Cell Biol. 2001;3:361–367. doi: 10.1038/35070042. [DOI] [PubMed] [Google Scholar]
- Cafferty P, Yu L, Long H, Rao Y. Semaphorin-1a functions as a guidance receptor in the Drosophila visual system. J Neurosci. 2006;26:3999–4003. doi: 10.1523/JNEUROSCI.3845-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, Gertler FB. The growth cone cytoskeleton in axon outgrowth and guidance. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a001800. a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijck AA, Van Erp S, Pasterkamp RJ. Semaphorin signaling: molecular switches at the midline. Trends Cell Biol. 2010;20:568–576. doi: 10.1016/j.tcb.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 2001;11:103–110. doi: 10.1016/s0959-4388(00)00180-x. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Egea J, Klein R. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol. 2007;17:230–238. doi: 10.1016/j.tcb.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Godenschwege TA, Hu H, Shan-Crofts X, Goodman CS, Murphey RK. Bidirectional signaling by Semaphorin 1a during central synapse formation in Drosophila. Nat. Neurosci. 2002;5:1294–1301. doi: 10.1038/nn976. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a001818. a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BJ, Robinson RA, Perez-Branguli F, Bell CH, Mitchell KJ, Siebold C, Jones EY. Structural basis of semaphorin-plexin signalling. Nature. 2010;467:1118–1122. doi: 10.1038/nature09468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann N, Wills ZP, Van Vactor D. Drosophila Rac1 controls motor axon guidance. Development. 1998;125:453–461. doi: 10.1242/dev.125.3.453. [DOI] [PubMed] [Google Scholar]
- Keshishian H, Broadie K, Chiba A, Bate M. The Drosophila neuromuscular junction: a model system for studying synaptic development and function. Annu. Rev. Neurosci. 1996;19:545–575. doi: 10.1146/annurev.ne.19.030196.002553. [DOI] [PubMed] [Google Scholar]
- Kim JE, Billadeau DD, Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J. Biol. Chem. 2005;280:5733–5739. doi: 10.1074/jbc.M409298200. [DOI] [PubMed] [Google Scholar]
- Klaes A, Menne T, Stollewerk A, Scholz H, Klambt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a001727. a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sweeney LB, Schuldiner O, Garcia KC, Luo L. Graded expression of semaphorin-1a cell-autonomously directs dendritic targeting of olfactory projection neurons. Cell. 2007;128:399–410. doi: 10.1016/j.cell.2006.12.028. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Bossing T, Technau GM, Bate M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. 1997;17:9642–9655. doi: 10.1523/JNEUROSCI.17-24-09642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Juo ZS, Shim AH, Focia PJ, Chen X, Garcia KC, He X. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142:749–761. doi: 10.1016/j.cell.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak ET, Meadows RP, Schkeryantz JM, Janowick DA, Harlan JE, Harris EA, et al. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- Luo L. Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Mann F, Chauvet S, Rougon G. Semaphorins in development and adult brain: Implication for neurological diseases. Prog Neurobiol. 2007;82:57–79. doi: 10.1016/j.pneurobio.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Nogi T, Yasui N, Mihara E, Matsunaga Y, Noda M, Yamashita N, Toyofuku T, Uchiyama S, Goshima Y, Kumanogoh A, et al. Structural basis for semaphorin signalling through the plexin receptor. Nature. 2010;467:1123–1127. doi: 10.1038/nature09473. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Ishikawa Y, Katoh H, Negishi M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- O'Keefe DD, Thor S, Thomas JB. Function and specificity of LIM domains in Drosophila nervous system and wing development. Development. 1998;125:3915–3923. doi: 10.1242/dev.125.19.3915. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, Brumby A, O'Keefe L, Prior L, He Y, Saint R, Bellen HJ. A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev. 1999;13:2301–2314. doi: 10.1101/gad.13.17.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, He Y, Lu Y, Bellen HJ. Mutations affecting the development of the peripheral nervous system in Drosophila: a molecular screen for novel proteins. Genetics. 2000;156:1691–1715. doi: 10.1093/genetics/156.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, Chen X, Lee CC, Lorenzi MV, Ohara N, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J. Biol. Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Shen K, Cowan CW. Guidance molecules in synapse formation and plasticity. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a001842. a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney LB, Chou YH, Wu Z, Joo W, Komiyama T, Potter CJ, Kolodkin AL, Garcia KC, Luo L. Secreted Semaphorins from degenerating larval ORN axons direct adult projection neuron dendrite targeting. Neuron. 2011;72:734–747. doi: 10.1016/j.neuron.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Settleman J. Rho family GTPases: more than simple switches. Trends Cell Biol. 2000;10:415–419. doi: 10.1016/s0962-8924(00)01832-8. [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Yabuki M, Harada K, Hori M, Kikutani H. Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat. Cell Biol. 2004;6:1204–1211. doi: 10.1038/ncb1193. [DOI] [PubMed] [Google Scholar]
- Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, Miyake T. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. In Vitro Cell. Dev. Biol. Anim. 1994;30A:209–216. doi: 10.1007/BF02632042. [DOI] [PubMed] [Google Scholar]
- Van Vactor D, Sink H, Fambrough D, Tsoo R, Goodman CS. Genes that control neuromuscular specificity in Drosophila. Cell. 1993;73:1137–1153. doi: 10.1016/0092-8674(93)90643-5. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal. 2012;5:ra6. doi: 10.1126/scisignal.2002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M, Goodman CS. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95:903–916. doi: 10.1016/s0092-8674(00)81715-8. [DOI] [PubMed] [Google Scholar]
- Wu Z, Sweeney LB, Ayoob JC, Chak K, Andreone BJ, Ohyama T, Kerr R, Luo L, Zlatic M, Kolodkin AL. A combinatorial semaphorin code instructs the initial steps of sensory circuit assembly in the Drosophila CNS. Neuron. 2011;70:281–298. doi: 10.1016/j.neuron.2011.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Terman JR. 14-3-3ε couples Protein Kinase A to Semaphorin signaling and silences Plexin RasGAP-mediated axonal repulsion. Neuron. 2012;74:108–121. doi: 10.1016/j.neuron.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Araj HH, Ralls SA, Kolodkin AL. The transmembrane Semaphorin Sema I is required in Drosophila for embryonic motor and CNS axon guidance. Neuron. 1998;20:207–220. doi: 10.1016/s0896-6273(00)80450-x. [DOI] [PubMed] [Google Scholar]
- Yu L, Zhou Y, Cheng S, Rao Y. Plexin a-semaphorin-1a reverse signaling regulates photoreceptor axon guidance in Drosophila. J. Neurosci. 2010;30:12151–12156. doi: 10.1523/JNEUROSCI.1494-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.