Abstract
Isotopic tracers have been used to examine lipid trafficking for many years, and data from those studies have typically yielded novel insight regarding the pathophysiology of dyslipidemia. Previous experimental designs were suitable for studies in humans because relatively large volumes of plasma could be regularly sampled. We have expanded on the earlier logic by applying high-throughput analytical methods that require reduced sample volumes. Specifically, we have examined the possibility of coupling gel-based separations of lipoproteins (e.g., lipoprint) with LC-MS/MS analyses of complex lipid mixtures as a way to routinely measure the labeling profiles of distinct lipids in discrete lipoprotein subfractions. We demonstrate the ability to measure the incorporation of [U-13C]oleate into triglycerides (TG), PLs (PL), and cholesterol esters (CE) in VLDL, LDL, and HDL particles in mice. Although rodent models of dyslipidemia are inherently different from humans because of alterations in enzyme activities and underlying metabolism, rodent models can be used to screen novel compounds for efficacy in altering a given biochemical pathway and therein enable studies of target engagement in vivo. We expect that it is possible to translate our approach for application in other systems, including studies in humans.
Keywords: stable isotopes, mass spectrometry, kinetic biomarker, cardiovascular disease
Investigators have demonstrated the ability to measure the incorporation of labeled fatty acids into circulating triglycerides (TG), PLs (PL), and cholesterol esters (CE). The movement of these complex lipids in specific lipoprotein fractions has been used to understand how production, clearance, and/or exchange reactions contribute to problems surrounding dyslipidemia. Although the seminal studies that were compiled by Berman, Grundy, and Howard (1) have proved valuable, application of the previous analytical methods is less than ideal for high-throughput studies and/or for use in rodent models in which sample volumes may be limiting. Advances in mass spectrometry and stable isotope tracer methods now permit routine analyses of specific analytes (2, 3). For example, whereas previous experiments studied the bulk pool of TG, it is now possible to examine individual species (2–6).
The advances noted above regarding instrumentation are expected to further impact and expand our understanding of the pathogenesis of disease, including the mechanism of action for novel therapeutics. For example, we recently demonstrated the ability to quantify TG kinetics using a high-throughput mass spectrometry-based assay, in which attention was directed toward an examination of whether the kinetics of an individual TG(s) would reflect the overall kinetics of the TG pool (5). In the current study, we have expanded on our earlier logic by directing attention toward an examination of the temporal labeling of distinct analytes in specific lipoprotein subfractions. Classical studies of Berman and colleagues (7) suggested that free fatty acids (FFA) are rapidly incorporated into complex lipids (e.g., TG, PL, and CE), which are then preferentially incorporated into very low density lipoprotein (VLDL) particles and that the labeling of other particles largely occurs via exchanges in the plasma compartment [e.g., the appearance of TG in high-density lipoprotein (HDL)] and/or via the delipidation of VLDL particles [e.g., the appearance of TG in low-density lipoprotein (LDL)].
In the current study, we have examined the suitability of using lipoprint as a method of separating lipoprotein subclasses, followed by an analysis of the isotopic labeling of specific lipids in a given fraction (8–10). Briefly, this gel-based approach is appealing for studies in rodents because it can be run using small sample volumes (e.g., ≤50 μl of plasma), and it can be used to obtain quantitative and high-resolution separations of VLDL, LDL, and HDL particles in ∼1 h. Specifically, we aimed to determine whether the use of a gel-based method (e.g., lipoprint) would be suitable in studies that examined the distribution of stable isotopically labeled lipids in VLDL, LDL, and HDL particles. To test our approach, we contrasted temporal changes in lipid labeling in wild-type versus cholesteryl ester transfer protein (CETP)-transgenic mice.
MATERIALS AND METHODS
Biological
All experimental protocols were reviewed and approved by the Merck Research Laboratories Institutional Animals Care and Use Committee (Rahway, NJ). Mice were maintained in a 12 h light/dark cycle with free access to food and water in group-housing conditions in a temperature-controlled environment (22°C). Male C57BL/6 and transgenic C57BL/6 mice expressing the human CETP gene, under the control of the human CETP promoter located in the natural flanking region of the human CETP gene (NFR-CETP) (11), were obtained from Taconic Farms, Inc. (Germantown, NY). Mice were maintained on regular chow (Teklad, 7012, 5% dietary fat; 3.75 kcal/g, Madison, WI) and studied at 12 weeks of age.
