Abstract
AIM: To study the polymorphisms of toll-like receptor 4 (TLR4) gene Asp299Gly, Thr399Ile and TLR2 gene Arg753Gln, Arg677Trp and susceptibility to inflammatory bowel disease (IBD) in the Zhuang population from Guangxi, China.
METHODS: A case-control study was performed from February 2007 to October 2011 which included 146 Zhuang patients with IBD in the experimental group and 164 healthy Zhuang subjects who acted as the control group. All patients and healthy subjects were from the Guangxi Zhuang Autonomous Region of China. Genomic DNA was extracted from intestinal tissue by the phenol chloroform method. TLR4 gene Asp299Gly, Thr399Ile and TLR2 gene Arg753Gln, Arg677Trp were amplified by polymerase chain reaction (PCR), and then detected by PCR-restriction fragment length polymorphism (RFLP).
RESULTS: The TLR4 gene Asp299Gly was digested using Nco I restriction enzyme, and a single band of 249 bp was observed which showed that it was a wild type (AA). The TLR4 gene Thr399Ile was digested using Hinf Irestriction enzyme and only the wild type (CC) was detected. In addition, the TLR2 gene Arg677Trp was digested using Aci I restriction enzyme and only the wild type (CC) was detected. The TLR2 gene Arg753Gln was digested using Pst I restriction enzyme. Only the wild type (GG) as a single band of 254 bp was observed during RFLP. Overall, no heterozygous or homozygous single nucleotide polymorphism mutations were found in patients with Crohn’s disease and ulcerative colitis both in the TLR4 gene Asp299Gly, Thr399Ile and the TLR2 gene Arg677Trp, Arg753Gln in the Zhuang population from the Guangxi Zhuang Autonomous Region of China.
CONCLUSION: The TLR4 gene Asp299Gly, Thr399Ile and TLR2 gene Arg753Gln, Arg677Trp polymorphisms may not be associated with IBD in the Zhuang population from the Guangxi Zhuang Autonomous Region of China.
Keywords: Toll-like receptor 2, Toll-like receptor 4, Inflammatory bowel disease, Gene polymorphism
INTRODUCTION
Inflammatory bowel disease (IBD) is a chronic disorder caused by multiple factors in genetically susceptible hosts, and includes ulcerative colitis (UC) and Crohn’s disease (CD). The molecular basis of the pathogenesis is not completely clear, but involves a complex interaction of factors such as genetics, immunology, environment and infection. The incidence of IBD in Western populations has increased with an estimated incidence of 0.10%-1.00% for UC and 0.35%-1.00% for CD during the past few decades[1]. NOD2/CARD15 was the first verified predisposing gene for CD, where three NOD2/CARD15 polymorphisms, Arg702Trp, Gly908Arg and Leu1007fsinsC, were found to be significantly associated with CD in Caucasian populations[2,3]. Nevertheless, these single nucleotide polymorphisms (SNPs) were not reported to be associated with CD in Japanese, and Hubei, Zhejiang, or Hong Kong populations in China[4-7], thus, their exact role in CD is controversial. Recently, genome wide association studies, provided evidence for several determinants including Toll-like receptor 2 (TLR2) and TLR4[8]. TLR4 is upregulated in intestinal epithelial cells, macrophages, and dendritic cells in patients with UC and CD. In contrast, the expression of TLR2 is unchanged. The Asp299Gly and Thr399Ile polymorphisms of the TLR4 gene were shown to be associated with lipopolysaccharide hyporesponsiveness and with both CD and UC in some studies[9,10]. The TLR2 gene Arg753Gln polymorphism frequency is approximately 1%-4% in the Caucasian population, significantly higher than that in Indian patients with IBD[11].
In view of the discrepant data regarding the association between key regulatory genes and IBD susceptibility, the purpose of our study was to investigate whether the known gene polymorphisms in TLR2 and TLR4 determine susceptibility to IBD in the Guangxi Zhuang population from the Guangxi Zhuang Autonomous Region. Guangxi has a large Zhuang population, where genetic diseases and gene polymorphisms are unique. Therefore, research on the relation between TLR2 and TLR4 polymorphisms and IBD in Chinese Zhuang patients from the Guangxi Zhuang Autonomous Region is needed.
