Abstract
Phosphatidylserine (PS), the major anionic phospholipid in eukaryotic cell membranes, is synthesized by the integral membrane enzymes PS synthase 1 (PSS1) and 2 (PSS2). PSS2 is highly expressed in specific tissues, such as brain and testis, where docosahexaenoic acid (DHA, 22:6n-3) is also highly enriched. The purpose of this work was to characterize the hydrocarbon-chain preference of PSS2 to gain insight on the specialized role of PSS2 in PS accumulation in the DHA-abundant tissues. Flag-tagged PSS2 was expressed in HEK cells and immunopurified in a functionally active form. Purified PSS2 utilized both PE plasmalogen and diacyl PE as substrates. Nevertheless, the latter was six times better utilized, indicating the importance of an ester linkage at the sn-1 position. Although no sn-1 fatty acyl preference was noted, PSS2 exhibited significant preference toward DHA compared with 18:1 or 20:4 at the sn-2 position. Preferential production of DHA-containing PS (DHA-PS) was consistently observed with PSS2 purified from a variety of cell lines as well as with microsomes from mutant cells in which PS synthesis relies primarily on PSS2. These findings suggest that PSS2 may play a key role in PS accumulation in brain and testis through high activity toward DHA-containing substrates that are abundant in these tissues.
Keywords: mass spectrometry, plasmalogen, membranes, omega-3 fatty acids, phospholipids
Cell membranes consist of phospholipids that maintain and regulate the structure and function of integral membrane proteins. Phosphatidylserine (PS), the major anionic phospholipid in mammalian cell membranes, plays critical biological roles (1–3), including intra- and intercellular molecular signaling (4). In the plasma membrane, PS is strictly localized in the inner leaflet under normal conditions, and its exposure on the outer membrane is an indication of apoptotic cell death (5–7). It has been also reported that PS becomes exposed on the surface of tumor endothelial cells (8, 9), and thus, it has become a potential marker for targeted drug delivery (10–12). PS is synthesized in mammalian cells by two integral membrane proteins, PS synthase 1 and 2 (PSS1 and PSS2). These enzymes catalyze the formation of PS by an exchange reaction in which serine replaces the head group of the corresponding substrate phospholipids, phosphatidylcholine (PC) for PSS1 and phosphatidylethanolamine (PE) for PSS2 (1, 2).
While PSS1 is expressed ubiquitously (13), PSS2 is expressed in a tissue-specific manner (14). A high degree of PSS2 expression was observed in Purkinje neurons in the brain and in Sertoli cells of the testis in mice (14). In brain and testis, PE and PS, the substrate and product of PSS2, are highly enriched with docosahexaenoic acid (DHA, 22:6n-3). The accumulation of PS containing DHA at the sn-2 position (DHA-PS) is often associated with the proper function of these tissues (15–20). DHA accounts for ∼60% of the acyl chains at the sn-2 position of PS specifically in the brain (21). However, the mechanism responsible for the accumulation of DHA-PS has not been elucidated.
PS is synthesized in mammalian cells in the endoplasmic reticulum and mitochondria-associated membranes (22–24). A fraction of the PS is decarboxylated to produce PE by the PS decarboxylation (PSD) reaction that takes place in the mitochondria (25). Previously, we demonstrated that DHA-PS species is the best substrate for PSD in isolated brain mitochondria (26), indicating that the substrate specificity of PSD is not a contributing factor for DHA-PS enrichment in the brain. Although PSS1 has also been shown to preferentially produce sn-2 DHA-PS from PC in isolated microsomes (27), PC containing DHA in the sn-2 position is far less abundant than sn-2 DHA-PE, the substrate for PSS2. Moreover, the ubiquitous expression of PSS1 does not support a major role of PSS1 in the specific enrichment of DHA-PS in the brain.
We have previously reported that brain microsomes convert sn-2 DHA-PE to DHA-PS more efficiently than they convert other PE molecular species (28). Because both PSS1 and PSS2 are present in the microsomal preparations and PSS1 can utilize not only PC but also PE in vitro (29, 30), these microsomal studies were unable to distinguish the contributions of PSS2 to the preferential production of DHA-PS. In addition, the high level of DHA esterified at sn-2 of the brain PE is coupled with different sn-1 saturated acyl or vinyl ether chains (28, 31, 32), suggesting possible utilization of these species by PSS2 for PS synthesis in the brain.
To determine hydrocarbon chain preference of PSS2 in synthesizing PS from PE containing palmitic acid (16:0), stearic acid (18:0), or a vinyl ether linkage at the sn-1 position as well as a variety of sn-2 fatty acids, we have immunopurified functional PSS2. We found that purified PSS2 produces diacyl PS much more efficiently compared with PS containing an sn-1 vinyl ether linkage. While purified PSS2 utilized PE containing 16:0 and 18:0 at the sn-1 position similarly, reaction efficiency of PSS2 was substantially dependent on the sn-2 fatty acyl chain. The highest selectivity was found for sn-2 DHA-PS. Microsomes isolated from Chinese hamster ovary (CHO) mutant cells (33), which lack PSS1 activity and in which PS synthesis relies on PSS2, also produced DHA-PS as the dominant product. These results indicate a significant role of PSS2 in enriching DHA-PS in brain.
EXPERIMENTAL PROCEDURES
Materials
Anti-FLAG M2 affinity gel, 3 × FLAG peptide, FLAG antibody, asolectin, calf serum, bovine brain PS, Dulbecco's Modified Eagle Medium (DMEM), and dithiothreitol (DTT) were purchased from Sigma-Aldrich (St. Louis, MO); sucrose monolaurate was from Sigma-Aldrich or Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Ham's F12 medium (F-12) and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA). Other phospholipids were purchased from Avanti Polar Lipids (Alabaster, AL). Serine, L-[3-3H] (15-30 Ci/mmol, 1 mCi/ml) and 13C3,15N-serine were purchased from American Radiolabeled Chemicals, Inc. (St Louis, MO) and Cambridge Isotope Laboratories, Inc. (Andover, MA), respectively. FuGENE 6 Transfection Reagent and Protease Inhibitor were obtained from Roche (Indianapolis, IN), and BCA protein assay kit was obtained from Thermo Fisher Scientific Inc. (Rockford, IL). All solvents were HPLC grade and purchased from Thermo Fisher Scientific Inc. or Burdick and Jackson (Muskegon, MI).
Construction of Flag-PSS2 plasmid
PSS2 cloned from mouse was inserted into p3XFLAG-CMV-7.1 EXPRESSION VECTOR (Sigma) at Not I and Sal I restriction sites.
Cell culture
CHO-K1 wild-type and mutant (PSA-3, a kind gift from Dr. O. Kuge, Kyushu University, Japan), and Neuro 2A cells (mouse neuroblastoma cells, from ATCC) were cultured at 37°C in F-12 and DMEM media, respectively, containing 5% FBS under a 5% CO2 atmosphere with saturated humidity. The mutant CHO cells were supplemented with approximately 30 µM bovine brain PS or 1-stearoyl-2-oleoyl-sn-glycero-3-phosphoserine (18:0,18:1-PS) (34). A variant of the HEK-293 (FreeStyle 293-F Cells, Invitrogen) were cultivated according to the manufacturer's instruction (FreeStyle MAX 293 Expression System).
