Abstract
Objective: Biomarker assay is a noninvasive method for the early detection of esophageal squamous cell carcinoma (ESCC). Searching for new biomarkers with high specificity and sensitivity is very important for the early detection of ESCC. Serum surface-enhanced laser desorption/ionization-time of flight mass spectrometry (SELDI-TOF-MS) is a high throughput technology for identifying cancer biomarkers using drops of sera. Methods: In this study, 185 serum samples were taken from ESCC patients in a high incidence area and screened by SELDI. A support vector machine (SVM) algorithm was adopted to analyze the samples. Results: The SVM patterns successfully distinguished ESCC from pre-cancerous lesions (PCLs). Also, types of PCL, including dysplasia (DYS) and basal cell hyperplasia (BCH), and healthy controls (HC) were distinguished with an accuracy of 95.2% (DYS), 96.6% (BCH), and 93.8% (HC), respectively. A marker of 25.1 kDa was identified in the ESCC patterns whose peak intensity was observed to increase significantly during the development of esophageal carcinogenesis, and to decrease obviously after surgery. Conclusions: We selected five ESCC biomarkers to form a diagnostic pattern which can discriminate among the different stages of esophageal carcinogenesis. This pattern can significantly improve the detection of ESCC.
Keywords: Biomarker, Esophageal squamous cell carcinoma, Surface-enhanced laser desorption/ionization, Support vector machine
1. Introduction
Among malignant tumors, the mortality rate of esophageal squamous cell carcinoma (ESCC) ranks sixth in the world (Parkin et al., 2005). The occurrence rate of this tumor greatly depends on geographic variation. The highest incidence is found in Linxian City in China. Patients with ESCC have a poor prognosis and the five-year survival rate is as low as 15% (Yang, 1980; Lu et al., 2002). Though esophageal endoscopy is a common method for screening for ESCC, in China it is more practical first to use a serum biomarker because of the large population and the unbalanced distribution of medical resources.
The application of serum biomarkers for the screening and diagnosis of ESCC was studied because of their advantages of less pain and wider accessibility (Handy, 2009). Among serum biomarkers, carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 19-9, and squamous cell carcinoma antigen (SCCA) are the three most commonly used for the early detection of esophageal cancer (Kosugi et al., 2004; Yilmaz et al., 2006; Parenti et al., 2007). However, the sensitivity of these tumor markers is very low, which limits their clinical utilization for early detection of esophageal cancer (Handy, 2009). Therefore, it is necessary to search for new biomarkers with high sensitivity and specificity. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) combined with ProteinChip technology is a high-throughput proteomic analysis approach which has been shown to be effective in the detection of biomarkers for early stage malignancy (Petricoin and Liotta, 2004). Using this combined technology, some new serum biomarkers with higher sensitivity were found for the early detection of different cancers, including prostate cancer (Okamoto et al., 2009; Yamamoto-Ishikawa et al., 2009), ovarian cancer (Høgdall et al., 2010; Tang et al., 2010), brain cancer (Liu et al., 2005), colorectal cancer (Yu et al., 2004; Helgason et al., 2010), breast cancer (Hu et al., 2005; Opstal-van Winden et al., 2011), lung cancer (Rathinam et al., 2011), and pancreatic cancer (Felix et al., 2011). However, reports on the protein profiling of different stages of ESCC development have not yet been published.
It was reported that dysplasia (DYS) and basal cell hyperplasia (BCH) are the precancerous lesions of ESCC (Wang et al., 2003). The purpose of this study is to identify support vector machine (SVM) patterns which can be used to distinguish ESCC from DYS, BCH, and healthy controls (HC), and to seek markers associated with esophageal carcinogenesis.