MATERIALS AND METHODS
Patients
The study group consisted of 146 IBD Zhuang patients without genetic kinship enrolled in the Gastroenterology Department, First Affiliated Hospital of Guangxi Medical University, from February 2007 to October 2011. The control group included 164 healthy Zhuang subjects without genetic kinship who were healthy individuals or patients with functional dyspepsia, and did not have liver or gastrointestinal diseases. All patients had a well-established diagnosis of UC or CD, according to standard clinical criteria based on endoscopic and histopathological examinations. There were no significant differences in age and sex between the study group and the control group. All patients and healthy controls gave informed consent and the study was approved by the ethical committee of the institute.
DNA extraction and genotyping of the TLR4 and TLR2 polymorphisms
Genomic DNA was extracted according to the modified protocol of Taggart. The primers used to amplify the TLR4 gene (Asp299Gly, Thr399 Ile) were designed according to the National Center for Biotechnology Information gene database, NM_138554, and the TLR2 gene (Arg677Trp, Arg753Gln) with NM _003264 shown in Table 1 (primers were synthesized in SHENGGONG Biological Technology Co., Ltd., Shanghai, China).
Table 1.
Specific polymerase chain reaction primers and restriction enzymes for each single nucleotide polymorphism
| Gene | Amino acid substitution | Sequence of primers( 5'→3') | Fragments (bp) | Enzymes |
| TLR4 | Asp299Gly | F-GATTAGCATACTTAGACTACTACCTCCATG | 249 | NcoI |
| R-GATCAACTTCTGAAAAAGCATTCCCAC | ||||
| Thr399Ile | F-GGTTGCTGTTCTCAAAGTGATTTTGGGAGAA | 406 | HinfI | |
| R-ACCTGAAGACTGGAGAGTGAGTTAAATGCT | ||||
| TLR2 | Arg677Trp | F-GCCTACTGGGTGGAGAACCTT | ||
| R-CCAGTTCATACTTGCACCACTC | 199 | AciI | ||
| Arg753Gln | F-CCTGGCAAGTGGATCATTGAC | |||
| R-GGCCACTCCAGGTAGGTCTT | 254 | PstI |
TLR: Toll-like receptor.
Amplification was performed using H2O2 10.5 μL, 2 × Taq polymerase chain reaction (PCR) Master-mix 11.5 μL (TIANGEN), DNA 1 μL, and 1 μL in addition to 10 μmol/L of each primer in a total volume of 25 μL. For Asp299Gly and Arg753Gln, cycle conditions were an initial denaturation for 5 min at 95 °C, followed by 28 cycles of denaturing at 95 °C for 40 s, annealing at 58 °C for 30 s, primer extension at 72 °C for 50 s, followed by a final extension at 72 °C for 10 min. For Thr399Ile and Arg677Trp, cycle conditions were an initial denaturation for 5 min at 95 °C, followed by 28 cycles of denaturing at 95 °C for 40 s, annealing at 62 °C for 40 s, primer extension at 72 °C for 50 s, followed by a final extension at 72 °C for 10 min (Thermo Electron Corporation, Waltham, MA, United States). All the PCR products were electrophoresed on a 1.5% agarose gel, with 1 × tris-borate-EDTA buffer, V = 100V for 30 min, and visualized under ultraviolet illumination (Bio-Rad Gel Doc-2000, United States).
Five μL PCR products of TLR4 gene (Asp299Gly, Thr399Ile) and TLR2 gene (Arg677Trp,Arg753Gln) were digested at 37 °C overnight with 0.5 μL (10 U) Nco I, Hinf I, Aci I, and Pst I restriction enzymes (Fermentas), respectively. After enzymatic digestion, the fragments were separated and visualized by gel electrophoresis (2% Yito Bio-Instrument Company Ltd., Shanghai, China).
PCR products of each SNP were purified with a PCR purification kit (QIAGEN, Hilden, Germany) and were sequenced using an ABI Prism 377 DNA sequencer (LIU HE HUA DA GENE Technology Co, Ltd., Beijing, China).
Statistical analysis
The genetic equilibrium was tested using Hardy-Weinberg. Allele and genotype frequencies in patients and in controls were compared using the χ2 test with SPSS 13.0, and P values were considered significant at a level of < 0.05.
RESULTS
RFLP analysis was performed to assess the status of SNPs in our samples after digestion with restriction enzymes (Figure 1).
Figure 1.