Preparation of microsomes from PSS2 overexpressed HEK and CHO mutant cells
Microsomes from the PSS2 overexpressed HEK and CHO-K1 mutant cells were prepared as described earlier (27, 28). Briefly, the microsomal fraction from the homogenized cells was isolated by differential centrifugation, and the microsomes were suspended in Hepes buffer (25 mM Hepes/NaOH, pH 7.4) containing 0.5 mM ethylene glycol-bis (β-aminoethylether)-N, N, N’, N’-tetraacetic acid (EGTA). In the case of PSS2 overexpressed HEK microsomes, HEK cells were transfected with the Flag-PSS2 plasmid for 48 h prior to the harvest. All procedures were carried out at 4°C or on ice.
Purification of FLAG-tagged PSS2
Purified Flag-PSS2 was prepared according to Kuge et al. (35) with slight modifications. After washing twice with cold buffer (PBS or Hepes buffer), cells transfected with the Flag-PSS2 plasmid for 48 h were harvested by scraping and brief centrifugation (1,000 g for 5 min). All procedures were performed at 4°C or on ice after harvesting the cells. The harvested cells were suspended with 0.25 M sucrose buffer containing 10 mM Hepes/NaOH (pH 7.4) and 1 mM EDTA and homogenized with a Teflon pestle. The homogenate was centrifuged at 1,000 g for 5 min, and the supernatant fraction was centrifuged at 100,000 g for 1 h. The resultant membrane fraction was suspended and incubated with sucrose monolaurate-containing buffer [3.5 mM Hepes/NaOH (pH 7.4), 0.07 mM L-serine, 14% (w/v) glycerol, 10 mg/ml asolectin, 150 mM NaCl, and 1% sucrose monolaurate] for 15 min, and then centrifuged at 100,000 g for 30 min.
The resulting solubilized membrane protein was subjected to immunopurification using anti-FLAG M2 affinity gel, which had been equilibrated with buffer A containing 10 mM Hepes/NaOH (pH 7.4), 0.1 mM L-serine, 15% (w/v) glycerol, 2 mg/ml asolectin (16:0,18:2 and 18:2,18:2 are the major species in PS, PE, and PC of asolectin), 150 mM NaCl, and 0.2% sucrose monolaurate. After either a 3 h or overnight incubation of the solubilized membrane protein and gel with gentle tilting, the gel was washed with buffer A five times. The washed gels were incubated with 0.2 mg/ml of 3XFLAG peptide in buffer A for 1 h and then briefly centrifuged. The supernatant fraction containing Flag-PSS2 protein was collected, dithiothreitol and protease inhibitor (Roche) were added, and the solution was stored at −70°C.
Analysis of purified PSS2
Fractions from each purification step were incubated with NuPAGE LDS (lithium dodecyl sulfate) sample buffer (4×) containing 5% β-mercaptoethanol at 90°C for 3 min or at 37°C for 1 h. The samples and protein marker (Bio-Rad, Hercules, CA) were loaded to the NuPAGE Novex 4-12% Bis-Tris PreCast Gel, and the proteins were separated by electrophoresis using the NuPAGE Electrophoresis System (Invitrogen). Visualization of the proteins was performed either by silver staining (Invitrogen) or by Western blotting. In Western blotting, the proteins separated on the gels were transferred to transfer membrane (Millipore, Billerica, MA), blocked with 5% skim milk in Tris buffered saline containing 0.1% Tween 20 (TBS-T), and incubated with primary (Anti-Flag antibody, 1:1,000 dilution, Sigma) and secondary (anti-mouse IgG-Peroxidase antibody, 1:5000 dilution, Sigma) antibodies. The proteins were visualized with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific Inc.) using the Gel Logic 440 Imaging System and Kodak 1D software.
The concentration of the purified PSS2 was estimated by band intensity of silver staining compared with the band intensity of known concentrations of standard BSA.
Preparation of unilamellar vesicles
Unilamellar vesicles were prepared by a slight modification of the extrusion method as described earlier (27, 28). Phospholipids in chloroform and 2,6-di-tert-butyl-p-cresol (BHT, 1 mol % of total lipids) were mixed and then dried under a nitrogen stream. The dried lipids were dissolved in cyclohexane, frozen, and then lyophilized under vacuum until only a lipid film or cake remained. This film or cake was hydrated with argon-purged Hepes buffer (25 mM Hepes/NaOH, pH 7.4) containing 0.5 mM EGTA under agitation to form large multilamellar vesicles (MLV). MLVs were disrupted by three to five freeze-thaw cycles and then converted to unilamellar vesicles by extrusion through polycarbonate filters containing 100 nm pores on a mini-extruder (Avanti Polar Lipids) 21 times in an argon atmosphere.
PSS2 activity assay
Purified Flag-PSS2 or microsomes were incubated at 37°C in a shaking water bath with unilamellar vesicles consisting of substrate PE or plasmalogen PE together with 16:0,18:1-PC and 16:0,18:1-PA at a molar ratio of 48/12/40 (PE/PC/PA). The reaction mixture also contained Hepes buffer, 5 mM CaCl2, and 200 µM serine (4 µCi/ml 3H serine or 13C3,15N-serine were utilized depending on the experiment). The total volume was 100 µl. The reaction was stopped by adding CHCl3/CH3OH/CH3OH-BHT (1:1:1), and the lipids were extracted by the Bligh-Dyer method (36). For the evaluation of PSS2 activity, incorporation of 3H-serine or 13C3,15N-serine into PS was measured by liquid scintillation or mass spectrometry, respectively. When analyzed by mass spectrometry, suitable internal standards were added before extraction.
Phospholipid molecular species analysis
Phospholipids were separated and detected using reversed-phase liquid chromatography-electrospray ionization/mass spectrometry (HPLC-ESI/MS) as described previously (27, 32). Phospholipids were separated on a C18 column (Prodigy or Gemini, 150 × 2.0 mm, 5 μm; Phenomenex, Torrance, CA) using a mobile phase consisting of water/0.5% ammonium hydroxide in methanol/hexane with a gradient changing from 12/88/0 to 0/88/12 (37). The flow rate was 0.4 ml/min delivered by the Agilent 1100 or 1290 HPLC system (Santa Clara, CA). HPLC/ESI-MS/MS analysis was performed using a Finnigan TSQ Quantum mass spectrometer (San Jose, CA) or Hewlett Packard HP 1100 Series LC/MSD system. Quantification of individual phospholipid molecular species was calculated by the area ratio against the added internal standards of the same phospholipid class.