To determine the isotopic labeling in various lipoprotein subfractions, C57BL/6 mice were given an intravenous bolus of [13C18]oleate (K+ salt, 50 mg/kg, Cambridge Isotope Labs, Cambridge, MA) in Intralipid 20 (Sigma, St. Louis, MO). The tracer was formulated by weighing an appropriate amount of [13C18]oleate and adding the required volume of Intralipid to achieve a concentration of 5.0 mg tracer per milliliter. The preparation was then alternately vortexed and sonicated in a warm water bath for 10 min to ensure dissolution of the tracer. The intravenous bolus was administered via a tail vein using a volume of 10 ml/kg. At regular intervals after the tracer was administered, mice were euthanized (n = 2 per time point) via CO2 asphyxiation, and blood was collected by cardiac puncture. Plasma was obtained via centrifugation (14,000 rpm for 10 min) and added to a dry tube containing a lipase inhibitor (Paraoxan di-ethyl-p-nitrophenyl phosphate, Sigma, adjusted so that it had a concentration of 2 mM on addition of plasma) and a protease inhibitor cocktail (Roche #04 693 116 001, 1 μl per 100 μl of plasma). Samples were gently mixed and then stored at −70°C until lipoprotein separation, which was done by using lipoprint (LDL kit, Quantimetrix, Redondo Beach, CA) or by ultracentrifugation. Followed by MgCl2:dextran sulfate precipitation of LDL, lipids were extracted from the various fractions and analyzed as described below.
To examine the effect of CETP on lipid labeling in different particles, C57BL/6 (wild-type) and NFR-CETP-transgenic mice were dosed with [13C18]oleate (the tracer was prepared and dosed as described above). Eight mice from each strain were bled via tail nick to collect 40 μl plasma at 0.5, 2, and 24 h post tracer administration; another eight mice from each strain were bled in the same fashion at 1, 6, and 24 h post tracer treatment. Lipoprotein fractions were separated by lipoprint (LDL kit), and lipids were extracted from the isolated lipoprotein fractions as described below.
Analytical
Lipoprotein isolation: gel-electrophoresis (lipoprint).
VLDL, LDL+IDL, and HDL were separated from 25 μl of plasma (collected in the presence of lipase and protease inhibitors) using lipoprint (LDL gel kit; a photopolymerized loading gel was used). Samples were subjected to electrophoresis for 60 min at 3 mA per gel tube (i.e., sample) set at maximum delivery of 500V. Gels were stained with Sudan Black dye. Bands containing VLDL, LDL+IDL, and HDL were identified based on migration distance, cut from the gel using a razor blade, and then homogenized for lipid analysis.
Lipoprotein isolation: ultracentrifugation and dextran sulfate precipitation.
VLDL was first separated from 100 μl of plasma by ultracentrifugation. Briefly, 100 μl of plasma was transferred to a mini-ultracentrifuge tube. Using an Hamilton syringe, 100 μl of PBS buffer (no Ca2+, Mg2+, standard culture buffer, 1×, HyClone Laboratories, Thermo Scientific) was slowly placed on top of the plasma sample (attention was directed to ensure that the two layers did not mix). Samples were centrifuged for 18 h (45,000 rpm at 4°C). The resulting top layer contained the VLDL fraction, of which 100 μl was carefully removed from the bottom of the tube (containing both LDL and HDL) and transferred to a regular microfuge tube where LDL was precipitated from the infranatant using MgCl2 mixed with dextran sulfate.
Lipid extraction.
