Abstract
Lipoprotein-associated phospholipase A2 (Lp-PLA2), specifically Group VIIA PLA2, is a member of the phospholipase A2 superfamily and is found mainly associated with LDL and HDL in human plasma. Lp-PLA2 is considered as a risk factor, a potential biomarker, a target for therapy in the treatment of cardiovascular disease, and evidence suggests that the level of Lp-PLA2 in plasma is associated with the risk of future cardiovascular and stroke events. The differential location of the enzyme in LDL/HDL lipoproteins has been suggested to affect Lp-PLA2 function and/or its physiological role and an abnormal distribution of the enzyme may correlate with diseases. Although a mutagenesis study suggested that a surface helix (residues 362–369) mediates the association between Lp-PLA2 and HDL, the molecular details and mechanism of association has remained unknown. We have now employed hydrogen deuterium exchange mass spectrometry to characterize the interaction between recombinant human Lp-PLA2 and human HDL. We have found that specific residues 113–120, 192–204, and 360–368 likely mediate HDL binding. In a previous study, we showed that residues 113–120 are important for Lp-PLA2-liposome interactions. We now find that residues 192–204 show a decreased deuteration level when Lp-PLA2 is exposed to apoA-I, but not apoA-II, the most abundant apoproteins in HDL, and additionally, residues 360–368 are only affected by HDL.The results suggest that apoA-I and phospholipid membranes play crucial roles in Lp-PLA2 localization to HDL.
Keywords: lipoprotein-associated phospholipase A2, high density lipoprotein, apolipoproteins, enzymology
Human lipoprotein-associated phospholipase A2 (Lp-PLA2), also known as plasma platelet activating factor acetylhydrolase, belongs to the phospholipase A2 superfamily (designated as Group VIIA PLA2) (1). Lp-PLA2 is found mainly associated with lipoproteins in human plasma, about 70% with LDL and another 30% with HDL (2). Lp-PLA2 was first identified as an enzyme that inactivates platelet activating factor (PAF) by hydrolyzing the acetyl group at the sn-2 position to produce lyso-PAF and acetate (3). Later, it was found that Lp-PLA2 has comparable activity toward phosphatidycholines (PCs) containing short acyl chains at the sn-2 position, oxidized PCs (oxPCs) and F2-isoprostanes esterified phospholipids, which are phospholipids with oxidized fatty acyl chains at the sn-2 position (4–6). Lp-PLA2 is secreted predominantly by macrophages (7) and it has been shown that its expression is significantly increased during activation of macrophages in atherosclerotic lesions (8). By inactivating PAF and hydrolyzing oxPC in the oxidized LDL particles, Lp-PLA2 could act as an anti-atherogenic enzyme (9). However, a large number of studies have suggested a pro-atherosclerotic role (10–12). This is because Lp-PLA2 generates the pro-inflammatory and pro-apoptotic lipid mediators lyso-PC and oxidized nonesterified fatty acids, which play an important role in the development of atherosclerotic necrotic cores.
Increased Lp-PLA2 mass or activity in plasma is believed to be associated with an increased risk of various cardiovascular diseases (13). Therefore, Lp-PLA2 is considered as a biomarker for cardiovascular diseases and it has been approved as a diagonostic for stroke and coronary disease risk by the US Food and Drug Administration (14). In addition to serving as a biomarker, Lp-PLA2 is also considered as a target for the treatment of cardiovascular disease. A GSK inhibitor, Darapladid or SB-40848, has been shown to be a good selective inhibitor and has been successfully applied in a diabetic and hypercholesterolemic swine model to reduce the development of advanced coronary atherosclerosis (15). Lp-PLA2 is found predominantly bound to lipoproteins in human plasma and a small amount may also be bound to other microparticles (16). The location of the enzyme on LDL or HDL may alter the protein's catalytic behavior. The enzyme associated with LDL appears to be more active than the same enzyme associated with HDL (17). In vitro assays using low PAF concentrations that mimic physiological levels showed that Lp-PLA2 associated with HDL particles is inactive (2). LDL-associated Lp-PLA2 plays a pro-inflammatory and pro-atherogenic role because it can hydrolyze oxPC to generate pro-atherogenic mediators, lyso-PC, and oxidized fatty acids. Additionally, Lp-PLA2 shows preferential association with dense LDL and the very dense lipoprotein subfraction in human plasma, which contains the most atherogenic LDL particles (18). Although only about 30% of Lp-PLA2 in normal human plasma associates with HDL, this ratio varies greatly among different animal models and growing evidence supports an anti-atherogenic role of HDL associated Lp-PLA2 (19, 20). The ratio of Lp-PLA2 associated with HDL or LDL to the total plasma enzyme may serve as a useful biomarker of atherogenicity in patients with dyslipidemias (21). Together, the distribution and location of Lp-PLA2 in LDL/HDL lipoproteins may regulate Lp-PLA2 catalytic activity, function, and/or its physiological roles. Therefore, a precise characterization of the interaction between Lp-PLA2 and lipoproteins is necessary. Previously, residues 115, 116, and 205 were suggested to contribute to the association of Lp-PLA2 with LDL (22). The C terminus of apoB-100 could be important for LDL binding to Lp-PLA2 (22). Although a domain on Lp-PLA2 containing residues 367–370 has been proposed to mediate Lp-PLA2/HDL association (23), the detailed molecular basis of Lp-PLA2/HDL association is not clear. The binding interactions between Lp-PLA2 and HDL may be involved in more than one binding region or accompany conformational changes.