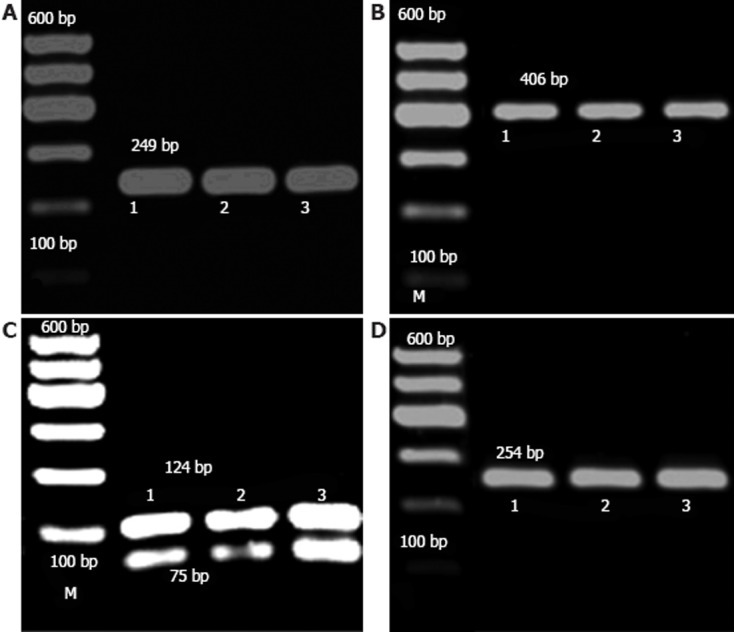
Polymerase chain reaction-restriction fragment length polymorphism analysis. A: Electrophoresis of Asp299Gly restriction fragment length polymorphism (AA type); B: Electrophoresis of Thr399Ile restriction fragment length polymorphism (CC type); C: Electrophoresis of Arg677Trp restriction fragment length polymorphism (CC type); D: Electrophoresis of Arg753Gln restriction fragment length polymorphism (GG type). M: Marker; 1: Ulcerative colitis; 2: Crohn’s disease; 3: Control.
TLR4 gene Asp299Gly PCR-RFLP
Nco I restriction enzyme identified the sequence of CCATGG. For the heterozygous type (AG), this could be generated when digested with Asp299Gly and three bands of 249 bp, 223 bp and 26 bp, where one chain was cut, and the others were not. For the homozygous type (GG), a transite to G, Nco I identified the mutated site, and the mutated DNA was visible as a double band of 223 bp and 26 bp, whereas a single band of 249 bp was observed in the wild type (AA), as the site was not cut. In this study, only the wild type (AA) was detected, and no other types (Figure 1A).
TLR4 gene Thr399Ile PCR-RFLP
Hinf I restriction enzyme identified the sequence of GANTC. For the heterozygous type (CT), this could be generated when digested with Thr399Ile and three bands of 406 bp, 377 bp and 29 bp, where only one chain was cut, and the others were not. For the homozygous type (TT), C transite to T, Hinf I identified the mutated site, and the mutated DNA was visible as a double band of 377 bp and 29 bp, whereas a single band of 406 bp was observed in the wild type (CC), as the site was not cut. In this study, only the wild type (CC) was detected, and no other types (Figure 1B).
TLR2 gene Arg677Trp PCR-RFLP
Aci I restriction enzyme identified the sequence of CCGC. For the heterozygous type (CT), this could be generated when digested with Arg677Trp and three bands of 199 bp, 124 bp and 75 bp, where one chain was cut, and the others were not. For the homozygous type (TT), C transite to T, the site was not cut by Aci I restriction enzyme, and a single band of 199 bp was observed, for the wild type (CC), it identified the site, and the DNA was visible as a double band of 124 bp and 75 bp. In this study, only the wild type (CC) was detected, and no other types (Figure 1C).
TLR2 gene Arg753Gln PCR-RFLP
Pst I restriction enzyme identified the sequence of CTGCAG. For the heterozygous type (GA), this could be generated when digested with Arg753Gln and three bands of 254 bp, 214 bp and 40 bp, where one chain was cut, and the others were not. For the homozygous type (AA), G transite to A, Pst I identified the mutated site, and the mutated DNA was visible as a double band of 214 bp and 40 bp, whereas a single band of 254 bp was observed in the wild type (GG), as the site was not cut. In this study, only the wild type (GG) was detected, and no other types (Figure 1D).
DISCUSSION
UC and CD are multifactorial diseases of unknown etiology. Despite being disorders of the gastrointestinal tract, an abnormal immune response directed against the gut microflora has been postulated as a possible explanation in genetically susceptible hosts[12].