Gel bands containing VLDL and HDL were first homogenized in 500 μl of PBS buffer, whereas the gel band containing LDL+IDL was homogenized in 1.0 ml of PBS buffer. The 150 μl of each gel homogenate was extracted by addition of 200 μl of methanol containing the following lipid internal standards (2,000 nM [2H6]cholesterol-[13C7]oleate, 400 nM [13C21]triolein, 500 nM [2H9]phosphatidyl choline (16:0/18:1), 500 nM [2H5]triglyceride (16:0/18:1/18:2), 500 nM [2H5]triglyceride (16:0/18:1/18:1), and 400 μl of dichloromethane. Note that the nomenclature used here to identify the fatty acids specifies the chain length and number of double bonds; for example, 16:0 is 16 carbon atoms with no double bonds. Following centrifugation to separate the phases, 150 μl of the lower, lipid-containing dichloromethane layer was removed, evaporated to dryness under N2, and reconstituted in 150 μl of injection solvent (65% acetonitrile:30% isopropyl alcohol:5% water, v/v/v).
LC-MS/MS analyses.
Samples were analyzed using an Acquity UPLC system interfaced with a Xevo triple quadrupole mass spectrometer (Waters, Milford, MA). Lipids were separated on a Hypersil Gold C18 column (2.1 × 50 mm; 1.9 μm) maintained at 60°C using a 5 μl injection volume. A binary gradient was used to elute free fatty acids, PL, TG, and CE. Solvent A was10 mM ammonium formate in 40% H2O:60% acetonitrile (v/v), and solvent B was 10% acetonitrile:90% isopropanol (v/v). The column was initially equilibrated at 20% solvent B at a flow of 600 μl/min. This condition was held for 0.2 min after injection, then the composition was linearly ramped to 70% solvent B over 2.0 min. This condition was held for 0.1 min, after which the composition was linearly ramped to 90% solvent B over 1.4 min and finally to 95% solvent B over 0.05 min. This condition was maintained for 0.3 min, then the composition was then returned to 20% solvent B to (re)equilibrate the column.
Lipids were analyzed using electrospray ionization in the positive ion mode for PL, TG (as NH4+ adducts), and CE (as NH4+ adducts). All analytes were detected using multiple reaction monitoring. For the specific lipids presented here, transitions were programmed as follows: phosphatidyl choline 34:1, M+H+ > 184.1; for TG 52:2, the M+NH4+ parent ion was fragmented via collision induced dissociation to produce a neutral loss of oleic acid (resulting in loss of 299.3 Da for unlabeled oleate or 317.3 for [13C18]oleate); for CE 18:1, M+NH4+ > 369.4. Transitions were monitored for the endogenous (unlabeled) form of each lipid, as well as for all possible isotopomers obtained via acquisition of the [13C18]oleic acid tracer. Quantitation was achieved using single-point calibration by reference to the matched stable, labeled internal standard.
RESULTS
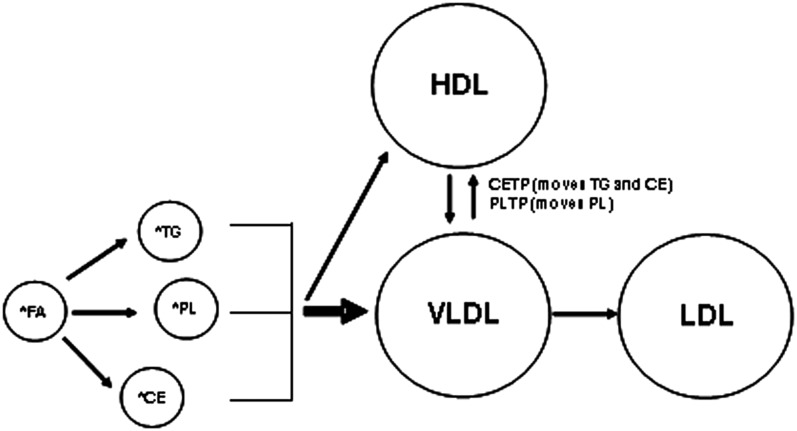
Tracer studies typically require a model to explain the movement of label from a pool of free fatty acids into various end-products (Fig. 1). Briefly, fatty acids are rapidly removed from the circulation and then either oxidized or (re)incorporated in various complex lipids, which are then transported in specific lipoprotein subfractions. Based on the studies described by Berman et al. (7), it appears that the potential for direct transfer of TG (which presumably happens in the liver) into HDL is relatively low, and thus the majority (e.g., ∼90%) of newly made TG is preferentially directed toward the VLDL particle.
Fig. 1.