2. Materials and methods
2.1. Patients
One hundred and eight-five serum samples were collected with the agreement of the patients. Thirty serum samples were taken preoperatively from ESCC patients. Twenty-five of these patients were selected for postoperative controls (PO) and serum samples were also taken from them one week after the operation. Samples from 63 HC (22 males, 41 females) and 67 pre-cancer lesion (PCL) patients, including those with DYS (27 cases, 13 males, 14 females) or BCH (40 cases, 15 males, 25 females), were collected from Linxian City, where the incidence of ESCC is the highest in China. The ESCC group consisted of early stage (n=2) and advanced stage (n=28) groups. The DYS group consisted of I stage (n=19), II stage (n=7), and III stage (n=1). The BCH group consisted of different clinical stages: low grade BCH (n=32) and medium grade BCH (n=8). The average ages of the ESCC, DYS, BCH, and HC groups collected from the high incidence area were 59 years (range 42–72 years), 49 years (range 35–70 years), 51 years (range 38–72 years), and 47 years (range 31–69 years), respectively (Table 1). Diagnoses were pathologically confirmed. Patients with acute infection, allergic disease, or autoimmune disease were excluded from the study because these conditions were considered likely to affect the expression of serum proteins. All blood samples were taken from the patients in the morning prior to food intake. The sera were first placed at room temperature for 30 min, and then centrifuged at 2 000 r/min for 20 min. The samples were collected and stored at −80 °C for further investigation.
Table 1.
Sample statistics of each group
| Group | Mean age (year) | Sample number |
|||||
| Men | Women | I stage/low grade | II stage/medium grade | III stage/high grade | IV stage | ||
| ESCC | 59 | 21 | 9 | ||||
| DYS | 49 | 13 | 14 | 19 | 7 | 1 | 0 |
| BCH | 51 | 15 | 25 | 32 | 8 | 0 | |
| HC | 47 | 22 | 41 | ||||
Stage for ESCC and DYS, grade for BCH. There were 25 ESCC postoperative control (PO) samples out of 30 ESCC patients. ESCC: esophageal squamous cell carcinoma; DYS: dysplasia; BCH: basal cell hyperplasia; HC: healthy control
2.2. SELDI ProteinChip analyses
All the samples were ordered randomly with quality control (QC) samples before the experiment to ensure that they were run in blind batches in the ProteinChips. After thawing on ice, the serum samples were centrifuged at 3 000 r/min for 5 min and the supernatants were collected. Then, 10 μl of serum was mixed with 20 μl of lysate (9 mol/L urea, 2% (20 g/L) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane (CHAPS), 0.1% (1 g/L) dithiothreitol (DTT), 50 mmol/L Tris-HCl, pH 9.0 (Sigma, USA)). After vortex-mixing, 360 μl of binding buffer (50 mmol/L sodium acetate, pH 3.5, containing 0.1% (1 g/L) Octyl-β-D-glucopyranoside (OGP) (Sigma)) were added to each serum sample. The weak cation exchange (CM10) chips were assembled in a bioprocessor (Ciphergen Biosystems, Fremont, CA, USA) and every spot on the chips was equilibrated twice with 150 μl of binding buffer for 5 min. Then, 100 μl of diluted serum sample was added to each well of the bioprocessor and agitated for 60 min to allow the protein to combine with the protein chips. After discarding the uncombined samples, each well was washed three times with 150 μl of binding buffer, and then twice with deionized water. After all the spots were air-dried, 1 μl of sinapinic acid (SPA) solution (a semi-saturated solution of sinapinic acid in 50% acetonitrile and 0.5% trifluoroacetic acid) was added to each spot and air-dried. All the chips were read on a protein biological system II mass spectrometer reader. The data for each spot was the average of 65 laser shots. The detection parameters were set as follows: the laser intensity was 170, the detector sensitivity was 6 and the mass range was between 2 and 50 kDa. The all-in-one peptide molecular mass standard was adopted to calibrate the mass accuracy to make the relative mass error less than 0.1%.
2.3. Bioinformatics analysis
All of the data processing was performed using ProteinChip Software 3.2 (Ciphergen Biosystems) to adjust the intensity and molecular weight. First, the baseline was subtracted. Then the spectra intensities of all samples were normalized. After the filtration of noise from the spectra, the automatic peak detection pass was used to detect the markers. Finally, the peaks in different samples were clustered. The values of intensities were standardized within the range from −1 to 1.