Numerous studies have been performed on the frequency of NOD2/CARD15 mutations in IBD populations in Western Europe and Northern America. In CD patients from Northern America or Western Europe, allele frequencies ranged from 9.1%-12.9%, 6.6%-16.0% and 3.3%-6.0% for Arg702Trp, 1007finsC, and Gly908Arg, respectively[13]. Interestingly, a recent study reported lower allele frequencies in Finnish IBD patients for all three NOD2/CARD15 variants in CD patients compared with the above-mentioned studies and similar frequencies in patients with UC[14]. Additionally, in Asian IBD populations, NOD2/CARD15 variants were not detected at all[4,5]. Thus, there is a need for more studies in different populations with IBD. Human TLRs participate in the innate immune response and signal the activation of adaptive immunity. Therefore, these TLRs may be important in autoimmune diseases such as IBD, rheumatoid arthritis and systemic lupus erythematosus[15].
This is the first study to report the TLR4 (Asp299Gly, Thr399Ile) and TLR2 (Arg677Trp, Arg753Gln) gene mutations in patients with IBD from the Guangxi Zhuang population of China, where the ethnic background is heterogeneous, and includes Han, Zhuang and Rao. In this study, the mutation genotypes of the TLR4 gene Asp299Gly, Thr399Ile and TLR2 gene Arg677Trp, Arg753Gln were not found in the Zhuang population with IBD. Our results are in agreement with those from studies on Asian patients from Hong Kong, Hubei, Zhejiang, Shanghai and Japan[4-7].
In recent years, several studies have reported divergent results[16]. The allele frequency of the Asp299Gly variant ranges from 0%-10% in UC, from 8%-13% in CD and from 3%-15% in healthy controls[17]. Franchimont et al[10] reported that the TLR4 SNP was associated with UC and CD in a Belgian study. This association was in accordance with Dutch, Greek, Australian and German populations with CD, and an association with colonic disease has been described[18-21]. In one German cohort, an association between the TLR4 Thr399Ile SNP and UC was demonstrated[22]. However, because of substantial heterogeneity between populations, no association was noted in Scottish CD patients[23]. The TLR2 gene Arg753Gln variant frequency was 9.4% in Germany, and 1% in Spain[24], the TLR2 gene Arg677Trp was 30.3% in Tunisia[25], where the difference was significant. In addition, Pierik et al[26] reported that an association was found between non-synonymous variants and extensive colonic disease with UC and CD in the TLR2 genes.
In our study, we did not find mutations in TLR4 gene Asp299Gly, Thr399Ile and TLR2 gene Arg677Trp, Arg753Gln in the Zhuang population with IBD. These findings were different from those in Tunisia, Germany and Spain. Our results showed that the TLR4 gene Asp299Gly, Thr399 Ile and TLR2 gene may not be associated with IBD patients from the Guangxi Zhuang Autonomous Region of China. This is possibly due to the existence of racial and geographic differences.
The present study supports the notion that genetics, immunology, environment and infection may be vital in the pathogenesis of IBD. However, the heterogeneity in the small number of available studies limited the ability to draw conclusions. Further studies using a larger cohort of patients with IBD are warranted to identity the risk factors and genetic susceptibility to IBD.
COMMENTS
Background
The pathogenesis of inflammatory bowel disease (IBD) is not completely clear, however, contributing factors may include immunology, genetics, environment and infection. It has been reported that the NOD2/CARD15 polymorphisms (single nucleotide polymorphisms, SNPs) were found to be associated with Crohn’s disease (CD) in Caucasian populations. However, the SNPs were not found to be associated with CD in Japan and China. In addition, the TLR2 and TLR4 were reported to provide evidence for several determinants. The study assessed whether the known gene polymorphisms in the toll-like receptor 2 (TLR2) and TLR4 genes determined susceptibility for IBD in the Guangxi Zhuang population. Since Guangxi has a large Zhuang population, genetic diseases and gene polymorphisms are unique.
Research frontiers
TLR4 (Asp299Gly, Thr399Ile) and TLR2 (Arg677Trp, Arg753Gln) were found to be associated with IBD in Tunisia, Germany and Spain, but not in Hong Kong, Hubei, Zhengjiang, Shanghai and Japan. This is possibly due to the existence of racial and geographic differences. This study determined whether the TLR4 (Asp299Gly, Thr399Ile) and TLR2 (Arg677Trp, Arg753Gln) were associated with IBD in the Guangxi Zhuang population from the Guangxi Zhuang Autonomous Region of China.