General model of lipid flux. Stable isotopically labeled fatty acids are incorporated into various complex lipids, including TG, PL, and CE, which are then transported in the circulation in VLDL, LDL, and/or HDL particles.
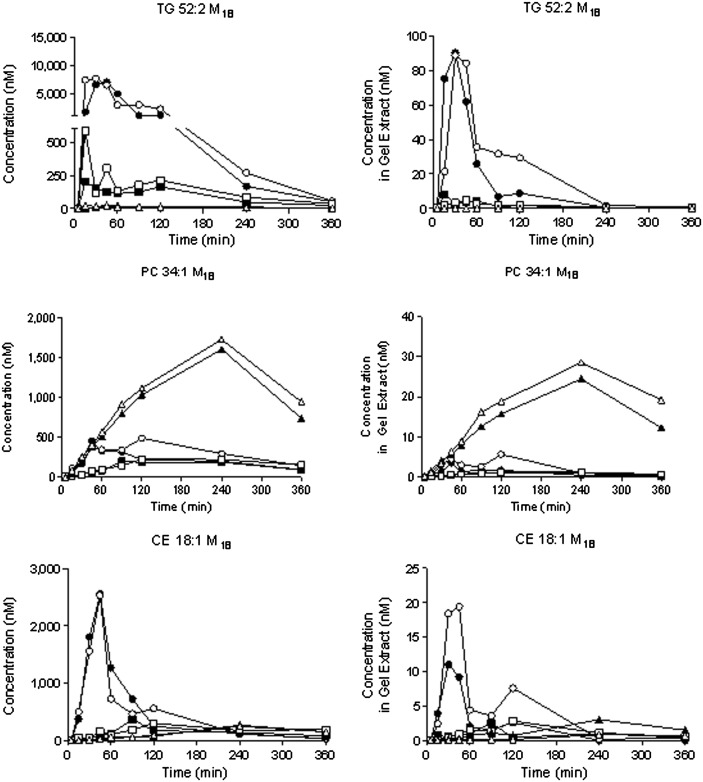
Various methods are available for isolating lipoprotein subfractions. Fig. 2 demonstrates a comparison of the separations that can be achieved by using lipoprint (on the LDL kit) or by combining ultracentrifugation (to isolate VLDL) with dextran sulfate precipitation (to obtain LDL and HDL in the pellet and supernatant, respectively). It is clear that the methods yield comparable data in relative terms. For example, the concentration of TG 52:2 is consistently greater in the VLDL fractions than in the LDL and/or HDL factions, regardless of the method used to separate the particles; this observation is expected given the distribution of TG in plasma. Noting the scale that is used on each y axis, it is clear that the magnitude is different when comparing the methods for isolating lipoproteins. This is to be expected given the different dilution factors that are used during the sample preparation. Consequently, it is not immediately possible to make definitive statements regarding absolute (or true) concentrations; nevertheless, it is possible to draw semiquantitative conclusions regarding differences between groups.
Fig. 2.
Comparison of lipoprotein separation achieved using different methods of sample preparation. Plasma was collected as various time points from wild-type mice following an intravenous bolus of [13C18]oleate; samples were used to contrast ultracentrifugation followed by dextran sulfate precipitation (left column) versus lipoprint (right column) as methods for separating lipoprotein classes. Data were obtained from 18 different mice (n = 2 per time point). Filled versus open symbols distinguish data from different mice at each time point: VLDL, open/filled circles; LDL, open/filled squares; and HDL, open/filled triangles, respectively. The notation M18 refers to the incorporation of a single [13C]oleate in the respective analyte; the mass is shifted by 18 because [13C18]oleate was administered.