2.4. SVM classifier
SVM has been used in various fields as a statistical learning theory system (Vapnik, 1995). It is a powerful analytical tool which is especially suitable for analyzing complex data such as that derived from SELDI-TOF-MS. To distinguish the data from different groups, we constructed a non-linear SVM classifier based on the shareware program OSU SVM V. 3.00 Toolbox, with a radial based function (RBF) kernel, with a parameter Gamma of 0.6 and a cost of the constrain violation of 19. The accuracy, specificity, and sensitivity of the model were estimated using the 1 000 fold cross-validation approach. In this approach, 4/5 of the samples were taken at random to form a training set to fit the parameters of the classifier, and 1/5 to form the test set to assess the performance of the specified classifier. The procedure was repeated 1 000 times. The accuracy, specificity, and sensitivity of the model were calculated based on the average of the 1 000 test results.
2.5. Feature selection and model establishment
The p value of each peak was calculated by nonparametric tests from the biomarker wizard application (Ciphergen Biosystems) and the power of each peak in discriminating different groups was estimated according to the p value. Receiver operating characteristic (ROC) curves were also generated to calculate the areas under the curves (AUCs) using SPSS 10.0. We selected the peaks that had statistically significant differences in their intensities (p≤0.01) as markers with higher individual diagnostic power. All combinations of these markers were used to establish SVM models in order to select the best set of candidate biomarkers. The SVM model with the highest diagnostic power (that with the highest accuracy) was selected to be the final model and the markers in this SVM model were selected as the set of potential biomarkers.
3. Results
3.1. Markers and patterns for discrimination between ESCC and HC
After filtering noises and clustering using Ciphergen ProteinChip Software 3.2, 179 peak clusters from 2 to 100 kDa were detected from all the spectra.
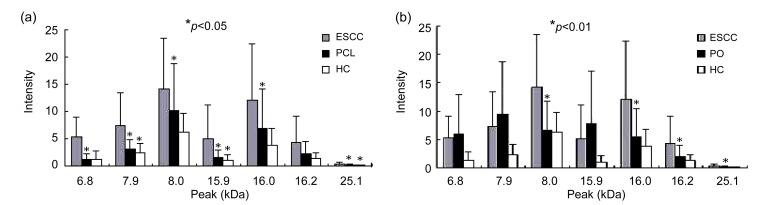
Data derived from ESCC and HC groups were analyzed firstly. The 179 qualifying peaks were ranked by their p values from nonparametric tests. The 12 top-scoring peaks (p<10−5) were selected and the AUCs of these markers were calculated (AUC>0.78). Then, changes in the 12 markers were evaluated in the ESCC, PCL, HC, and PO groups. The markers 6 814 (6.8 kDa), 7 945 (7.9 kDa), 7 989 (8.0 kDa), 15 869 (15.9 kDa), 15 969 (16.0 kDa), 16 172 (16.2 kDa), and 25 127 (25.1 kDa) were up-regulated in ESCC compared with PCL and HC (p<0.05). The intensities of the 7.9, 8.0, 16.0, 15.9, 16.2, and 25.1 kDa markers increased gradually in the HC, PCL, and ESCC groups. The expressions of the 7.9, 15.9, and 25.1 kDa markers also showed significant differences (p<0.05) between the PCL and HC groups (Fig. 1a). In the PO group, the expressions of the 8.0, 16.0, 16.2, and 25.1 kDa markers were much lower than those in the ESCC group (p<0.01) (Fig. 1b).
Fig. 1.