The large size of HDL particles and their heterogeneity (24) and the complexity of lipoprotein-enzyme interactions preclude the use of many standard biophysical analytical techniques including X-ray crystallography and solution NMR. Previously, we have used peptide amide deuterium exchange mass spectrometry (DXMS) to investigate Lp-PLA2 /liposome interactions (25). DXMS has been widely used to analyze protein-protein, protein-substrate, and protein-inhibitor interactions, as well as protein dynamics and protein conformational changes (26–28). Moreover, the interactions between membrane proteins and recombinant HDL like nanodiscs have been successfully studied with DXMS (29). Herein, we employ DXMS to characterize the association of Lp-PLA2 with HDL.
EXPERIMENTAL METHODS
Materials
D2O (99.6%) was obtained from Cambridge Isotope Laboratories. [3H-acteyl] PAF was purchased from Perkin Elmer. Unlabeled PAF and 1,2-dimyristoyl-sn-glycero-3-phosphocholine were from Avanti Polar Lipids (Alabaster, AL). Human human HDL, human apoA-I human apoA-II, and BSA were purchased from Sigma-Aldrich. All other reagents were analytical reagent grade or better.
Preparation of recombinant protein and activity assay
The recombinant human Lp-PLA2 protein including residues 47-441 with an additional alanine residue at the N terminus was prepared as described previously (25). Purified Lp-PLA2 was concentrated to 2.5 mg/ml in protein buffer (20 mM Tris, 150 mM NaCl, 10% glycerol, pH 7.5, 1 mM DTT) with an Amicon Ultra-15 (Millipore) with the protein concentration determined by bicinchoninic acid protein assay. The enzyme activity was determined by a radiometric assay described elsewhere (30). The assay was performed in 100 mM HEPES buffer (pH = 7.5, 1 mM EGTA, 1 mM DTT, 2 mM CHAPS) by using 0.1 mM [3H-acteyl] PAF as substrate in a final volume of 500 µl. The reaction was initiated by adding 1 ng purified Lp-PLA2 and then was incubated at 37°C for 20 min. The reaction was quenched using 500 µl 10 M acetic acid. The released 3H-acetate was separated by C18 reversed-phase cartridges (Phenomenex) and quantitated by scintillation counting.
Preparation of samples for DXMS experiments
D2O buffer consisted of 12 mM Tris, 50 mM NaCl in 99% D2O, pDread 7.5, as previously described (31–35). Exchange experiments were initiated by mixing 20 µL of Lp-PLA2 (2.5 mg/ml) in protein buffer, or Lp-PLA2 preincubated with human HDL, apoA-I, apoA-II, or BSA with 60 µL of D2O buffer to a final concentration of 50% D2O. The mixtures were incubated at 0°C for 10, 30, 100, 300, 1,000, 3,000, or 10,000 s and then the exchange reaction was quenched by adding 120 µl of ice-cold quench solution (0.96% formic acid, 0∼0.8 M guanidine hydrochloride) resulting in samples with final concentrations of 0.58% formic acid and 0∼0.5 M guanidine hydrochloride, pH 2.5. The samples were then immediately frozen on dry ice and stored at −80°C.
Proteolysis: LC/MS analysis of samples
All steps were performed at 0°C as described previously (25). Data processing of DXMS experiments utilized specialized software as previously described (DXMS Explorer, Sierra Analytics Inc.) (25).
RESULTS
Identification of HDL/Lp-PLA2 binding sites by DXMS
The Lp-PLA2 protein peptide digestion was optimized as described previously and gave a 94% coverage of recombinant protein (25).
However, due to interference by the peptic peptides from the apoproteins in the HDL, in the studies with HDL, we identified 56 unique peptides with the least number of overlapping residues, which gave an 84% coverage in the present Lp-PLA2/HDL DXMS study. These peptides were used to determine the deuteration level of Lp-PLA2. According to a reported Lp-PLA2/HDL binding assay (23), 800 µg of purified Lp-PLA2 was incubated with 10 mg HDL particles for at least 30 min to reach maximum binding, so we carried out a similar incubation before applying DXMS to our Lp-PLA2/HDL association experiments. Lp-PLA2 alone or previously admixed with HDL particles was then supplemented with D2O buffer to a final concentration of 50% D2O and aliquots were exchange-quenched in formic acid without guanidine hydrochloride at intervals from 10 s to 10,000 s at 0°C. We utilized three different batches of human HDL purchased from Sigma and ran DXMS experiments on each batch. The absolute deuteration values of Lp-PLA2 peptides identified in Lp-PLA2/HDL interaction experiments with each batch of HDL were substantially different from one another; however, the relative differences in deuteration values of Lp-PLA2 peptides in the presence and absence of HDL were similar for all three human HDL samples.