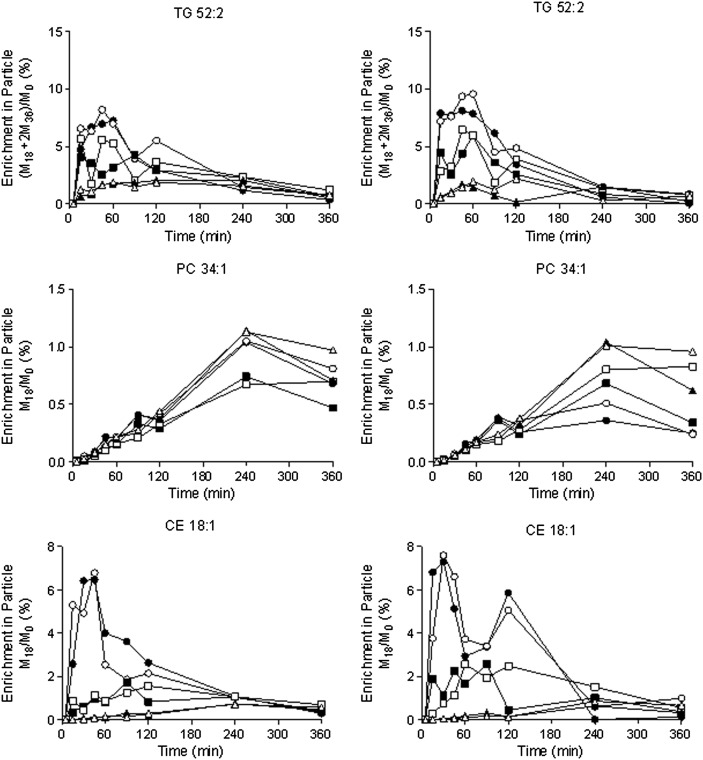
It does not appear that the method for isolating lipoprotein subfractions impacts the isotopic-labeling profiles (Fig. 3). For example, when we examined the temporal labeling of different lipids in various lipoproteins following the administration of [13C18]oleate to control mice, we observed a rapid and transient wave of labeling in TG and CE in VLDL particles and a much slower labeling of PL. In contrast to the different scales noted on the y axis in Fig. 2, the enrichment of the different lipids shown in Fig. 3 is virtually identical regardless of the method used to isolate the lipoproteins. That observation is expected, as the method of extracting the lipids should not differentiate between labeled and unlabeled molecules.
Fig. 3.
Isotopic labeling of different lipids in specific particles. Wild-type mice were given an intravenous injection of [13C18]oleate, and lipoprotein fractions were separated using ultracentrifugation followed by dextran sulfate precipitation (left column) or lipoprint (right column). Data were obtained from 18 different mice (n = 2 per time point). Filled versus open symbols distinguish data from different mice at each time point: VLDL, open/filled circles; LDL, open/filled squares; and HDL, open/filled triangles, respectively. The isotopic labeling of TG, PL, and CE was determined in the various fractions. Data are shown as enrichments, where the notation M18 and M36 refer to the number of labeled oleates that are incorporated; the mass is shifted 18 and 36 units, respectively.
Note that, in certain cases, we have observed scatter between duplicate measurements made at a single time point (Fig. 2). In some cases, this represents true biological variability, which is expected from sampling two different animals. For example, comparable scatter is observed in the measurements for TG 52:2 M18 in the VLDL fraction at the 15 min time point, regardless of whether ultracentrifugation or lipoprint was used to isolate the particle. A similar statement can be made regarding the CE 18:1 M18 in VLDL at the 120 min time point. In other cases, there appears to be some method dependence to the data. Whereas the measurements for TG 52:2 M18 in the VLDL fraction are somewhat scattered at the 90 and 120 min time points in the lipoprint-separated samples, there appears to be better agreement in the samples processed by ultracentrifugation. Nonetheless, when enrichment of the different lipids is plotted (as shown in Fig. 3), the scatter is substantially reduced, and this reinforces the fact that studies of lipid enrichment can be successfully conducted using either ultracentrifugation or lipoprint to isolate the fractions.