Differences in the expressions of seven markers in ESCC, PCL, and HC groups (a) and in ESCC, PO, and HC groups (b)
(a) Markers showed significantly higher expressions in ESCC than in PCL and HC groups (p<0.05). The expressions of the 7.9, 15.9, and 25.1 kDa markers also showed significant differences (p<0.05) between PCL and HC groups; (b) Expressions of 8.0, 16.0, 16.2, and 25.1 kDa markers in the PO group were significantly lower than those in the ESCC group (p<0.01) (* p<0.05 or * p<0.01 significant difference versus the groups before)
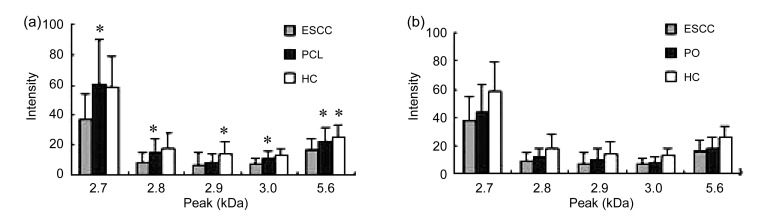
In contrast, the expressions of the 2 749 (2.7 kDa), 2 770 (2.8 kDa), 2 940 (2.9 kDa), 2 956 (3.0 kDa), and 5 646 (5.6 kDa) markers were down-regulated in the ESCC group compared with the HC group. The intensities of the 2.8, 2.9, 3.0, and 5.6 kDa markers decreased gradually in the HC, PCL, and ESCC groups. The expression of the 5.6 kDa marker showed significant differences not only between the ESCC and PCL groups but also between the PCL and HC groups (p<0.05) (Fig. 2a). All these markers showed a tendency to increase after operation (Fig. 2b).
Fig. 2.
Differences in the expressions of five markers in ESCC, PCL, and HC groups (a) and in ESCC, PO, and HC groups (b)
(a) Expressions of the 2.7, 2.8, 3.0, and 5.6 kDa markers decreased significantly in the ESCC group compared with in the PCL and HC groups (p<0.05). The expressions of the 2.9 and 5.6 kDa markers decreased significantly in the PCL group compared with the HC group (p<0.05). * p<0.05 significant difference versus the groups before. (b) All these markers tended to increase expression after operation
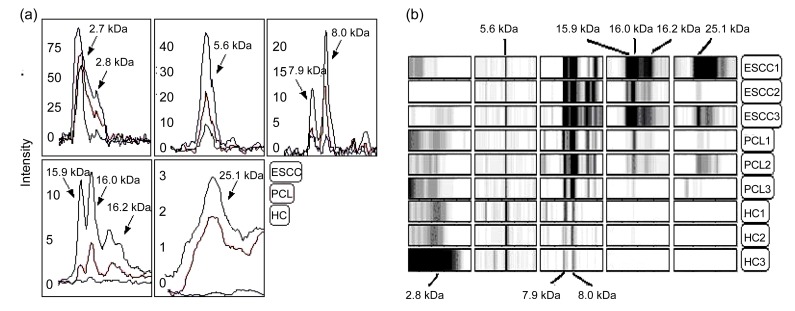
A representative gel view and spectral overlay of these markers are shown in Fig. 3. The 12 peaks from the 74 training set samples were selected and put into SVM models randomly. After the model accuracy calculation, the peak with the highest Youden’s index was selected and used as the optimal diagnosis model to distinguish the different groups. This diagnosis model consisted of five biomarkers with m/z values of 5.6, 2.8, 16.2, 25.1, and 7.9 kDa, respectively. The combination of these five biomarkers generated a higher AUC (0.94) value than the best individual biomarker (0.82; 5.6 kDa). The biomarkers 7.9, 25.1, and 5.6 kDa changed gradually in all the ESCC, PCL, and HC groups (p<0.05) (Figs. 1–3). According to the evaluation of results by leave-one cross-validation, the five-peak SVM model had a specificity of 100% and a sensitivity of 100%.
Fig. 3.
Differential expressions of serum markers
(a) Representative spectral overlays of the serum markers from ESCC (black line), PCL (red line), and HC (blue line) individuals. The overlays show that expressions of the markers 7.9, 8.0, 15.9, 16.0, 16.2, and 25.1 kDa gradually increase from the HC, PC to ESCC group, whereas concurrently the expressions of the markers 2.7, 2.8, and 5.6 kDa decrease in the ESCC group. Numbers in the mass spectra represent the observed mass of the marker in the particular sample; (b) Detection of differentially expressed ovarian cancer associated serum proteins. A representative gel view of ESCC was associated serum markers. Gels from three representative ESCC serum samples are shown at the top, three representative PCL samples in the middle, and three HC spectral profiles at the bottom (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
The remaining 19 serum samples were tested as a blind group and analyzed according to the five-peak SVM model. The results showed that the specificity and sensitivity of the five-peak SVM model for the blind test were 96.8% and 87.1%, respectively.