Innovations and breakthroughs
Only a few studies have investigated the TLR2 and TLR4 gene polymorphisms in China. Moreover, all the subjects in these studies were from the Han population. This is the first study to report that TLR4 gene Asp299Gly, Thr399Ile and TLR2 gene Arg753Gln, Arg753Gln polymorphisms may not be associated with IBD in the Zhuang population from the Guangxi Zhuang Autonomous Region of China.
Applications
TLR2 and TLR4 variants may be rare or non-existent in the Zhuang population from the Guangxi Zhuang Autonomous Region of China.
Terminology
TLRs play a key role in microbial recognition in innate immunity and control the adaptive immune responses. Among the TLRs, TLR2 recognizes bacterial components such as lipoprotein, lipoteichoic acids, peptido-glycan and zymozan. TLR4 requires CD14 to form the lipopolysaccharide (LPS) receptor. LPS, found in the outer membrane of Gram-negative bacteria, is opsonized by LPS-binding protein, and recognized by CD14. The LPS-LPS binding protein-CD14 complex activates TLR4, which results in the activation of NF-κB. TLR2 and TLR4 gene mutations or deletions can induce abnormal immune responses.
Peer review
This study demonstrates no association of TLR2 and/or TLR4 polymorphism with IBD in a patient cohort within a restricted chinese population. Although this is a “negative” result, it adds to the impact of genetic and/or ethnical predisposition to IBD.
Footnotes
Supported by The Natural Science Foundation of Guangxi Zhuang Autonomous Region, China, No. 0832009; the Chinese Traditional Medicine Science Foundation of Guangxi Zhuang Autonomous Region, China, No. GZKZ10-108
Peer reviewer: Dr. Dieter Kabelitz, Professor, Department of Immunology, Kiel University (CAU). Michaelisstraße 5, 24105 Kiel, Germany
S- Editor Song XX L- Editor A E- Editor Xiong L
References
- 1.Branch CMAD. Guidelines for diagnosis and management of inflammatory bowel disease. Zhonghua Xiaohua Zazhi. 2001;21:236–239. [Google Scholar]
- 2.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 3.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson AJ, et al. Association between insertion mutation in NOD2 gene and Crohn’s disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 4.Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y, et al. Lack of common NOD2 variants in Japanese patients with Crohn’s disease. Gastroenterology. 2002;123:86–91. doi: 10.1053/gast.2002.34155. [DOI] [PubMed] [Google Scholar]
- 5.Leong RW, Armuzzi A, Ahmad T, Wong ML, Tse P, Jewell DP, Sung JJ. NOD2/CARD15 gene polymorphisms and Crohn’s disease in the Chinese population. Aliment Pharmacol Ther. 2003;17:1465–1470. doi: 10.1046/j.1365-2036.2003.01607.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang ZW, Ji F, Teng WJ, Yuan XG, Ye XM. Risk factors and gene polymorphisms of inflammatory bowel disease in population of Zhejiang, China. World J Gastroenterol. 2011;17:118–122. doi: 10.3748/wjg.v17.i1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie ZQ, Lv B, Li-Jun Zou, Xue-Lan Shang, Yi-Kai Zhou. Genotyping of Toll-like Receptor 4 in Hubei Population of Chinese. J Huazhong Univ Sci Technol. 2004;33:630–632. [Google Scholar]
- 8.Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat Rev Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 9.Okayama N, Fujimura K, Suehiro Y, Hamanaka Y, Fujiwara M, Matsubara T, Maekawa T, Hazama S, Oka M, Nohara H, et al. Simple genotype analysis of the Asp299Gly polymorphism of the Toll-like receptor-4 gene that is associated with lipopolysaccharide hyporesponsiveness. J Clin Lab Anal. 2002;16:56–58. doi: 10.1002/jcla.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Devière J, et al. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn’s disease and ulcerative colitis. Gut. 2004;53:987–992. doi: 10.1136/gut.2003.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas D, Gupta SK, Sindhwani G, Patras A. TLR2 polymorphisms, Arg753Gln and Arg677Trp, are not associated with increased burden of tuberculosis in Indian patients. BMC Res Notes. 2009;2:162. doi: 10.1186/1756-0500-2-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182–205. doi: 10.1016/s0016-5085(98)70381-6. [DOI] [PubMed] [Google Scholar]