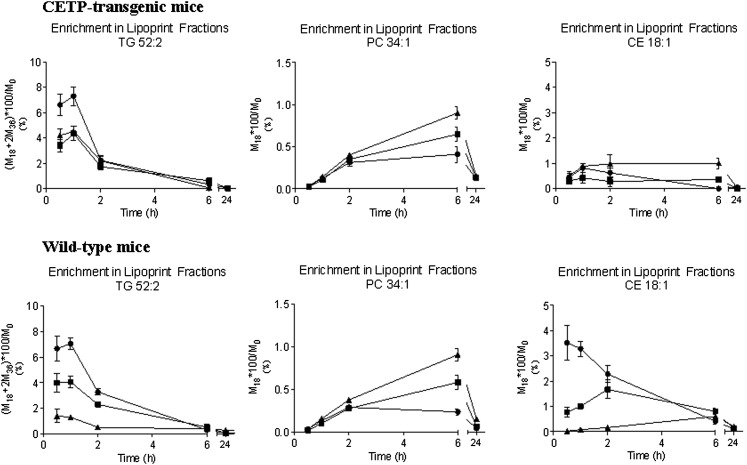
Given the observations noted above, it was possible to contrast the time-dependent labeling of a specific TG, PL, and CE in different lipoprotein fractions in wild-type versus NFR-CETP-transgenic mice (in Fig. 4, data are shown as the enrichment in a given analyte). Although these mice were given a single bolus of [13C18]oleate, there is a substantial change in the labeling of VLDL-TG, which is comparable in magnitude in wild-type and NFR-CETP-transgenic mice. The labeling (or enrichment) of the oleate containing PL 34:1 and CE 18:1 is dramatically lower than that that of the TG 52:2, which is consistent with the data shown in Fig. 3. In addition, the temporal change in labeling in the various lipids is different depending on the class (i.e., TG, PL, or CE); however, these observations are consistent with the data shown in Fig. 3. A notable point in Fig. 4 concerns the degree of equilibration between the labeling of TG and CE between VLDL and HDL in NFR-CETP-transgenic mice versus wild-type controls; this is somewhat expected given the presence and absence of CETP, respectively. In contrast, there is a high degree of equilibration in PL labeling between the two mouse models; again, this is reasonable considering the fact that both groups of mice have PL transfer protein (PLTP).
Fig. 4.
Differential labeling of lipids in wild-type versus CETP-transgenic mice. The isotopic enrichment of specific TG, PL, and CE in distinct lipoprotein fractions was measured in wild-type versus CETP-transgenic mice following a bolus injection of [13C18]oleate. There is marked equilibration of label in the PL in all lipoprotein fractions regardless of the model. However, there is a clear alteration in the equilibration between VLDL and HDL or LDL in the case of TG and/or CE transfer. The TG and CE labeling is generally greater in VLDL (filled circles) than in either HDL (filled triangles) or LDL (filled squares) in wild-type mice, which is consistent with the absence of CETP.
DISCUSSION
The ability to rapidly and reliably study the movement of circulating lipids should help to explain the underlying pathophysiology of dyslipidemia while yielding novel insight regarding drug development. Although tracer studies are useful in understanding lipid biology, the methods that have been utilized in humans are not immediately translatable for application in rodent models (especially in areas related to the development of pharmaceutical interventions in which one expects to process large numbers of samples). Fortunately, advances in mass spectrometry and other areas now permit high-throughput, high-resolution measurements. For example, we, and others recently demonstrated the ability to measure the incorporation of [13C18]oleate into TG following a single bolus administration of tracer (5, 12). We demonstrated the utilization of that approach in quantifying TG synthesis in vivo. We now demonstrate the ability to couple those analyses to lipoprotein subfractions and to follow the movement of specific lipids in individual lipoprotein classes.
An important aspect in developing this work was to first determine whether gel-based separation of lipoproteins would yield comparable data as measured against a more traditional separation method (e.g., a density-gradient separation). We determined that one would obtain comparable temporal labeling profiles in mice following the injection of [13C18]oleate (Fig. 3). Those data support the idea that the method of lipoprotein separation is not immediately critical in studies of kinetics, at least in terms of enrichment profiles and/or labeling ratios. In addition to comparing the different lipoprotein separation methods, our data demonstrate the general time-dependent profile for the turnover of specific lipids. For example, samples were obtained at numerous points within a 6 h window after administering the tracer. Those data clearly demonstrate faster turnover of TG versus PL and CE. In addition, the data are comparable with the literature, which demonstrated the rapid incorporation of labeled fatty acids into various end-products (1).