RESULTS
Purification of PSS2
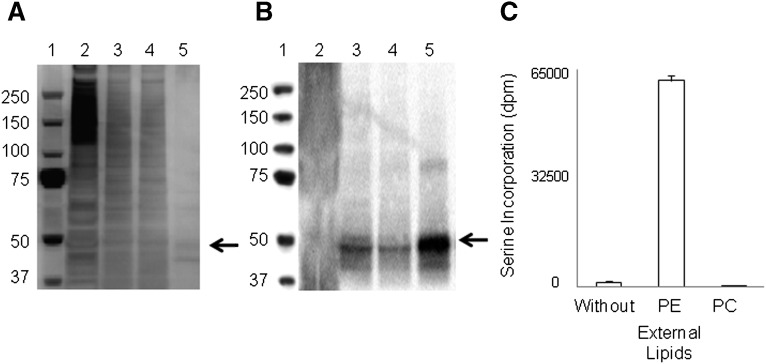
To investigate the acyl chain preference for PS synthesis by PSS2, we have prepared functionally active PSS2 by immunopurification of Flag-PSS2 overexpressed in a variant of the HEK-293 cell line as described previously with slight modifications (30, 35). The cell homogenate was fractionated to isolate the membrane fraction containing PSS2. Most of the high molecular weight proteins over ∼100 kDa in the membrane fraction (Fig. 1A, lane 2) were effectively removed in the detergent solubilization and ultracentrifugation steps (Fig. 1A, lane 3, solubilized membrane proteins). By immunopurification using anti-Flag M2 affinity gel, most of the solubilized membrane proteins other than Flag-PSS2 were removed (Fig. 1A, lane 4, unbound proteins to anti-Flag affinity gel), and the Flag-PSS2 (55 kDa) retained on the affinity gel was eluted and detected at ∼50 kDa with minimal background (Fig. 1A, lane 5, purified PSS2).
Fig. 1.
Purification of PSS2. Representative fractions from the PSS2 purification procedure were analyzed by SDS-PAGE, followed by silver staining (A) and Western blot analysis using anti-Flag antibody (B). Lanes 1–5 in (A) and (B) represent following: Lane 1, protein marker; 2, membrane fraction; 3, solubilized membrane proteins; 4, unbound proteins to anti-Flag affinity gel; and 5, purified PSS2. For evaluating PSS2 activity, purified Flag-PSS2 was incubated for 2.5 h with or without PE (18:0,22:6-PE / 16:0,18:1-PC / 16:0,18:1-PA, at a molar ratio of 48/12/40) or PC liposomes (18:0,22:6-PC / 16:0,18:1-PC / 16:0,18:1-PA, at a molar ratio of 48/12/40) in the presence of 200 µM serine (4 µCi/ml 3H serine) and 5 mM CaCl2. The incorporation of 3H serine into PS was measured by scintillation counting (C). Results in (C) are expressed as mean ± SD for four samples, representing two independent experiments.
Western blot data using anti-Flag antibody showed successful enrichment of the ∼50 kDa protein (Fig. 1B). Analysis of the 50 kDa band by mass spectrometry detected 41% of the PSS2 amino acid sequence (data not shown). The purified Flag-PSS2 maintained functional integrity as indicated by the serine base-exchange reaction measured by 3H-serine incorporation (Fig. 1C) as well as by conversion of PE-d35 to the corresponding PS detected by HPLC-ESI/MS (see Fig. 3). The PS synthetic activity was enhanced >620-fold by the purified enzyme compared with the cell homogenate before purification. Purified PSS2 converted PE but not PC to PS when these lipids were supplied (Fig. 1C), in agreement with the previous results (30, 35, 38, 39). Purified PSS2 without the PE supplementation (control) was 35-fold less active than when the incubation was supplemented with 18:0,22:6-PE. The basal activity detected in the control incubation was due to minor presence of PE in soybean phospholipids that were included to stabilize PSS2 during purification. The purified PSS2 was used in the subsequent experiments to study its functional specificity.
Fig. 3.
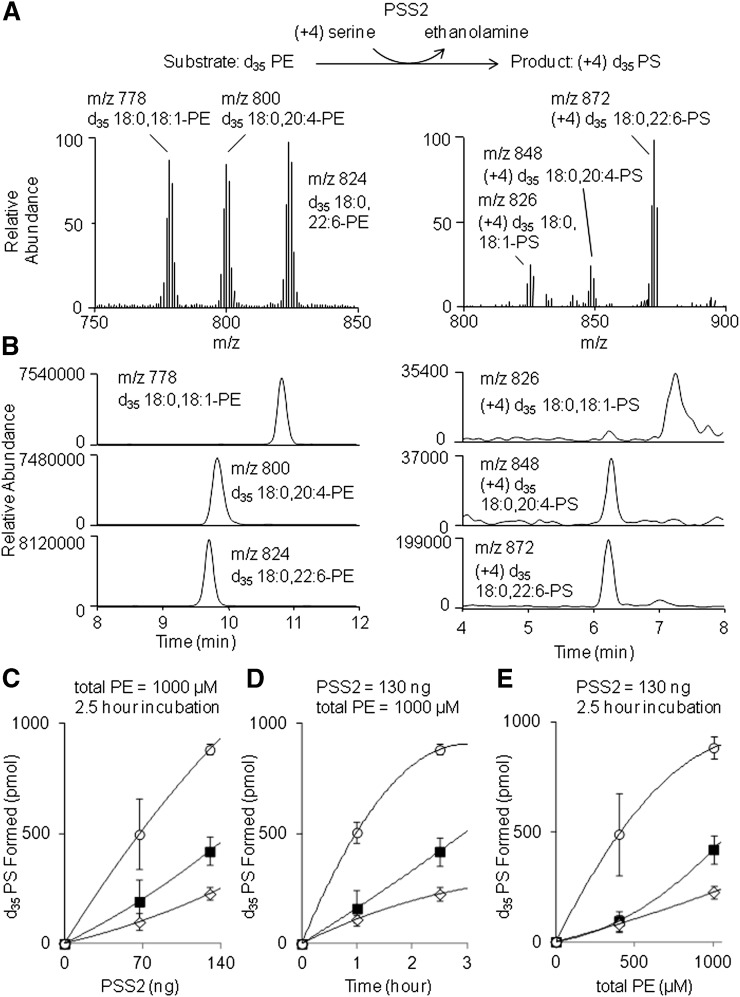
Effect of sn-2 fatty acyl chain on PS synthesis by purified PSS2 monitored by LC/MS. Representative mass spectra (A) and mass chromatograms (B) were obtained before and after incubation of purified PSS2 with liposomes containing equimolar d35 PE species (d35 18:0,18:1-PE, d35 18:0,20:4-PE, and d35 18:0,22:6-PE). Activity of purified PSS2 was assayed as a function of PSS2 concentration (C), incubation period (D), and substrate PE concentration (E) by incubation with the liposomes containing equimolar concentrations of d35 PE species in the presence of 200 µM serine and 5 mM CaCl2. The absolute amount of PS produced was calculated as the area ratio of each species relative to standard 16:0,16:0-PS in the HPLC-ESI/MS analyses. Filled squares, open diamonds, and open circles represent d35 18:0,18:1-PS, d35 18:0,20:4-PS, and d35 18:0,22:6-PS, respectively. The data in (C–E) are expressed as mean ± SD from 2–4 independent measurements for each data point.