3.2. Discrimination of esophageal PCL
Esophageal PCL is a very important stage in the carcinogenesis of ESCC. The SVM patterns distinguishing PCL from ESCC and HC groups were also established. The pattern discriminating ESCC from DYS based on five markers had a specificity of 92.2% and a sensitivity of 97.6%. The pattern discriminating ESCC from BCH based on ten markers had a specificity of 97.3% and a sensitivity of 95.6%. The pattern discriminating DYS from HC based on four markers had a specificity of 90.6% and a sensitivity of 84.7%, and the pattern discriminating BCH from HC based on three markers had a specificity of 96.3% and a sensitivity of 59.5% (Table 2). Except for the pattern discriminating BCH from HC, all patterns had high specificity and sensitivity. The PO group was also compared with the ESCC group. The pattern based on two markers had a specificity of 83.3% and a sensitivity of 84.7% (Table 2).
Table 2.
Patterns discriminating ESCC, DYS, BCH, HC, and PO
| Distinguishing pattern | Marker (Da) | Specificity (%) | Sensitivity (%) | Accuracy (%) |
| ESCC vs. HC | 2 770, 5 646, 7 945, 16 172, 25 127 | 96.8 | 87.1 | 93.8 |
| ESCC vs. DYS | 2 749, 3 944, 6 642, 15 869, 28 061 | 92.2 | 97.6 | 95.2 |
| ESCC vs. BCH | 2 162, 2 696, 3 896, 3 944, 4 393 | 97.3 | 95.6 | 96.6 |
| 5 347, 5 925, 6 814, 23 376, 23 527 | ||||
| DYS vs. HC | 2 944, 13 768, 13 978, 28 061 | 90.6 | 84.7 | 84.6 |
| BCH vs. HC | 9 304, 7 781, 8 159 | 96.3 | 59.5 | 82.3 |
| ESCC vs. PO | 2 780, 3 825 | 83.3 | 84.7 | 83.9 |
3.3. Reproducibility
Twelve serum samples from the same healthy person, collected and applied to chips at random, were used to determine the reproducibility of the protein chips. Thirty-five proteins in the range of 4–10 kDa observed on spectra were selected to calculate the coefficient of variance (CV). The CV for the normalized intensity of the 35 selected peaks was 17.95% (<20%) and the CV for the mass of the 35 selected peaks was 0.01% (<0.05%).
3.4. Esophageal carcinoma tumor marker levels
Cytokeratin-19 fragment (CYFRA 21-1), CA 19-9, and CEA have been reported as common tumor markers of esophageal carcinoma. In this paper, these markers were measured in 29 esophageal carcinoma patients and in 56 healthy people using enzyme-linked immunosorbent assay (ELISA), and the sensitivity and specificity of each individual biomarker were also studied. The cut-off points were at 2.0 ng/ml for CYFRA 21-1, 30 U/ml for CA 19-9, and 5 ng/ml for CEA. The analysis results showed that the diagnostic sensitivities of CYFRA 21-1, CA 19-9, and CEA were 17.2% (5/29), 17.2% (5/29), and 27.6% (8/29), respectively. The specificities for CYFRA 21-1, CA 19-9, and CEA were 96.4% (54/56), 89.3% (50/56), and 91.1% (51/56), respectively. The SVM model based on proteomics serum biomarkers had much higher sensitivity and specificity than CYFRA 21-1, CA 19-9, and CEA.
4. Discussion
Early detection is one of the greatest challenges in the study of oncology. The five-year survival rate is more than 90% for early esophageal cancer patients, but only 10%–15% for patients in late or advanced stages (Yang, 1980; Lu et al., 1988). Therefore, prevention and early detection are both very important for improving the prognosis of ESCC. Recent advances in protein profiling technologies for identifying candidate novel tumor biomarkers have raised great interest in searching for cancer biomarkers. New cancer biomarkers could be used as indicators of early-stage disease.