We next turned our attention toward an examination of whether it would be possible to detect a change in interparticle lipid flux. As it is well known that wild-type mice are deficient in CETP, one might predict a reduced transfer of labeled TG and CE between VLDL and HDL but normal transfer of labeled PLs because wild-type mice have PLTP (13, 14). Therefore, we administered a comparable dose of [13C18]oleate to wild-type mice and NFR-CETP-transgenic mice, and then measured the isotopic enrichment of specific lipids in the lipoprotein particles (Fig. 4). As expected, we observed a marked difference in the movement of TG and CE but not PL between VLDL and HDL (13, 14). Overall, the presence and/or absence of CETP has a pronounced impact on the transfer of neutral lipids (e.g., TG and CE) but not PL.
In summary, our data support the hypothesis of coupling high-throughput, high-resolution measurements of isotope flux in lipoproteins in vivo in rodent models. The data shown herein should provide a useful framework for future studies, most notably the time-course for labeling in different fractions is considerably different. We expect that the approach outlined here can be used to support studies of lipoprotein kinetics in novel in vivo models; although we have directed our attention to rodents in the current report, our previous work implies that the logic is immediately translatable to higher species. Our interest in the specific lipids reported here arose for different reasons. First, because these represent major analytes within the respective lipid subclasses, we assumed that their movement would largely reflect the general pool of each type of lipid. Second, using the current methods, it is not possible to obtain clean signals for all lipid analytes. For example, although one can ascribe exact masses and retention times to virtually all of the analytes, the ability to reliably quantify the isotopic-labeling patterns in complex mixtures presents a difficult challenge (2, 3).
We conclude with a few remarks regarding where we expect other investigators might expand on the logic outlined here. First, although our example considers the use of [13C]oleate, it should be possible to utilize other isotopically substituted tracers and therein quantify additional parameters (e.g., administer [2H]oleate and estimate fatty acid oxidation) (15). Second, although our studies have considered a relatively large physiological perturbation that results from the way in which we administer the tracer (i.e., the oleate tracer was delivered in ∼300 μl of Intralipid), in our experience, it is possible to administer labeled fatty acids in smaller volumes of Intralipid and/or use alternative formulations. Specifically, we have had success dosing the oleate tracer in both a 4-fold diluted Intralipid formulation (volumes of saline and Intralipid combined to achieve a relative TG content of 5% versus the standard 20% used in these studies) and an 8% solution of BSA. In both cases, the kinetics of appearance of the labeled fatty acid bound in complex lipids such as TG were somewhat different than observed when a 20% Intralipid emulsion was used as the delivery vehicle (unpublished observations). Nevertheless, in our experience, this would not preclude use of either of these formulations for studies such as we have presented here in which the purpose is to track the appearance of the labeled complex lipids as they move between different lipoprotein particles. We note that one of our goals here was to examine lipid kinetics under a perturbed state; thus, we used a relatively large volume of Intralipid. Third, presumably one could use a strategy such as we have outlined and administer [2H5]glycerol. Given its solubility in water, one would expect no confounding variables regarding vehicle effects. However, that approach immediately eliminates the possibility of quantifying the kinetics of CE and other end-products that do not incorporate [2H5]glycerol (4, 6). Finally, we recognize that our sampling schedule was not adjusted to optimize the calculations of any specific flux rate(s); we chose to collect samples at the respective intervals to simply establish the general temporal labeling profiles of the various lipids (4). Presumably, future studies will likely need to consider when to obtain samples to best fit the desired kinetic curve(s); we suspect that those efforts will largely be directed by the hypothesis that is tested. We expect that our understanding of the biochemical models will likely evolve as the level of data quality improves. For example, it appears that there is a biphasic change in the labeling of VLDL cholesterol oleate in mice (Figs. 2 and 3); the underlying physiology that might explain this observation in rodent models is not clear, nor is it known whether this will be observed in higher species.
Footnotes
Abbreviations:
- CE
- cholesterol ester
- CETP
- cholesteryl ester transfer protein
- PL
- PL
- TG
- triglyceride
REFERENCES
- 1.Berman, Mones. 1982. Lipoprotein Kinetics and Modeling. Academic Press, New York.