sn-1 chain preference for PS production by PSS2
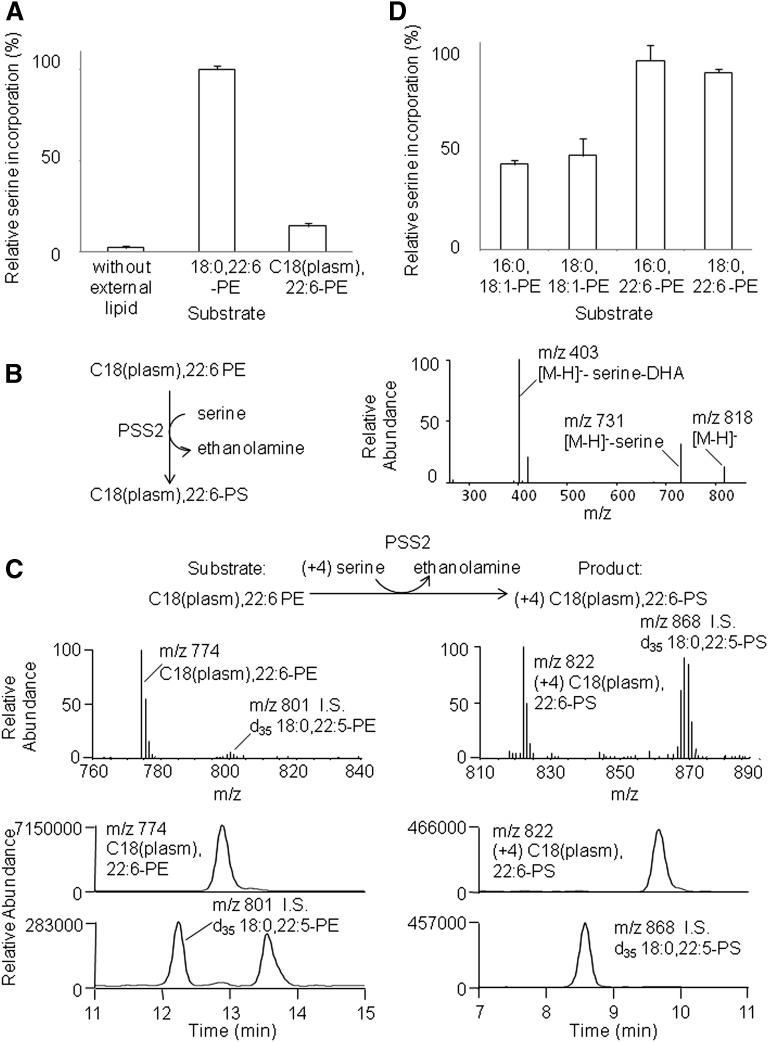
To investigate the influence of sn-1 chain on PS production by purified PSS2, 3H serine incorporation into diacyl-PE (ester linkage at sn-1) and plasmalogen PE (vinyl ether linkage at sn-1) was compared. Both PE substrates contained a hydrocarbon chain of 18 carbons in the sn-1 position and 22:6 in the sn-2 position [18:0,22:6-PE and C18(plasm),22:6-PE]. Considerable incorporation of 3H serine was observed when purified PSS2 was incubated with PE plasmalogen (Fig. 2A). Nevertheless, plasmalogen PE was six times less effective as a substrate than the corresponding ester-linked PE species. The production of PS plasmalogen [C18(plasm),22:6-PS] from C18(plasm),22:6-PE by purified PSS2 was confirmed by HPLC/ESI-MS/MS (Fig. 2B). The MS/MS spectrum of precursor ion of C18(plasm),22:6-PS at m/z 818 produced characteristic fragments m/z 731 and 403, which resulted from dissociation of serine moiety and further elimination of sn-2 fatty acid (DHA). We further confirmed the production of PS plasmalogen in a more native environment by using microsomes obtained from PSS2 overexpressed HEK cells (PSS2 microsomes). When PSS2 microsomes were incubated with C18(plasm),22:6-PE and 13C3,15N-serine (+4 serine), a chromatographic peak appeared at 9.85 min with m/z 822 that was 4 mass units higher than that of the unlabeled C18(plasm),22:6-PS (m/z 818) (Fig. 2C). Six times more effective production of diacyl PS compared with plasmalogen PS (Fig. 2A) indicated that a carbonyl group next to the glycerol backbone in the substrate-binding pocket substantially increased the catalytic activity of PSS2.
Fig. 2.
Sn-1 chain preference for PS synthesis by PSS2. Purified PSS2 was incubated with or without PE liposomes in the presence of 200 µM serine (4 µCi/ml 3H serine) and 5 mM CaCl2 for 2.5 h. The incorporation of 3H serine was measured by scintillation counting (A, D). The composition of the PE liposomes was PE / 16:0,18:1-PC / 16:0,18:1-PA, at a molar ratio of 48/12/40. PE contained either ester (18:0,22:6) or vinyl ether (C18(plasm),22:6) (A) or different fatty acyl chains (D). Identification of C18(plasm),22:6-PS production by MS/MS after incubation of purified PSS2 with C18(plasm),22:6-PE in the presence of 200 µM serine and 5 mM CaCl2 (B). Typical mass chromatograms and mass spectra of C18(plasm),22:6-PE and C18(plasm),22:6-PS, the reaction product of PSS2 microsomes after incubation with C18(plasm),22:6-PE in the presence of 200 µM 13C3,15N-serine and 5 mM CaCl2 (C). Results in (A) and (D) are expressed as mean ± SD for four samples, representing two independent experiments.
The preference for sn-1 chain length by purified PSS2 was also investigated by comparison of 3H serine incorporation into PE substrates containing either 16:0 or 18:0 at the sn-1 position (Fig. 2D). 16:0,18:1-PE and 18:0,18:1-PE were equally converted into PS. Similarly, equivalent 3H serine incorporation was observed with substrates containing 22:6 at the sn-2 position, 16:0,22:6-PE and 18:0,22:6-PE. These results are in contrast to those reported for PSS1, in which a 2-fold higher preference for sn-1 18:0 compared with 16:0 was observed when the sn-2 position contains 18:1, and a 10-fold preference for sn-1 18:0 when 22:6 is esterified at the sn-2 position (27).
sn-2 acyl chain preference for PS production
To investigate the sn-2 fatty acyl chain preference by purified PSS2, PS synthesis was examined using deuterium-labeled PE species containing different sn-2 chains. The substrates used for comparison were PE species containing 18:0 at sn-1 and either oleic acid (OA; 18:1n-9), DHA (DHA; 22:6n-3), or arachidonic acid (AA; 20:4n-6) at sn-2. Formation of the deuterium-labeled PS was monitored by HPLC-ESI/MS. Figure 3 shows representative mass spectra (Fig. 3A) and mass ion chromatograms (Fig. 3B) of the substrate PE and product PS. Purified PSS2 produced these PS molecular species from all of the PE substrates tested. The synthesis of 18:0,22:6-PS was most efficient compared with 18:0,18:1-PS and with 18:0,20:4-PS.
The sn-2 acyl chain preference was evaluated as a function of protein concentration (Fig. 3C), incubation time (Fig. 3D), and concentration of the PE-d35 substrate (Fig. 3E). Under all conditions examined, the production of 18:0,22:6-PS was two to five times higher than that of either the 18:0,18:1-PS or 18:0,20:4-PS species. This finding clearly demonstrates that the catalytic activity of PSS2 is significantly influenced by the molecular structure of the sn-2 chain of the PE substrate. The preferential production of DHA-PS by PSS2 was also observed with PE substrates containing 16:0 at the sn-1 position as indicated in Fig. 2D in which about twice more 16:0,22:6-PE was converted to PS than 16:0,18:1-PE.