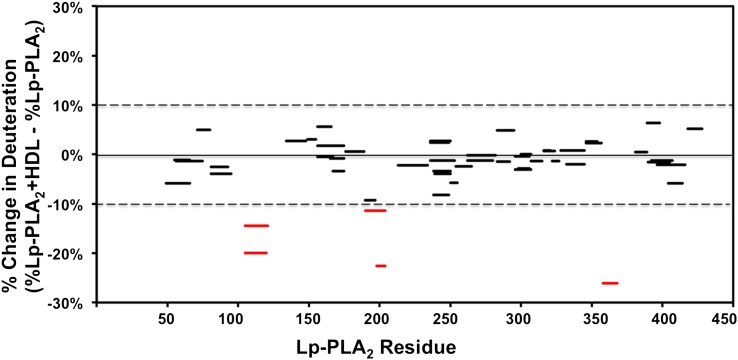
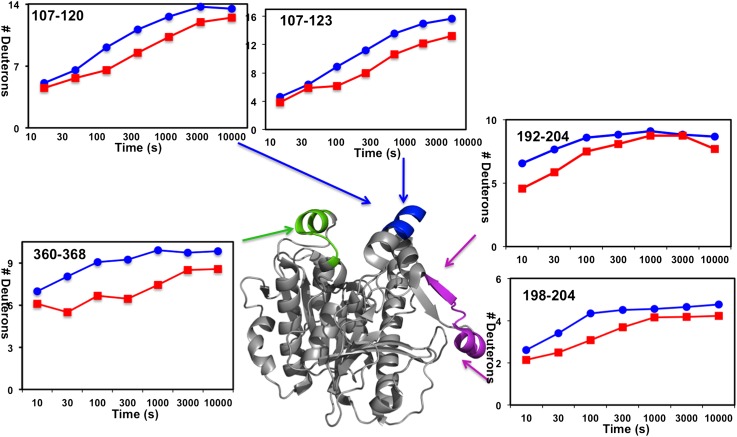
The on-exchange results of Lp-PLA2 alone and Lp-PLA2 associated with HDL for one typical sample of human HDL are shown in Fig. 1 and supplementary Fig. I. Five peptides from three distinct regions of Lp-PLA2 had over a 10% reduction when incubated in the presence of HDL at 100 s (Fig. 1). Similar peptides, identified using two other batches of human HDL, showed similar deuteration changes as shown in in supplementary Fig. II. These three regions (containing peptides 107–120, 192–204, and 360–368) showed a significant decrease in deuteron incorporation throughout the entire time course in the presence of HDL. The time courses for Hydrogen/Deuterium (H/D) exchange in the five probe peptides that cover these three regions are shown in Fig. 2. In the region 107–120, peptides 107–120 and 107–123 both showed a slow H/D exchange at the beginning of the 10 s and 30 s time points and both incorporated two deuterons less at later time points through 10,000 s in the presence of HDL. This region has been reported to mediate Lp-PLA2/liposome interactions in our previous work (25). Considering that HDL contains about 20–35% phospholipids as a monolayer on its surface, it is reasonable that Lp-PLA2 binds to the phospholipids on the HDL surface when associating with HDL at its liposome binding site. Peptide 360–368, a surface α-helix, which had previously been demonstrated to be important for Lp-PLA2 /HDL interactions (23), showed an average of 2 deuterons (22%) decrease of deuteration after 10 s of H/D exchange. Amides 192–204 showed a two deuteron decrease at the first four time points and increased deuteration after 1,000 s. Amides 198–204 showed a similar deuteration pattern with amides 192–204. These two peptides showed a one deuteron decrease at earlier time points and showed no difference with HDL after 1,000 s. Comparing the deuteration change of amides 192–204 and 198–204, we conclude that the one deuteron decrease is caused by one of the amides in 192–197 and that amides 198–204 contribute to the rest of the deuteron decrease. Peptide probes immediately flanking the regions 107–120, 192–204, and 360–368 showed identical deuterium incorporation in the presence or absence of HDL, as did all other peptides assessed throughout Lp-PLA2 (see supplementary Fig. I).
Fig. 1.
Deuterium exchange of Lp-PLA2 in the presence of human HDL. The difference in the percentage between deuterium incorporation for individual peptides of Lp-PLA2 in the presence (red) or absence (blue) of HDL at 100 s is shown. Peptides are shown as bars spanning over the indicated sequence on the x axis. All changes greater than 10% are shown in red.
Fig. 2.
Deuterium exchange of Lp-PLA2 upon association with human HDL. The number of incorporated deuterons at each of the seven time points from 10 to 10,000 s in Lp-PLA2 alone (blue) and Lp-PLA2 in the presence of HDL (red) are plotted for Lp-PLA2 peptides containing amides 107–120, 107–123, 192–204, 198–204, and 360–368.