- 13.Bonen DK, Cho JH. The genetics of inflammatory bowel disease. Gastroenterology. 2003;124:521–536. doi: 10.1053/gast.2003.50045. [DOI] [PubMed] [Google Scholar]
- 14.Heliö T, Halme L, Lappalainen M, Fodstad H, Paavola-Sakki P, Turunen U, Färkkilä M, Krusius T, Kontula K. CARD15/NOD2 gene variants are associated with familially occurring and complicated forms of Crohn’s disease. Gut. 2003;52:558–562. doi: 10.1136/gut.52.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Pope RM. Toll-like receptor signaling: a potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J Leukoc Biol. 2010;88:253–262. doi: 10.1189/jlb.0310126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riis L, Vind I, Vermeire S, Wolters F, Katsanos K, Politi P, Freitas J, Mouzas IA, O’Morain C, Ruiz-Ochoa V, et al. The prevalence of genetic and serologic markers in an unselected European population-based cohort of IBD patients. Inflamm Bowel Dis. 2007;13:24–32. doi: 10.1002/ibd.20047. [DOI] [PubMed] [Google Scholar]
- 17.Russell RK, Nimmo ER, Satsangi J. Molecular genetics of Crohn’s disease. Curr Opin Genet Dev. 2004;14:264–270. doi: 10.1016/j.gde.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Brand S, Staudinger T, Schnitzler F, Pfennig S, Hofbauer K, Dambacher J, Seiderer J, Tillack C, Konrad A, Crispin A, et al. The role of Toll-like receptor 4 Asp299Gly and Thr399Ile polymorphisms and CARD15/NOD2 mutations in the susceptibility and phenotype of Crohn’s disease. Inflamm Bowel Dis. 2005;11:645–652. doi: 10.1097/01.mib.0000168372.94907.d2. [DOI] [PubMed] [Google Scholar]
- 19.Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, Archimandritis A, Ikonomopoulos J, Gorgoulis VG. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol. 2005;11:681–685. doi: 10.3748/wjg.v11.i5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hume GE, Fowler EV, Doecke J, Simms LA, Huang N, Palmieri O, Griffiths LR, Florin TH, Annese V, Radford-Smith GL. Novel NOD2 haplotype strengthens the association between TLR4 Asp299gly and Crohn’s disease in an Australian population. Inflamm Bowel Dis. 2008;14:585–590. doi: 10.1002/ibd.20362. [DOI] [PubMed] [Google Scholar]
- 21.Oostenbrug LE, Drenth JP, de Jong DJ, Nolte IM, Oosterom E, van Dullemen HM, van der Linde K, te Meerman GJ, van der Steege G, Kleibeuker JH, et al. Association between Toll-like receptor 4 and inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:567–575. doi: 10.1097/01.mib.0000161305.81198.0f. [DOI] [PubMed] [Google Scholar]
- 22.Török HP, Glas J, Tonenchi L, Mussack T, Folwaczny C. Polymorphisms of the lipopolysaccharide-signaling complex in inflammatory bowel disease: association of a mutation in the Toll-like receptor 4 gene with ulcerative colitis. Clin Immunol. 2004;112:85–91. doi: 10.1016/j.clim.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Cuthbert AP, Fisher SA, Mirza MM, King K, Hampe J, Croucher PJ, Mascheretti S, Sanderson J, Forbes A, Mansfield J, et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122:867–874. doi: 10.1053/gast.2002.32415. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez E, Orozco G, López-Nevot MA, Jiménez-Alonso J, Martín J. Polymorphisms of toll-like receptor 2 and 4 genes in rheumatoid arthritis and systemic lupus erythematosus. Tissue Antigens. 2004;63:54–57. doi: 10.1111/j.1399-0039.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Ali M, Barbouche MR, Bousnina S, Chabbou A, Dellagi K. Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin Diagn Lab Immunol. 2004;11:625–626. doi: 10.1128/CDLI.11.3.625-626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierik M, Joossens S, Van Steen K, Van Schuerbeek N, Vlietinck R, Rutgeerts P, Vermeire S. Toll-like receptor-1, -2, and -6 polymorphisms influence disease extension in inflammatory bowel diseases. Inflamm Bowel Dis. 2006;12:1–8. doi: 10.1097/01.mib.0000195389.11645.ab. [DOI] [PubMed] [Google Scholar]