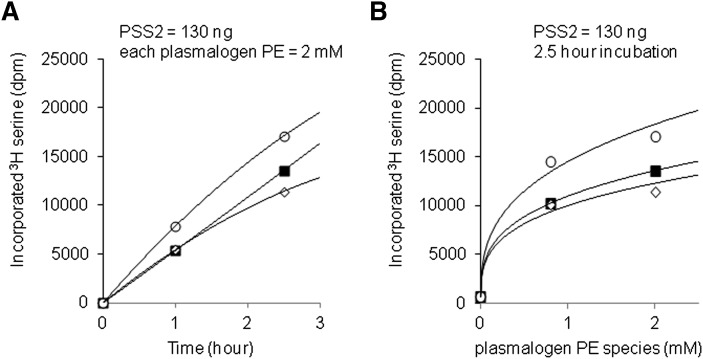
We investigated whether PSS2 similarly prefers DHA at sn-2 position when PE plasmalogen is used as the substrate. The utilization of C18(plasm),18:1-PE, C18(plasm),20:4-PE, or C18(plasm),22:6-PE for PS synthesis was compared by 3H serine incorporation. Purified PSS2 was incubated with unilamellar vesicles containing PC, PA, and one of these PE plasmalogen species in the presence of 3H serine. Production of PS plasmalogen was monitored as a function of incubation period (Fig. 4A) and PE plasmalogen concentration (Fig. 4B). Under all conditions tested, C18(plasm),22:6-PS was produced more than either of the other PS plasmalogen molecular species, the same preference observed in the case of diacyl PS synthesis (see Fig. 3). Although the preference for 22:6(plasm)-PS synthesis was statistically significant under all conditions tested, the degree of preference was appreciably smaller (1.3∼1.5-fold) compared with that found for diacyl DHA-PS synthesis (2∼5-fold, see Fig. 3).
Fig. 4.
Plasmalogen sn-2 acyl chain preference of PSS2. Activity and sn-2 acyl chain reference of purified PSS2 was assayed by incubation with PE plasmalogen liposomes (plasmalogen PE / 16:0,18:1-PC / 16:0,18:1-PA, at a molar ratio of 48/12/40, PE plasmalogen contains either C18(plasm),18:1-PE, C18(plasm),20:4-PE, or C18(plasm),22:6-PE species) in the presence of 200 µM serine and 5 mM CaCl2. The effects of time of incubation (A) and substrate PE plasmalogen concentration (B) were evaluated. 3H serine incorporation was measured with each PE plasmalogen species. Filled squares, open diamonds, and open circles represent C18(plasm),18:1-PS, C18(plasm),20:4-PS, and C18(plasm),22:6-PS, respectively. Data are presented as means of at least duplicated preparations, with a variation of <10%. The preference for C18(plasm),22:6-PS synthesis was statistically significant at all condition (<0.05 by Student t-test).
Substrate preference of PSS2 purified from a variety of cell systems
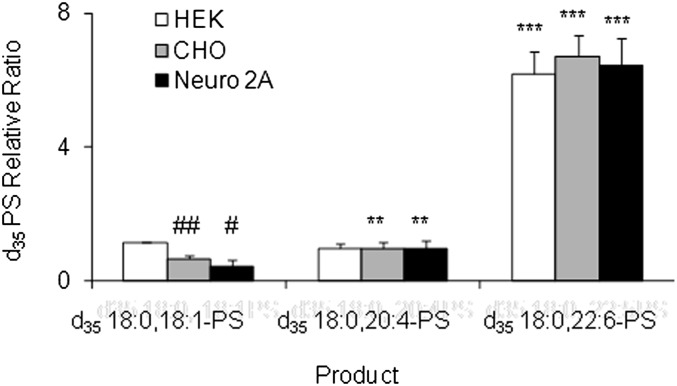
Cell-specific posttranslational modifications might affect the acyl-chain selectivity of PSS2. To test generality of the observed acyl-chain preference, PS synthesis was investigated using immunopurified Flag-PSS2 that was overexpressed in different cell types. In addition to HEK, a human cell line, Flag-PSS2 was expressed in two rodent cell lines, Neuro 2A (murine) and CHO (hamster), which represented neuronal and nonneuronal cell lines. PS synthesis by PSS2 purified from these cell lines was tested using deuterium-labeled PE species with different sn-2 chains (18:0,18:1-PE, 18:0,20:4-PE, and 18:0,22:6-PE). The corresponding PS-d35 product was detected by HPLC-ESI/MS.
Figure 5 shows the relative ratio of the product PS normalized to the amount of d35-18:0,20:4-PS formed. PSS2 purified from each of these cell lines consistently showed that the synthesis of 18:0,22:6-PS was by far (>5 times) the most efficient. Slightly less preference for 18:0,18:1-PS production was noted by the PSS2 purified from CHO or Neuro 2A cells compared with HEK cells. However, the amounts formed were relatively small and the differences were minor. Therefore, it was concluded that regardless of the cell lines in which PSS2 is expressed, a remarkable preference for DHA-PS synthesis by PSS2 is preserved.
Fig. 5.
Preferential synthesis of 18:0,22:6-PS by purified Flag-PSS2 expressed in a variety of cell lines. Flag-PSS2 overexpressed in HEK (white bars), CHO wild-type (gray bars), or Neuro 2A (filled bars) cells was purified by immunopurification. Each purified Flag-PSS2 preparation (∼130 ng) was incubated with the liposomes containing equal amount of each d35 PE species (total PE = 400 µM) in the presence of 200 µM serine and 5 mM CaCl2 for 2.5 h. The acyl-chain preference of the purified PSS2 is shown as the relative area ratio to d35 18:0,20:4-PS species in the HPLC-ESI/MS analyses. The statistical significance between different cell lines (#Neuro 2A versus HEK, ##CHO versus HEK) or different PS species (**d3518:0,20:4-PS versus d3518:0,18:1-PS, ***d3518:0,22:6-PS versus d3518:0,18:1-PS) was evaluated by Student t-test, #P < 0.05; ##P < 0.01; **P < 0.01; ***P < 0.001. The data are expressed as mean ± SD of triplicates, representing two independent experiments.