ApoA-I protein mediates human HDL binding to Lp-PLA2
As we identified three regions of Lp-PLA2 that associate with HDL particles and one region among them is the Lp-PLA2/liposome binding site, we next tested the possibility of the involvement of apolipoproteins in Lp-PLA2/HDL interactions. It seemed likely that apoA-I protein would be the protein component that mediates human HDL/Lp-PLA2 interaction because apoA-I protein constitutes about 70% of the HDL apoprotein component and is the major protein component in HDL. Furthermore, two molecules of apoA-I protein form a double-belt HDL skeleton. Lp-PLA2 is preferentially associated with dense HDL in human plasma, which contains a higher apoA-I protein to lipid ratio (18). Overexpression of apoA-I in apoE-deficient mice increases HDL associated Lp-PLA2 activity (36). These all suggest that apoA-I may play an important role in the association of Lp-PLA2 with HDL. To determine the structural basis of Lp-PLA2/apoA-I interactions, we employed DXMS to characterize the association between Lp-PLA2 and apoA-I.
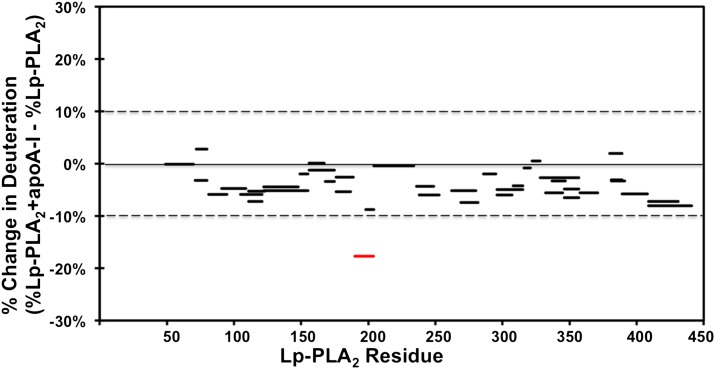
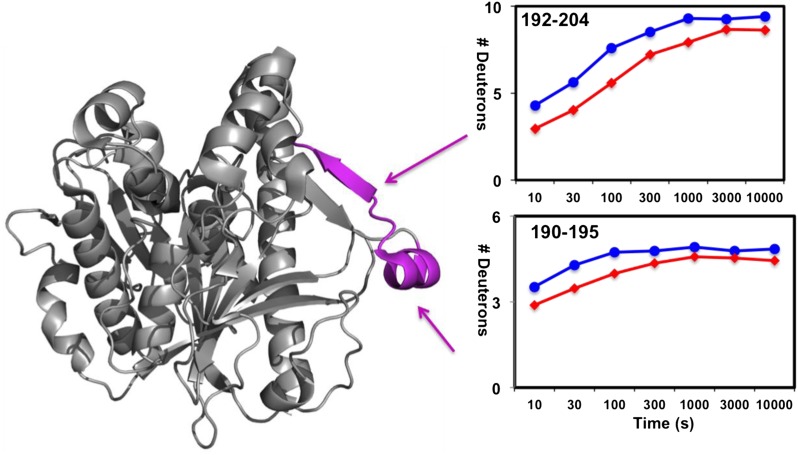
Purified Lp-PLA2 was incubated with lipid-free apoA-I protein for at least 30 min to reach maximum binding before carrying out Lp-PLA2/apoA-I DXMS experiments. Lp-PLA2 alone or previously admixed with apoA-I was then incubated with D2O buffer to a final concentration of 50% D2O at 0°C. The aliquots at intervals from 10 s to 10,000 s were exchange-quenched in formic acid with 0.5M guanidine hydrochloride. The on-exchange results of Lp-PLA2 alone and apoA-I bound Lp-PLA2 are shown in Figs. 3, 4, and supplementary Fig. III. The only region that shows a significant difference in deuteration between Lp-PLA2 and apoA-I bound Lp-PLA2 was at a surface region including amides 192–204 (Fig. 3). Two probed peptides in this region, as shown in Fig. 4, demonstrated a similar pattern with the peptides we found in the same region in Lp-PLA2/HDL experiments. Amides 192–204 show a two deuteron decrease in the presence of apoA-I. Amides 190–195 showed a one deuteron decrease. The amino acid sequence of this region contains mostly polar or hydrophilic residues. Apparently, the interaction between Lp-PLA2 and apoA-I is predominantly by electrostatic interactions as the cytosolic apoA-I protein has a hydrophilic surface. Actually, electrostatic interactions were proposed to occur between Lp-PLA2 and apoB-100 in LDL (37). Although apoA-I has been suggested to exist in different conformations between the lipid-free and lipid-bound forms (38, 39), we used only the lipid-free form in our DXMS study. It is likely that Lp-PLA2 binds the same hydrophilic surface in both conformations. Region 192–204 was also identified to interact with HDL in Lp-PLA2/HDL DXMS experiments. This indicates that Lp-PLA2 binds apoA-I when interacting with HDL particles. The current results strongly support our hypothesis that apoA-I plays an important role in Lp-PLA2/HDL interactions.
Fig. 3.
Deuterium exchange information on Lp-PLA2 in the presence of human apoA-I. The difference in percentage between deuterium incorporation for individual peptides of Lp-PLA2 in the presence (red) or absence of apoA-I (blue) at 100 s is shown. Peptides are shown as bars spanning over the indicated sequence on the x axis. All changes greater than 10% are shown in red.