Tumor biomarkers such as CEA, CA 19-9, and SCCA have been widely investigated in the treatment of esophageal cancer patients (Tanaka et al., 2010). However, the application of these markers to the clinical diagnosis of esophageal cancer is still limited by their low sensitivity and specificity. As a soluble CYFRA 21-1, the probability of clinical utilization of CYFRA 21-1 in esophageal cancer has also been tested. CYFRA 21-1 shows higher sensitivity than CEA, CA 19-9, and SCCA (Brockmann et al., 2000; Kawaguchi et al., 2000). However, the sensitivity of all these markers is still less than 10%, and therefore they do not meet the high requirements for early esophageal cancer detection (Nakamura et al., 1998). In our results, we also found that the sensitivity and specificity of individual CEA, CA 19-9, and CYFRA 21-1 biomarkers were very low in screening for esophageal cancer.
ESCC has a multi-factorial nature. An effective detection method can be achieved if we choose a combined diagnosis model instead of using single biomarkers. A combination of SELDI-MS and ProteinChip technology could provide a high-throughput proteomic profiling tool (Adam et al., 2002). The “fingerprints” of ESCC and a unique diagnostic model can also be established if a sophisticated bioinformatics tool is adopted for complex data analysis.
In our study, sera from many groups were collected to build the specific protein profiling model. Markers showing differential expression in the ESCC and HC groups were the first focus. We identified many markers that changed gradually in the ESCC, PCL, and HC groups. The markers 5.6, 7.9, 15.9, and 25.1 kDa showed significant differences among all three groups (p<0.05). The markers 8.0, 16.0, 16.2, and 25.1 kDa that showed increased expression in the ESCC group showed significantly decreased expression (p<0.05) in the postoperative patients. The marker 25.1 kDa was of particular interest because it increased significantly and progressively in the HC, PCL, and ESCC groups and decreased significantly after operation. This marker was selected to build the SVM pattern for the diagnosis of ESCC. The role of biomarker 25.1 kDa is very important in esophageal carcinogenesis.
To estimate the sensitivity and specificity of the combination model established here, 1 000 tests and training sets were compiled by random selection. The sensitivity and specificity were assessed according to the average of the 1 000 tests. Every combination of the markers screened out in the first step was considered. This method may be more time-consuming compared to a stepwise approach, but it can find the best marker combination.
Using ProteinChip technology, different groups of serum protein biomarkers for ESCC were identified with different expression characteristics in the HC, BCH, DYS, and ESCC groups. The aim of our study is to identify biomarkers for ESCC diagnosis as well as to screen the biomarkers associated with the carcinogenesis of ESCC, and to profile the differential protein expression patterns before and after operation. We created SVM patterns distinguishing ESCC from DYS, BCH, and HC groups with high sensitivity, specificity, and overall accuracy. The pattern discriminating BCH from HC yielded unsatisfactory results.
We carried out purification of these protein biomarkers. Other researchers have also announced successful purification of biomarkers using tryptic peptide mapping (Rai et al., 2002) or amino acid sequencing technology (Klade et al., 2001). The marker 28 061 Da, which showed a marked decrease in the expression in the DYS group, was identified as apolipoprotein A-1 by our primary bioinformatics analysis. The down-regulation of this protein in ovarian cancer was reported by Hu et al. (2005). However, many biomarkers of low abundance need further purification.