Acyl chain preference for PS production by microsomal PSS2
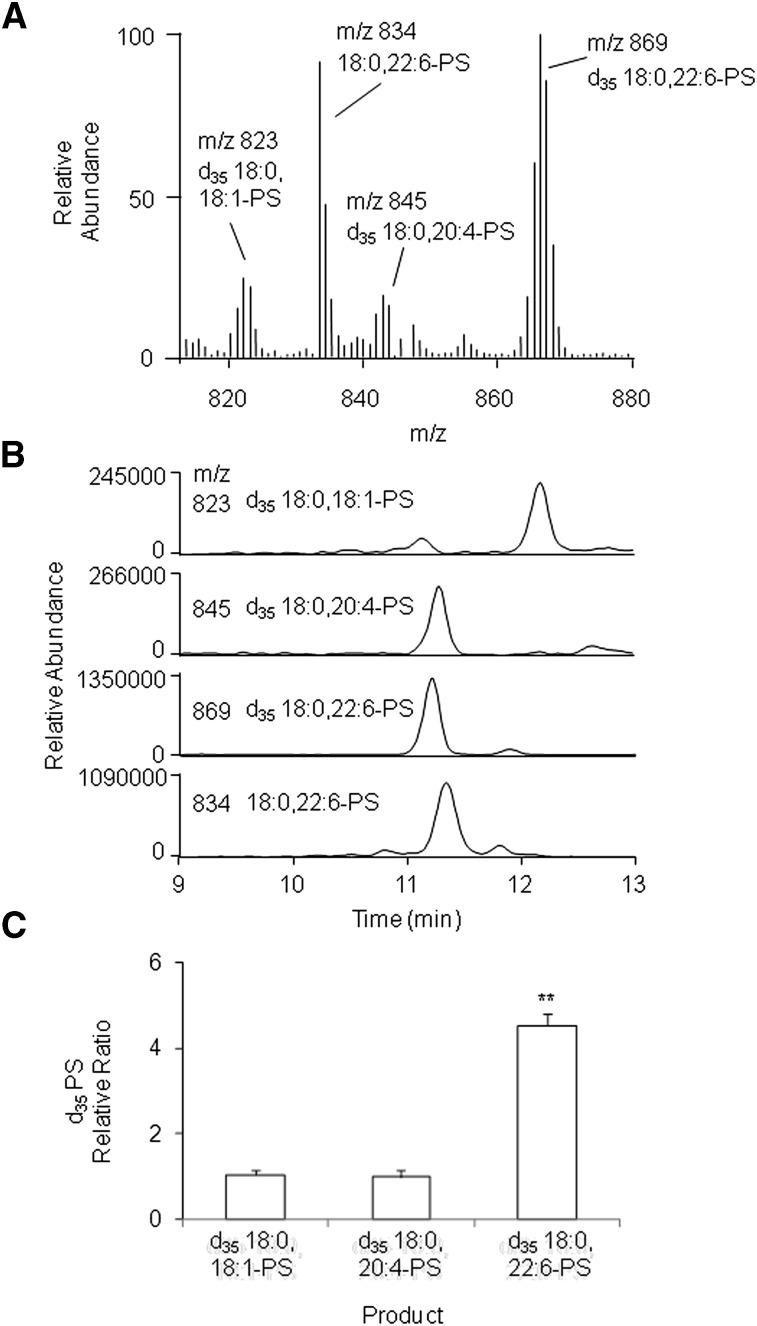
The substrate preference of PSS2 was further investigated in a more native environment by using microsomes isolated from mutant CHO cells in which PSS1 activity is lacking and PS synthesis primarily relies on PSS2. Unilamellar vesicles containing equal amounts of deuterium-labeled 18:0,18:1-PE, 18:0,20:4-PE, and 18:0,22:6-PE species were provided as the substrates, and the corresponding PS products were monitored by HPLC-ESI/MS. Fig. 6 shows a representative mass spectrum (Fig. 6A) and mass ion chromatograms (Fig. 6B) of the PS products, as well as the relative ratio normalized to the d35-18:0,20:4-PS (Fig. 6C). The mutant microsomes produced deuterium-labeled PS species (m/z 823, 845, and 869) from all the PE substrates tested. In addition to the reaction products, endogenous 18:0,22:6-PS (m/z 834) derived from microsomes was detected in the mass spectrum (Fig. 6A). The synthesis of d35-18:0,22:6-PS was most efficient, and its production was about five times higher than that of the other species in these microsomes. Thus, the selectivity for sn-2 DHA-PE observed with the purified PSS2 was confirmed with PSS2 present in a natural membrane.
Fig. 6.
Preferential synthesis of 18:0,22:6-PS by microsomes from CHO-K1 mutant cells. Microsomes (50 µg of total protein) were incubated for 1 h with liposomes containing equimolar concentrations of d35 PE species (total PE = 1 mM) in the presence of 200 µM serine and 5 mM CaCl2. Conversion of d35 PE to d35 PS was monitored by HPLC-ESI/MS. Representative mass spectrum (A) and mass chromatograms (B) of the products are presented. The acyl-chain preference of the microsomes is shown as the relative area ratio to d35 18:0,20:4-PS species (C). The statistical significance was evaluated by Student t-test. **P < 0.01. The data in (C) are expressed as mean ± SD of three samples, representing four independent experiments.
DISCUSSION
We have purified functionally active integral protein PSS2 and used it to investigate the hydrocarbon-chain preference of PSS2. We found that PSS2 synthesizes DHA-PS much more efficiently than PS containing either 18:1 or 20:4 at the sn-2 position. Preferential use of PE containing DHA over other fatty acids at the sn-2 position was consistently observed in PSS2-catalized PS synthesis, regardless of the type of linkage (Figs. 3 and 4) or acyl chain length at the sn-1 position (Fig. 2D). This remarkable selectivity for DHA-PS synthesis was common for PSS2 purified from human, murine, hamster, neuronal, or nonneuronal cell lines (Fig. 5). The preferential DHA-PS synthesis together with low PSS2 activity toward sn-2 20:4-PS production is consistent with the phospholipid profile of animal tissues in which 18:0,20:4-PE is a dominant PE species (28, 32), but the amount of 18:0,20:4-PS present is much less compared with 18:0,22:6-PS.
Substrate preference of PSS2 depends on availability of substrates as well as on the reactivity on each of the substrates. For example, if DHA-PE has better access to PSS2 compared with other species, it is likely converted more efficiently. Such a situation might mislead the conclusion that PSS2 has high preference for DHA-PE. In other experiments, we have designed the system to exclude the factor of substrate availability by reconstituting PSS2 in liposomes containing an equimolar concentration of PE substrates along with other phospholipid matrices. Regardless of the membrane composition, PSS2 consistently showed highest preference for DHA-PS production (data not shown), supporting that the substrate specificity toward DHA-PE is indeed a unique characteristics of PSS2.
The difference in the chain length between 16:0 and 18:0 at the sn-1 position of PE did not affect PSS2 activity (Fig. 2D). This finding differs from what has been observed with PSS1, a 2∼10 times higher preference for sn-1 18:0 than for 16:0 (27). This finding implies that the structure of the binding pocket of PSS1 is sufficiently different from that of PSS2 to recognize the sn-1 acyl-chain composition of substrate in addition to the headgroup difference between PC and PE. The fact that PS containing sn-1 18:0 is much more abundant than PS containing sn-1 16:0 in brain (28, 32) may be explained in part by the difference in acyl-chain preferences of PSS1 and PSS2 as well as by substrate availability. Despite a higher level of DHA-PC or AA-PC with sn-1 16:0 compared with the corresponding PC species with sn-1 18:0, low PSS1 activity toward the sn-1 16:0-PC may prevent the accumulation of sn-1 16:0-PS. In contrast, PS production by PSS2 may be impinged upon the availability of PE species containing either 16:0 or 18:0 at the sn-1 position as both forms are equally utilized by PSS2. Of course, further metabolism or refinement beyond PS synthesis such as PS decarboxylation and deacylation/reacylation reactions (40, 41) can also contribute to the eventual phospholipid profile in tissues.