Fig. 4.
Deuterium exchange of Lp-PLA2 upon association with human apoA-I. The number of incorporated deuterons at each of the seven time points from 10 to 10,000 s in Lp-PLA2 alone (blue) and Lp-PLA2 in the presence of apoA-I (red) are plotted for Lp-PLA2 peptides containing amides 190–195 and 192–204.
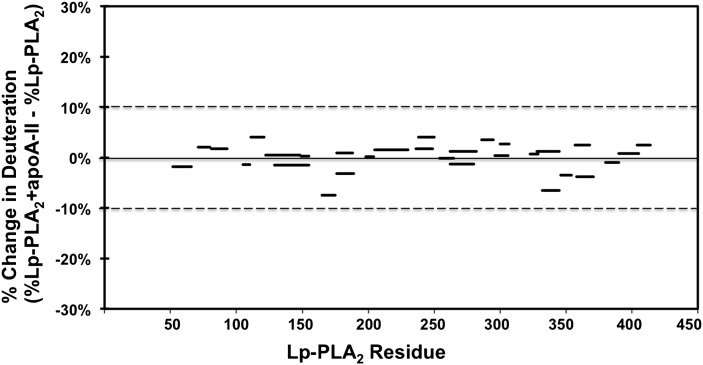
Although the apoA-I protein constitutes over 70% of the total protein component of HDL, we cannot exclude the possibility that other apolipoproteins might be involved in Lp-PLA2/HDL association. To resolve this question, we carried out a similar DXMS experiment to test the interaction between Lp-PLA2 and apoA-II protein, which is the second most abundant protein component in HDL. As shown in both Fig. 5 and supplementary Fig. IV, Lp-PLA2 did not show any deuteration difference in the absence or presence of apoAII. Based on our DXMS results, it appears that apoA-I but not apoA-II affects the interaction between Lp-PLA2 and HDL.
Fig. 5.
Deuterium exchange information on Lp-PLA2 in the presence of human apoA-II. The difference in percentage between deuterium incorporation for individual peptides of Lp-PLA2 in the presence or absence of apoA-II at 100 s is shown. Peptides are shown as black bars spanning over the indicated sequence on the x axis. All changes greater than 10% are considered significant.
As a further control for the specificity of interaction between apoA-I and Lp-PLA2, BSA as a representative of a typical plasma protein was incubated with purified Lp-PLA2 and DXMS experiments were carried out analogously to those on apoA-I and apoA-II described above. Lp-PLA2 peptides detected in the BSA control experiment did not show any deuteron difference in the presence or absence of BSA (see supplementary Fig. V). This result unambiguously indicates that apoA-I and Lp-PLA2 interact specifically.
DISCUSSION
Lp-PLA2 has attracted considerable attention during the past decade. It has been considered as a biomarker and a therapeutic target for atherosclerosis. Lp-PLA2 is mainly associated with LDL and HDL in human plasma. The protein associated with these two different lipoproteins has been shown to have opposite functions in atherosclerosis development. Lp-PLA2 associated with LDL has shown a much higher enzymatic activity toward PAF than the same enzyme bound to HDL (17). The HDL may sequester the Lp-PLA2 in a manner that the active site is inaccessible to substrate and thereby the HDL-bound form may serve as a reservoir whenever excess enzyme is present. Therefore, the interaction of Lp-PLA2 and LDL could be different than with HDL. Earlier studies have indicated that Lp-PLA2 uses one of the surface α-helicies to bind to the C terminus of apoB-100 when associated with LDL (22). Although a recent study identified residues 367–370 as mediating Lp-PLA2/HDL association (23), the structural characteristics that determine the distribution of lipoproteins are still not well understood.