In conclusion, we identified and selected five biomarker protein patterns to construct a five-peak SVM model for early detection of ESCC. This SVM diagnosis model was used to discriminate among the different stages of esophageal carcinogenesis. The results showed that the specificity and sensitivity of the five-peak SVM model for a blind test were 96.8% and 87.1%, respectively. The advantages of the five-peak SVM diagnosis model compared to a single biomarker indicate that there is a great potential to improve the detection of ESCC by using this kind of combined biomarker model.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30901731) and the Fundamental Research Funds for the Central Universities (No. 2012FZA7004), China
References
- 1.Adam B, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmens OJ, Schellhammer PF, Yasui Y, Ziding F, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62(13):3609–3614. [PubMed] [Google Scholar]
- 2.Brockmann JG, Nottberg H, Glodny B, Sprakel B, Senninger N. Analysis of serum CYFRA 21-1 concentrations in patients with esophageal cancer. Anticancer Res. 2000;20(6D):4899–4904. [PubMed] [Google Scholar]
- 3.Felix K, Fakelman F, Hartmann D, Giese NA, Gaida MM, Schnolzer M, Flad T, Buchler MW, Werner J. Identification of serum proteins involved in pancreatic cancer cachexia. Life Sci. 2011;88(5-6):218–225. doi: 10.1016/j.lfs.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Handy B. The clinical utility of tumor markers. Labmedicine. 2009;40(2):99–103. [Google Scholar]
- 5.Helgason HH, Engwegen J, Zapatka M, Vincent A, Cats A, Boot H, Beijnen JH, Schellens JH. Identification of serum proteins as prognostic and predictive markers of colorectal cancer using surface enhanced laser desorption ionization-time of flight mass spectrometry. Oncol Rep. 2010;24(1):57–64. doi: 10.3892/or_00000828. [DOI] [PubMed] [Google Scholar]
- 6.Høgdall E, Fung ET, Christensen IJ, Yip C, Nedergaard L, Engelholm SA, Risum S, Petri AL, Lundvall L, Lomas L, et al. Proteomic biomarkers for overall and progression-free survival in ovarian cancer patients. Proteom Clin Appl. 2010;4(12):940–952. doi: 10.1002/prca.200900171. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Zhang SZ, Yu JK, Liu J, Zheng S. SELDI-TOF-MS: the proteomics and bioinformatics approaches in the diagnosis of breast cancer. Breast. 2005;14(4):250–255. doi: 10.1016/j.breast.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi H, Ohno S, Miyazaki M, Hashimoto K, Egashira A, Saeki H, Watanabe M, Sugimachi K. CYFRA 21-1 determination in patients with esophageal squamous cell carcinoma: clinical utility for detection of recurrences. Cancer. 2000;89(7):1413–1417. doi: 10.1002/1097-0142(20001001)89:7<1413::AID-CNCR1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 9.Klade CS, Voss T, Krysteck E, Ahorn H, Zatloukal K, Pummer K, Adolf GR. Identification of tumor antigens in renal cell carcinoma by serological proteome analysis. Proteomics. 2001;1(7):890–898. doi: 10.1002/1615-9861(200107)1:7<890::AID-PROT890>3.3.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Kosugi S, Nishimaki T, Kanda T, Nakagawa S, Ohashi M, Hatakeyama K. Clinical significance of serum carcinoembryonic antigen, carbohydrate antigen 19-9, and squamous cell carcinoma antigen levels in esophageal cancer patients. World J Surg. 2004;28(7):680–685. doi: 10.1007/s00268-004-6865-y. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Zheng S, Yu JK, Zhang JM, Chen Z. Serum protein fingerprinting coupled with artificial neural network distinguishes glioma from healthy population or brain benign tumor. J Zhejiang Univ Sci. 2005;6B(1):4–10. doi: 10.1631/jzus.2005.B0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu JB, Yang WX, Zu SK, Chang QL, Sun XB, Lu WQ, Quan PL. Cancer mortality and mortality trends in Henan, China, 1974–1985. Cancer Detect Prev. 1988;13:167–173. [PubMed] [Google Scholar]
- 13.Lu JB, Sun XB, Dai DX, Zu SK, Chang QL. Cancer mortality and mortality trends in Henan Province, China, 1974–1999. Int J Cancer. 2002;13:238. [Google Scholar]