Using microsomes from the mutant cells expressing PSS2 but lacking PSS1 activity, along with deuterium-labeled PE substrate, effectively isolated the PSS2 activity by avoiding confounding effects, such as PS decarboxylation that occurs in the mitochondria. Also, microsomal PSS2 remains in the natural membrane environment in contrast to the purified PSS2 reconstituted in artificial membrane matrices. The preferential production of DHA-PS observed with both purified PSS2 and PSS1-deficienct microsomes (Fig. 6) suggests that preferential synthesis of DHA-PS represents the properties of PSS2.
Our finding that purified PSS2 is capable of converting PE plasmalogen to PS plasmalogen is intriguing in that PS plasmalogen is rarely found in mammalian tissues, although a recent report indicated the presence of PS plasmalogens in the retina and the optic nerve (42). Due to the unique double bond of vinyl ether group, plasmalogens are reported to function as an antioxidant by scavenging reactive oxygen species, thereby protecting polyunsaturated fatty acids, including DHA, from oxidative stress and preserving membrane function (43). While PE plasmalogen is abundant in certain tissues, e.g., brain, especially in the myelin sheath (31), the physiological significance of PS plasmalogen synthesis is uncertain. The 6-fold less efficient synthesis of PS plasmalogen by PSS2 compared with diacyl PS (Fig. 2A) may explain difficulties in finding PS plasmalogen even in tissues such as brain where PE plasmalogen content is relatively high.
Both PSS2 and DHA appear to colocalize in brain and testis. In brain, DHA is highly enriched, especially in the sn-2 position of PS and PE (44, 45), and such enrichment of DHA has been shown to be essential for normal brain function and development (15–17). Although critical need for PSS2 in the brain has not been demonstrated, higher expression and activity of PSS2 occur in the fetal brain during embryonic development where DHA is rapidly accumulating, compared with the adult brain (46). An important role of PSS2 is apparent for proper function of the testis, where the PSS2 expression is highest (14). Approximately 10% of PSS2 knockout male mice are infertile, and the testes of these knockout mice are on average smaller than in wild-type controls, whereas PSS1 knockout mice do not exhibit this phenotype. Furthermore, delta-6 desaturase-null mice that were unable to synthesize highly unsaturated fatty acids were infertile (18, 19), and only DHA supplementation restored fertility and spermatogenesis (20). These findings suggest an important role of PSS2 and DHA for the proper development and function of testis, probably by synthesizing DHA-containing PS molecular species.
In conclusion, we found that PSS2 preferentially utilizes DHA-containing substrate for PS synthesis. We propose that PSS2 plays a specialized role in synthesizing DHA-PS in tissues including brain where PSS2 is abundantly expressed. The efficient DHA-PS synthesis by PSS2 found in this study is likely to play a critical role in maintaining the essential functions of these tissues.
Acknowledgments
The authors thank Dr. Osamu Kuge and Dr. Shiho Tomohiro (Kyushu University) for helpful comments on the enzyme purification; Dr. Bill X. Huang and Mr. Karl Kevala for mass spectrometric analysis of purified PSS2 and plasmalogen PS, respectively; Ms. Theresa Hwang for silver staining; and Dr. Arthur A. Spector for editing the manuscript.
Footnotes
Abbreviations:
- AA
- (20:4 n-6) arachidonic acid
- CHO
- Chinese hamster ovary
- DHA
- (22:6 n-3) docosahexaenoic acid
- DHA-PS
- PS species containing DHA at sn-2
- HEK
- FreeStyle 293-F cell, a variant of the human embryonic kidney 293 (HEK-293) cell line
- Neuro 2A cell
- mouse neuroblastoma cell
- OA
- (18:1 n-9) oleic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PEMT
- phosphatidylethanolamine N-methyltransferase
- plasmalogen PE
- PE containing vinyl ether linkage at sn-1
- plasmalogen PS
- PS containing vinyl ether linkage at sn-1
- PS
- phosphatidylserine
- PSD
- phosphatidylserine decarboxylase
- PSS
- phosphatidylserine synthase
This work was supported by the Intramural Research Program of the National Institute of Alcohol Abuse and Alcoholism, National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Vance J. E. 2008. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J. Lipid Res. 49: 1377–1387 [DOI] [PubMed] [Google Scholar]
- 2.Kuge O., Nishijima M. 1997. Phosphatidylserine synthase I and II of mammalian cells. Biochim. Biophys. Acta. 1348: 151–156 [DOI] [PubMed] [Google Scholar]
- 3.Leventis P. A., Grinstein S. 2010. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39: 407–427 [DOI] [PubMed] [Google Scholar]
- 4.Huang B. X., Akbar M., Kevala K., Kim H. Y. 2011. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 192: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emoto K., Toyama-Sorimachi N., Karasuyama H., Inoue K., Umeda M. 1997. Exposure of phosphatidylethanolamine on the surface of apoptotic cells. Exp. Cell Res. 232: 430–434 [DOI] [PubMed] [Google Scholar]
- 6.Fadok V. A., Bratton D. L., Henson P. M. 2001. Phagocyte receptors for apoptotic cells: recognition, uptake, and consequences. J. Clin. Invest. 108: 957–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadok V. A., Voelker D. R., Campbell P. A., Cohen J. J., Bratton D. L., Henson P. M. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol. 148: 2207–2216 [PubMed] [Google Scholar]
- 8.Ran S., Downes A., Thorpe P. E. 2002. Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res. 62: 6132–6140 [PubMed] [Google Scholar]
- 9.Ran S., He J., Huang X., Soares M., Scothorn D., Thorpe P. E. 2005. Antitumor effects of a monoclonal antibody that binds anionic phospholipids on the surface of tumor blood vessels in mice. Clin. Cancer Res. 11: 1551–1562 [DOI] [PubMed] [Google Scholar]
- 10.He J., Yin Y., Luster T. A., Watkins L., Thorpe P. E. 2009. Antiphosphatidylserine antibody combined with irradiation damages tumor blood vessels and induces tumor immunity in a rat model of glioblastoma. Clin. Cancer Res. 15: 6871–6880 [DOI] [PubMed] [Google Scholar]
- 11.Stafford J. H., Thorpe P. E. 2011. Increased exposure of phosphatidylethanolamine on the surface of tumor vascular endothelium. Neoplasia. 13: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran S., Thorpe P. E. 2002. Phosphatidylserine is a marker of tumor vasculature and a potential target for cancer imaging and therapy. Int. J. Radiat. Oncol. Biol. Phys. 54: 1479–1484 [DOI] [PubMed] [Google Scholar]