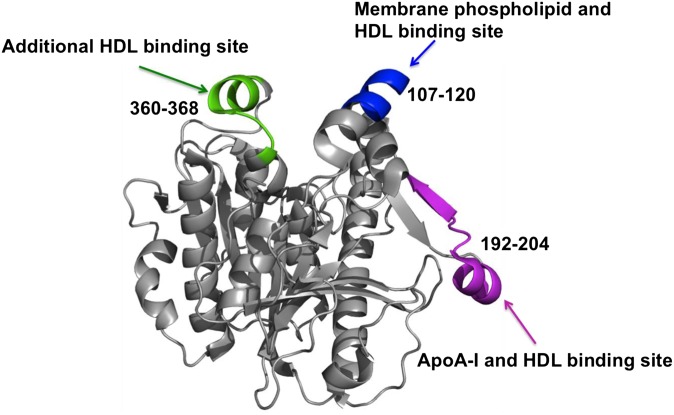
In our previous work, we employed DXMS to identify a domain mediating Lp-PLA2/liposome interactions (25). Specifically, residues Trp115 and Leu116 were found to be crucial for the interaction and most likely penetrate into the membrane phospholipid to mediate Lp-PLA2/liposome association (25). In the present study, we used DXMS to explore the domains involved in Lp-PLA2/HDL interactions and identified the component in HDL that interacts with Lp-PLA2. HDL is the smallest lipoprotein in human plasma and has the highest proportion of protein to cholesterol. ApoA-I and apoA-II comprise the majority of the HDL protein mass and they consititute about 70% and 20%, respectively. Besides the lipoproteins, the HDL surface also contains about 30% phospholipids. Considering the protein and lipid components of HDL and the properties of Lp-PLA2, it is reasonable to hypothesze that Lp-PLA2 binds to the phospholipids and/or lipoproteins. Moreover, early studies have shown that overexpression of apoA-I in apoE-deficient mice increases HDL associated Lp-PLA2 activity (36), which indicates the important role of apoA-I in Lp-PLA2/HDL interactions. Our results showed that regions 107–120, 192–204, and 360–368 contribute to Lp-PLA2/HDL association, region 192–204 contributes to Lp-PLA2/apoA-I interactions, and apoA-II is not involved in Lp-PLA2/HDL association. As shown in Fig. 6, all three regions identified in the current study are located on the Lp-PLA2 surface. ApoA-I and HDL share the same Lp-PLA2 binding region 192–204, which unambiguously indicates that Lp-PLA2 could bind to apoA-I when associated with HDL. Region 107–120 was also found to be critical in Lp-PLA2/liposome interactions (25), which suggests that the phospholipid on the monolayer surface is also involved in Lp-PLA2/HDL association. Region 360–368 is part of a surface α-helix and was found to be important for Lp-PLA2/HDL interactions (23). Although we have found this domain is important in Lp-PLA2/HDL interactions, we could not identify the exact HDL component responsible for interactions in this region so it could be some sort of combined phospholipid/protein site.
Fig. 6.
Peptide regions (107–120, 192–204, and 360–368) that interact with HDL are mapped on the Lp-PLA2 crystal structure.
It has been proposed that the differential locations of Lp-PLA2 in LDL/HDL lipoproteins affect Lp-PLA2 function and/or its physiological role and abnormal distribution of the enzyme may correlate with diseases (40). Lp-PLA2 has been shown to be more active when it is localized in LDL versus HDL particles (17). Assays performed at low PAF concentrations that mimic physiological levels showed that enzyme associated with HDL particles is inactive (2). This suggests that Lp-PLA2 interaction with HDL may form a noncatalytic or inhibitory conformation but that could be different when the same enzyme associates with LDL. Our DXMS experiments showed that three domains mediate the association of Lp-PLA2 with HDL and two of these three domains did not interact significantly with phospholipid membrane surfaces. In contrast, residues Trp115, Leu116, and Met117, which were shown by mutagenesis to be important for Lp-PLA2/LDL interactions, were all included in the Lp-PLA2/liposome interaction surface we saw with DXMS (22, 25). The enzyme associated with HDL may require a conformational change to access the lipid membrane surface for the hydrolysis reaction. Alternatively, Lp-PLA2 may have to be regulated to dissociate from HDL to regain its activity. This may explain why Lp-PLA2 associated with HDL appears to not be as active as the protein associated with LDL. The C terminus of apoB-100 was suggested to be involved in the binding of Lp-PLA2 with LDL (22). In the present study, we have demonstrated that apoA-I plays a very important role in Lp-PLA2/HDL association. The distribution of Lp-PLA2 between LDL and HDL may be closely related to its binding affinity with apoB-100 and apoA-I.
The work reported in this paper represents the first study of the molecular basis of Lp-PLA2/HDL interactions in solution. We employed DXMS to characterize the interaction between Lp-PLA2 and human HDL and apoA-I protein. We found that specific residues 113–120 and 192–206 likely mediate HDL binding and ApoA-I may play an important role in Lp-PLA2 and HDL association.
Supplementary Material
Acknowledgments
The authors thank Drs. Ewa Ninio and John Chapman, Université Pierre et Marie Curie, Paris, for insightful discussions about the association of Lp-PLA2 with lipoproteins.