- 2.Castro-Perez J., Previs S. F., McLaren D. G., Shah V., Herath K., Bhat G., Johns D. G., Wang S. P., Mitnaul L., Jensen K., et al. 2011. In vivo D2O labeling to quantify static and dynamic changes in cholesterol and cholesterol esters by high resolution LC/MS. J. Lipid Res. 52: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castro-Perez J. M., Roddy T. P., Shah V., McLaren D. G., Wang S. P., Jensen K., Vreeken R. J., Hankemeier T., Johns D. G., Previs S. F., et al. 2011. Identifying static and kinetic lipid phenotypes by high resolution UPLC-MS: unraveling diet-induced changes in lipid homeostasis by coupling metabolomics and fluxomics. J. Proteome Res. 10: 4281–4290 [DOI] [PubMed] [Google Scholar]
- 4.Magkos F., Patterson B. W., Mittendorfer B. 2007. Reproducibility of stable isotope-labeled tracer measures of VLDL-triglyceride and VLDL-apolipoprotein B-100 kinetics. J. Lipid Res. 48: 1204–1211 [DOI] [PubMed] [Google Scholar]
- 5.McLaren D. G., He T., Wang S. P., Mendoza V., Rosa R., Gagen K., Bhat G., Herath K., Miller P. L., Stribling S., et al. 2011. The use of stable-isotopically labeled oleic acid to interrogate lipid assembly in vivo: assessing pharmacological effects in preclinical species. J. Lipid Res. 52: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patterson B. W., Mittendorfer B., Elias N., Satyanarayana R., Klein S. 2002. Use of stable isotopically labeled tracers to measure very low density lipoprotein-triglyceride turnover. J. Lipid Res. 43: 223–233 [PubMed] [Google Scholar]
- 7.Berman M., Beltz W. F., Riemke R., Sedaghat A., Grundy S. M. 1982. HDL triglyceride kinetics and exchanges with VLDL in vivo. In Lipoprotein Kinetics and Modeling. M. Berman, S. M. Grundy, and B. V. Howard, editors. Academic Press, New York. 299–305.
- 8.Oravec S., Dostal E., Dukat A., Gavornik P., Kucera M., Gruber K. 2011. HDL subfractions analysis: a new laboratory diagnostic assay for patients with cardiovascular diseases and dyslipoproteinemia. Neuroendocrinol. Lett. 32: 502–509 [PubMed] [Google Scholar]
- 9.Varady K. A., Lamarche B. 2011. Lipoprint adequately estimates LDL size distribution, but not absolute size, versus polyacrylamide gradient gel electrophoresis. Lipids. 46: 1163–1167 [DOI] [PubMed] [Google Scholar]
- 10.Vandermeersch A., Ameye S., Puype D., Petitjean D., De Buyzere M., Langlois M. R. 2010. Estimation of the low-density lipoprotein (LDL) subclass phenotype using a direct, automated assay of small dense LDL-cholesterol without sample pretreatment. Clin. Chim. Acta. 411: 1361–1366 [DOI] [PubMed] [Google Scholar]
- 11.Jiang X. C., Agellon L. B., Walsh A., Breslow J. L., Tall A. 1992. Dietary cholesterol increases transcription of the human cholesteryl ester transfer protein gene in transgenic mice. Dependence on natural flanking sequences. J. Clin. Invest. 90: 1290–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi J., Lang W., Giardino E., Caldwell G. W., Smith C., Minor L. K., Darrow A. L., Willemsens G., Dewaepenaert K., Roevens P., et al. 2010. High-content assays for evaluating cellular and hepatic diacylglycerol acyltransferase activity. J. Lipid Res. 51: 3559–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masson D., Jiang X. C., Lagrost L., Tall A. R. 2009. The role of plasma lipid transfer proteins in lipoprotein metabolism and atherogenesis. J. Lipid Res. 50(Suppl.): S201–S206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tall A. 1995. Plasma lipid transfer proteins. Annu. Rev. Biochem. 64: 235–257 [DOI] [PubMed] [Google Scholar]
- 15.Mahsut A., Wang S. P., McLaren D. G., Bhat G., Herath K., Miller P. L., Hubbard B. K., Johns D. G., Previs S. F., Roddy T. P. 2011. Headspace analyses of (2)H labeling of acetone: enabling studies of fatty acid oxidation in vivo. Anal. Biochem. 408: 351–353 [DOI] [PubMed] [Google Scholar]