- 14.Nakamura T, Ide H, Eguchi R, Hayashi K, Takasaki K, Watanabe S. CYFRA 21-1 as a tumor marker for squamous cell carcinoma of the esophagus. Dis Esophagus. 1998;11(1):35–39. [PubMed] [Google Scholar]
- 15.Okamoto A, Yamamoto H, Imai A, Hatakeyama S, Iwabuchi I, Yoneyama T, Hashimoto Y, Kote T, Kamimura N, Mori K, et al. Protein profiling of post-prostatic massage urine specimens by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to discriminate between prostate cancer and benign lesions. Oncol Rep. 2009;21(1):73–79. doi: 10.3892/or_00000191. [DOI] [PubMed] [Google Scholar]
- 16.Opstal-van Winden AW, Krop EJ, Karedal MH, Gast MC, Lindh CH, Jeppsson MC, Jonsson BAG, Grobbee DE, Peeters PHM, Beijnen JH, et al. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer. 2011;11:381. doi: 10.1186/1471-2407-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parenti A, Porzionato A, Pizzi S, Guzzardo V, Fassina G, Macchi V, Ninfo V, de Caro R. Expression pattern of squamous cell carcinoma antigen in oesophageal dysplasia and squamous cell carcinoma. Histol Histopathol. 2007;22(9):989–995. doi: 10.14670/HH-22.989. [DOI] [PubMed] [Google Scholar]
- 18.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 19.Petricoin EF, Liotta LA. SELDI-TOF-based serum proteomic pattern diagnostics for early detection of cancer. Curr Opin Biotech. 2004;15(1):24–30. doi: 10.1016/j.copbio.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Rai A, Zhang Z, Rosenzweig J, Shih L, Pham T, Fung E, Sokoll L, Chan D. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126(12):1518–1526. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 21.Rathinam S, Ward DG, James ND, Rajesh PB. Proteomic analysis of resectable non-small cell lung cancer: post-resection serum samples may be useful in identifying potential markers. Interact Cardiovasc Thorac Surg. 2011;13(1):3–6. doi: 10.1510/icvts.2010.260.166. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka K, Yano M, Motoori M, Kishi K, Miyashiro I, Shingai T, Gotoh K, Noura S, Takahashi H, Ohue M, et al. CEA-antigen and SCC-antigen mRNA expression in peripheral blood predict hematogenous recurrence after resection in patients with esophageal cancer. Ann Surg Oncol. 2010;17(10):2779–2786. doi: 10.1245/s10434-010-1075-3. [DOI] [PubMed] [Google Scholar]
- 23.Tang KL, Li TH, Xiong WW, Chen K. Ovarian cancer classification based on dimensionality reduction for SELDI-TOF data. BMC Bioinformatics. 2010;11:109. doi: 10.1186/1471-2105-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vapnik V. The Nature of Statistical Learning Theory. New York: Springer-Verlag; 1995. [Google Scholar]
- 25.Wang LD, Zheng S, Liu B, Zhou JX, Li YJ, Li JX. CYP1A1, GSTs and mEH polymorphisms and susceptibility to esophageal carcinoma: study of population from a high-incidence area in north China. World J Gastroenterol. 2003;9(7):1394–1397. doi: 10.3748/wjg.v9.i7.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto-Ishikawa K, Suzuki H, Nezu M, Kamiya N, Imamoto T, Komiya A, Sogawa K, Tomonaga T, Nomura F, Ichikawa T. The isolation and identification of apolipoprotein C-I in hormone-refractory prostate cancer using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Asian J Androl. 2009;11(3):299–307. doi: 10.1038/aja.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CS. Research on esophageal cancer in China: a review. Cancer Res. 1980;40(8):2633–2644. [PubMed] [Google Scholar]
- 28.Yilmaz O, Eroglu A, Dag E, Karaoglanoglu N, Yilmaz A. Serum levels of IGF-I and IGFBP-III and their relation with carcinoembryonic antigen and carbohydrate antigen 19-9 in cases of esophageal cancer. Int J Clin Pract. 2006;60(12):1604–1608. doi: 10.1111/j.1742-1241.2006.00854.x. [DOI] [PubMed] [Google Scholar]
- 29.Yu JK, Chen YD, Zheng S. An integrated approach to the detection of colorectal cancer utilizing proteomics and bioinformatics. World J Gastroenterol. 2004;10(21):3127–3131. doi: 10.3748/wjg.v10.i21.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]