Footnotes
Abbreviations:
- DXMS
- deuterium exchange mass spectrometry
- Lp-PLA2
- lipoprotein-associated phospholipase A2
- PAF
- platelet activating factor
- PC
- phosphatidycholine
- oxPC
- oxidized phosphatidycholine
This study was supported by the National Institutes of Health grant RO1-GM20501 (E.A.D.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Dennis E. A., Cao J., Hsu Y. H., Magrioti V., Kokotos G. 2011. Phospholipase a(2) enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111: 6130–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stafforini D. M., McIntyre T. M., Carter M. E., Prescott S. M. 1987. Human plasma platelet-activating factor acetylhydrolase. Association with lipoprotein particles and role in the degradation of platelet-activating factor. J. Biol. Chem. 262: 4215–4222 [PubMed] [Google Scholar]
- 3.Tjoelker L. W., Eberhardt C., Unger J., Trong H. L., Zimmerman G. A., McIntyre T. M., Stafforini D. M., Prescott S. M., Gray P. W. 1995. Plasma platelet-activating factor acetylhydrolase is a secreted phospholipase A2 with a catalytic triad. J. Biol. Chem. 270: 25481–25487 [DOI] [PubMed] [Google Scholar]
- 4.Stremler K. E., Stafforini D. M., Prescott S. M., McIntyre T. M. 1991. Human plasma platelet-activating factor acetylhydrolase. Oxidatively fragmented phospholipids as substrates. J. Biol. Chem. 266: 11095–11103 [PubMed] [Google Scholar]
- 5.Stafforini D. M., Sheller J. R., Blackwell T. S., Sapirstein A., Yull F. E., McIntyre T. M., Bonventre J. V., Prescott S. M., Roberts L. J., 2nd 2006. Release of free F2-isoprostanes from esterified phospholipids is catalyzed by intracellular and plasma platelet-activating factor acetylhydrolases. J. Biol. Chem. 281: 4616–4623 [DOI] [PubMed] [Google Scholar]
- 6.Davis B., Koster G., Douet L. J., Scigelova M., Woffendin G., Ward J. M., Smith A., Humphries J., Burnand K. G., Macphee C. H., et al. 2008. Electrospray ionization mass spectrometry identifies substrates and products of lipoprotein-associated phospholipase A2 in oxidized human low density lipoprotein. J. Biol. Chem. 283: 6428–6437 [DOI] [PubMed] [Google Scholar]
- 7.Stafforini D. M., Elstad M. R., McIntyre T. M., Zimmerman G. A., Prescott S. M. 1990. Human macrophages secret platelet-activating factor acetylhydrolase. J. Biol. Chem. 265: 9682–9687 [PubMed] [Google Scholar]
- 8.Elstad M. R., Stafforini D. M., McIntyre T. M., Prescott S. M., Zimmerman G. A. 1989. Platelet-activating factor acetylhydrolase increases during macrophage differentiation. A novel mechanism that regulates accumulation of platelet-activating factor. J. Biol. Chem. 264: 8467–8470 [PubMed] [Google Scholar]
- 9.Noto H., Hara M., Karasawa K., Iso O. N., Satoh H., Togo M., Hashimoto Y., Yamada Y., Kosaka T., Kawamura M., et al. 2003. Human plasma platelet-activating factor acetylhydrolase binds to all the murine lipoproteins, conferring protection against oxidative stress. Arterioscler. Thromb. Vasc. Biol. 23: 829–835 [DOI] [PubMed] [Google Scholar]
- 10.Thompson A., Gao P., Orfei L., Watson S., Di Angelantonio E., Kaptoge S., Ballantyne C., Cannon C. P., Criqui M., Cushman M., et al. 2010. Lipoprotein-associated phospholipase A(2) and risk of coronary disease, stroke, and mortality: collaborative analysis of 32 prospective studies. Lancet. 375: 1536–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casas J. P., Ninio E., Panayiotou A., Palmen J., Cooper J. A., Ricketts S. L., Sofat R., Nicolaides A. N., Corsetti J. P., Fowkes F. G., et al. 2010. PLA2G7 genotype, lipoprotein-associated phospholipase A2 activity, and coronary heart disease risk in 10 494 cases and 15 624 controls of European ancestry. Circulation. 121: 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilensky R. L., Macphee C. H. 2009. Lipoprotein-associated phospholipase A(2) and atherosclerosis. Curr. Opin. Lipidol. 20: 415–420 [DOI] [PubMed] [Google Scholar]
- 13.Packard C. J. 2009. Lipoprotein-associated phospholipase A2 as a biomarker of coronary heart disease and a therapeutic target. Curr. Opin. Cardiol. 24: 358–363 [DOI] [PubMed] [Google Scholar]
- 14.Saenger A. K., Christenson R. H. 2010. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin. Chem. 56: 21–33 [DOI] [PubMed] [Google Scholar]
- 15.Wilensky R. L., Shi Y., Mohler E. R., 3rd, Hamamdzic D., Burgert M. E., Li J., Postle A., Fenning R. S., Bollinger J. G., Hoffman B. E., et al. 2008. Inhibition of lipoprotein-associated phospholipase A2 reduces complex coronary atherosclerotic plaque development. Nat. Med. 14: 1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsios J. V., Vini M. P., Stengel D., Ninio E., Tselepis A. D. 2006. Human platelets secrete the plasma type of platelet-activating factor acetylhydrolase primarily associated with microparticles. Arterioscler. Thromb. Vasc. Biol. 26: 1907–1913 [DOI] [PubMed] [Google Scholar]
- 17.Stafforini D. M., Carter M. E., Zimmerman G. A., McIntyre T. M., Prescott S. M. 1989. Lipoproteins alter the catalytic behavior of the platelet-activating factor acetylhydrolase in human plasma. Proc. Natl. Acad. Sci. USA. 86: 2393–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tselepis A. D., Dentan C., Karabina S. A., Chapman M. J., Ninio E. 1995. PAF-degrading acetylhydrolase is preferentially associated with dense LDL and VHDL-1 in human plasma. Catalytic characteristics and relation to the monocyte-derived enzyme. Arterioscler. Thromb. Vasc. Biol. 15: 1764–1773 [DOI] [PubMed] [Google Scholar]