- 13.Sturbois-Balcerzak B., Stone S. J., Sreenivas A., Vance J. E. 2001. Structure and expression of the murine phosphatidylserine synthase-1 gene. J. Biol. Chem. 276: 8205–8212 [DOI] [PubMed] [Google Scholar]
- 14.Bergo M. O., Gavino B. J., Steenbergen R., Sturbois B., Parlow A. F., Sanan D. A., Skarnes W. C., Vance J. E., Young S. G. 2002. Defining the importance of phosphatidylserine synthase 2 in mice. J. Biol. Chem. 277: 47701–47708 [DOI] [PubMed] [Google Scholar]
- 15.Kim H. Y. 2007. Novel metabolism of docosahexaenoic acid in neural cells. J. Biol. Chem. 282: 18661–18665 [DOI] [PubMed] [Google Scholar]
- 16.Salem N., Jr, Litman B., Kim H. Y., Gawrisch K. 2001. Mechanisms of action of docosahexaenoic acid in the nervous system. Lipids. 36: 945–959 [DOI] [PubMed] [Google Scholar]
- 17.Kim H. Y. 2008. Biochemical and biological functions of docosahexaenoic acid in the nervous system: modulation by ethanol. Chem. Phys. Lipids. 153: 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., et al. 2009. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50: 1870–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoffel W., Holz B., Jenke B., Binczek E., Gunter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roqueta-Rivera M., Stroud C. K., Haschek W. M., Akare S. J., Segre M., Brush R. S., Agbaga M. P., Anderson R. E., Hess R. A., Nakamura M. T. 2010. Docosahexaenoic acid supplementation fully restores fertility and spermatogenesis in male delta-6 desaturase-null mice. J. Lipid Res. 51: 360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murthy M., Hamilton J., Greiner R. S., Moriguchi T., Salem N., Jr, Kim H. Y. 2002. Differential effects of n-3 fatty acid deficiency on phospholipid molecular species composition in the rat hippocampus. J. Lipid Res. 43: 611–617 [PubMed] [Google Scholar]
- 22.Saito K., Kuge O., Akamatsu Y., Nishijima M. 1996. Immunochemical identification of the pssA gene product as phosphatidylserine synthase I of Chinese hamster ovary cells. FEBS Lett. 395: 262–266 [DOI] [PubMed] [Google Scholar]
- 23.Stone S. J., Vance J. E. 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 275: 34534–34540 [DOI] [PubMed] [Google Scholar]
- 24.Vance J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265: 7248–7256 [PubMed] [Google Scholar]
- 25.Shiao Y. J., Lupo G., Vance J. E. 1995. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270: 11190–11198 [DOI] [PubMed] [Google Scholar]
- 26.Kevala J. H., Kim H. Y. 2001. Determination of substrate preference in phosphatidylserine decarboxylation by liquid chromatography-electrospray ionization mass spectrometry. Anal. Biochem. 292: 130–138 [DOI] [PubMed] [Google Scholar]
- 27.Kim H. Y., Bigelow J., Kevala J. H. 2004. Substrate preference in phosphatidylserine biosynthesis for docosahexaenoic acid containing species. Biochemistry. 43: 1030–1036 [DOI] [PubMed] [Google Scholar]
- 28.Wen Z., Kim H. Y. 2007. Inhibition of phosphatidylserine biosynthesis in developing rat brain by maternal exposure to ethanol. J. Neurosci. Res. 85: 1568–1578 [DOI] [PubMed] [Google Scholar]
- 29.Vance J. E., Steenbergen R. 2005. Metabolism and functions of phosphatidylserine. Prog. Lipid Res. 44: 207–234 [DOI] [PubMed] [Google Scholar]
- 30.Tomohiro S., Kawaguti A., Kawabe Y., Kitada S., Kuge O. 2009. Purification and characterization of human phosphatidylserine synthases 1 and 2. Biochem. J. 418: 421–429 [DOI] [PubMed] [Google Scholar]
- 31.Farooqui A. A., Horrocks L. A. 2001. Plasmalogens: workhorse lipids of membranes in normal and injured neurons and glia. Neuroscientist. 7: 232–245 [DOI] [PubMed] [Google Scholar]
- 32.Wen Z., Kim H. Y. 2004. Alterations in hippocampal phospholipid profile by prenatal exposure to ethanol. J. Neurochem. 89: 1368–1377 [DOI] [PubMed] [Google Scholar]
- 33.Kuge O., Nishijima M., Akamatsu Y. 1986. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. II. Isolation and characterization of phosphatidylserine auxotrophs. J. Biol. Chem. 261: 5790–5794 [PubMed] [Google Scholar]
- 34.Kuge O., Nishijima M., Akamatsu Y. 1985. Isolation of a somatic-cell mutant defective in phosphatidylserine biosynthesis. Proc. Natl. Acad. Sci. USA. 82: 1926–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuge O., Hasegawa K., Ohsawa T., Saito K., Nishijima M. 2003. Purification and characterization of Chinese hamster phosphatidylserine synthase 2. J. Biol. Chem. 278: 42692–42698 [DOI] [PubMed] [Google Scholar]
- 36.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- 37.Ma Y. C., Kim H. Y. 1995. Development of the on-line high-performance liquid chromatography/thermospray mass spectrometry method for the analysis of phospholipid molecular species in rat brain. Anal. Biochem. 226: 293–301 [DOI] [PubMed] [Google Scholar]
- 38.Voelker D. R., Frazier J. L. 1986. Isolation and characterization of a Chinese hamster ovary cell line requiring ethanolamine or phosphatidylserine for growth and exhibiting defective phosphatidylserine synthase activity. J. Biol. Chem. 261: 1002–1008 [PubMed] [Google Scholar]
- 39.Kuge O., Nishijima M., Akamatsu Y. 1986. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. III. Genetic evidence for utilization of phosphatidylcholine and phosphatidylethanolamine as precursors. J. Biol. Chem. 261: 5795–5798 [PubMed] [Google Scholar]
- 40.Sprecher H. 2000. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta. 1486: 219–231 [DOI] [PubMed] [Google Scholar]
- 41.Lands W. E. 1958. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 231: 883–888 [PubMed] [Google Scholar]
- 42.Nagy K., Brahmbhatt V. V., Berdeaux O., Bretillon L., Destaillats F., Acar N. 2012. Comparative study of serine-plasmalogens in human retina and optic nerve: identification of atypical species with odd carbon chains. J. Lipid Res. 53: 776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lessig J., Fuchs B. 2009. Plasmalogens in biological systems: their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Curr. Med. Chem. 16: 2021–2041 [DOI] [PubMed] [Google Scholar]
- 44.O'Brien J. S., Fillerup D. L., Mead J. F. 1964. Quantification and fatty acid and fatty aldehyde composition of ethanolamine, choline, and serine glycerophosphatides in human cerebral grey and white matter. J. Lipid Res. 5: 329–338 [PubMed] [Google Scholar]
- 45.Yabuuchi H., O'Brien J. S. 1968. Positional distribution of fatty acids in glycerophosphatides of bovine gray matter. J. Lipid Res. 9: 65–67 [PubMed] [Google Scholar]
- 46.Steenbergen R., Nanowski T. S., Nelson R., Young S. G., Vance J. E. 2006. Phospholipid homeostasis in phosphatidylserine synthase-2-deficient mice. Biochim. Biophys. Acta. 1761: 313–323 [DOI] [PubMed] [Google Scholar]