- 19.Theilmeier G., De Geest B., Van Veldhoven P. P., Stengel D., Michiels C., Lox M., Landeloos M., Chapman M. J., Ninio E., Collen D., Himpens B., Holvoet P.2000. HDL-associated PAF-AH reduces endothelial adhesiveness in apoE−/− mice. FASEB J. 14: 2032–2039. [DOI] [PubMed]
- 20.Okamura K., Miura S., Zhang B., Uehara Y., Matsuo K., Kumagai K., Saku K.2007. Ratio of LDL- to HDL-associated platelet-activating factor acetylhydrolase may be a marker of inflammation in patients with paroxysmal atrial fibrillation. Circ. J.71: 214–219. [DOI] [PubMed]
- 21.Tsimihodimos V., Karabina S. A., Tambaki A. P., Bairaktari E., Miltiadous G., Goudevenos J. A., Cariolou M. A., Chapman M. J., Tselepis A. D., Elisaf M. 2002. Altered distribution of platelet-activating factor- acetylhydrolase activity between LDL and HDL as a function of the severity of hypercholesterolemia. J. Lipid Res. 43: 256–263 [PubMed] [Google Scholar]
- 22.Stafforini D. M., Tjoelker L. W., McCormick S. P., Vaitkus D., McIntyre T. M., Gray P. W., Young S. G., Prescott S. M. 1999. Molecular basis of the interaction between plasma platelet-activating factor acetylhydrolase and low density lipoprotein. J. Biol. Chem. 274: 7018–7024 [DOI] [PubMed] [Google Scholar]
- 23.Gardner A. A., Reichert E. C., Topham M. K., Stafforini D. M. 2008. Identification of a domain that mediates association of platelet-activating factor acetylhydrolase with high density lipoprotein. J. Biol. Chem. 283: 17099–17106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kontush A. C., John M.2011. High-density lipoproteins: structure, metabolism, function and therapeutics John Wiley & Sons, Inc., Hoboken, New Jersey.
- 25.Cao J., Hsu Y. H., Li S., Woods V. L., Dennis E. A. 2011. Lipoprotein-associated phospholipase A(2) interacts with phospholipid vesicles via a surface-disposed hydrophobic alpha-helix. Biochemistry. 50: 5314–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods V. L., Jr, Hamuro Y.2001. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell Biochem. Suppl.Suppl 37: 89–98.
- 27.Konermann L., Tong X., Pan Y. 2008. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass Spectrom. 43: 1021–1036 [DOI] [PubMed] [Google Scholar]
- 28.Engen J. R. 2009. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 81: 7870–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebling C. M., Morgan C. R., Stafford D. W., Jorgenson J. W., Rand K. D., Engen J. R. 2010. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal. Chem. 82: 5415–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stafforini D. M., McIntyre T. M., Prescott S. M. 1990. Platelet-activating factor acetylhydrolase from human plasma. Methods Enzymol. 187: 344–357 [DOI] [PubMed] [Google Scholar]
- 31.Burke J. E., Hsu Y. H., Deems R. A., Li S., Woods V. L., Jr, Dennis E. A. 2008. A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2. J. Biol. Chem. 283: 31227–31236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burke J. E., Karbarz M. J., Deems R. A., Li S., Woods V. L., Jr, Dennis E. A. 2008. Interaction of group IA phospholipase A2 with metal ions and phospholipid vesicles probed with deuterium exchange mass spectrometry. Biochemistry. 47: 6451–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu Y. H., Burke J. E., Stephens D. L., Deems R. A., Li S., Asmus K. M., Woods V. L., Jr, Dennis E. A. 2008. Calcium binding rigidifies the C2 domain and the intradomain interaction of GIVA phospholipase A2 as revealed by hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 283: 9820–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burke J. E., Babakhani A., Gorfe A. A., Kokotos G., Li S., Woods V. L., Jr, McCammon J. A., Dennis E. A. 2009. Location of inhibitors bound to group IVA phospholipase A2 determined by molecular dynamics and deuterium exchange mass spectrometry. J. Am. Chem. Soc. 131: 8083–8091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu Y. H., Burke J. E., Li S., Woods V. L., Jr, Dennis E. A. 2009. Localizing the membrane binding region of Group VIA Ca2+-independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 284: 23652–23661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Geest B., Stengel D., Landeloos M., Lox M., Le Gat L., Collen D., Holvoet P., Ninio E. 2000. Effect of overexpression of human apo A-I in C57BL/6 and C57BL/6 apo E-deficient mice on 2 lipoprotein-associated enzymes, platelet-activating factor acetylhydrolase and paraoxonase. Comparison of adenovirus-mediated human apo A-I gene transfer and human apo A-I transgenesis. Arterioscler. Thromb. Vasc. Biol. 20: E68–E75 [DOI] [PubMed] [Google Scholar]
- 37.Srinivasan P. a. B., B 2010. Molecular model of plasma PAF acetylhydrolase-lipoprotein association: insights from the structure. Pharmaceuticals. 3: 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253 [DOI] [PubMed] [Google Scholar]
- 40.Tellis C. C., Tselepis A. D. 2009. The role of lipoprotein-associated phospholipase A2 in atherosclerosis may depend on its lipoprotein carrier in plasma. Biochim. Biophys. Acta. 1791: 327–338 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.