Abstract
Mutations in the ATP-binding cassette transporter A1 (ABCA1) are a major cause of decreased HDL cholesterol (HDL-C), which infers an increased risk of cardiovascular disease (CVD). Many ABCA1 mutants show impaired localization to the plasma membrane. The aim of this study was to investigate whether the chemical chaperone, sodium 4-phenylbutyrate (4-PBA) could improve cellular localization and function of ABCA1 mutants. Nine different ABCA1 mutants (p.A594T, p.I659V, p.R1068H, p.T1512M, p.Y1767D, p.N1800H, p.R2004K, p.A2028V, p.Q2239N) expressed in HEK293 cells, displaying different degrees of mislocalization to the plasma membrane and discrete impacts on cholesterol efflux, were subject to treatment with 4-PBA. Treatment restored localization to the plasma membrane and increased cholesterol efflux function for the majority of mutants. Treatment with 4-PBA also increased ABCA1 protein expression in all transfected cell lines. In fibroblast cells obtained from low HDL-C subjects expressing two of the ABCA1 mutants (p.R1068H and p.N1800H), 4-PBA increased cholesterol efflux without any increase in ABCA1 expression. Our study is the first to investigate the effect of the chemical chaperone, 4-PBA on ABCA1 and shows that it is capable of restoring plasma membrane localization and enhancing the cholesterol efflux function of mutant ABCA1s both in vitro and ex vivo. These results suggest 4-PBA may warrant further investigation as a potential therapy for increasing cholesterol efflux and HDL-C levels.
Keywords: ATP-binding cassette transporter, high density lipoprotein, atherosclerosis, cholesterol efflux
Low levels of HDL cholesterol (HDL-C) are an independent risk factor for the development of cardiovascular disease (CVD). ABCA1 facilitates the rate-limiting step in HDL formation, the efflux of cellular lipid onto apoA-I. Over 150 unique ABCA1 mutations have been reported and impaired ABCA1 activity associated with mutations in ABCA1 is a major genetic cause of low HDL-C levels (1, 2). The reduced function of many of these has been attributed to intracellular retention and prolonged residence in the endoplasmic reticulum (ER), leading to mislocalization to the plasma membrane (1, 3, 4). Mutations that cause misfolding are an established cause of mislocalization for other membrane transporter proteins (5). Treatment with chemical chaperones can improve the folding, trafficking, and function of such proteins (5). Here, we investigate the effect of the chemical chaperone, sodium 4-phenylbutyrate (4-PBA) on the localization and function of ABCA1.
4-PBA is a short-chain fatty acid that can rescue the detrimental phenotype of misfolded, mislocalized proteins in vitro and in vivo (6). In particular, 4-PBA can restore functionality to mutant ATP-binding cassette transporters including ABCA3, ABCD2, ABCC6, ATPB7, and CFTR (7–11), which prompted its use here. 4-PBA has been used in a wide range of clinical applications including routine treatment of patients with cystic fibrosis, thalassemias, urea cycle disorders, and cancer (6). Treatment of CFTR mutants responsible for cystic fibrosis has shown that 4-PBA ameliorates ER retention and is sufficient to restore protein function in vitro and in vivo (11, 12). Mechanisms for its action include the ability to act as a chaperone (13), reduce ER stress (14), transcriptionally upregulate chaperone proteins (15), provide histone deactylase inhibitor activity (16), and scavenge ammonia (17).
The application of chemical chaperones to ABCA1 has not been described; however, mutant ABCA1 proteins with aberrant cellular localization are suitable candidates for such treatment. We investigated the impact of 4-PBA on nine naturally occurring ABCA1 missense mutants. Six of the ABCA1 mutations were previously identified in low HDL-C subjects and included three uncharacterized mutations, p.I659V, p.R2004K, and p.A2028V (18) and three variants, p.R1068H, p.T1512M, and p.N1800H, known to have reduced localization and cholesterol efflux (19, 20). We identified a further three novel mutations, p.A594T, p.Y1767D, and p.Q2239N, and these were also included in this study. We hypothesized that efflux function would be improved for mutants that are dysfunctional as a result of protein mislocation. The results presented here provide the first evidence that chaperone-mediated restoration of mutant ABCA1 cellular localization improves efflux function.
MATERIALS AND METHODS
ABCA1 gene analysis
The study was approved by the Lower South Regional Ethics Committee. All subjects gave informed consent and all procedures were in accordance with institutional guidelines. Screening of the ABCA1 gene in 10 low HDL-C subjects (<0.8 mmol/L) was performed by PCR amplification and sequencing of all 50 exons of the ABCA1 gene (20). Primer sequences are provided in supplementary Table I. All PCR reactions contained 10 ng DNA, 14 pmol each of the forward and reverse primers, 1× Roche Faststart Taq PCR buffer (2 mM MgCl2), 200 μM each dNTP, 1 U Faststart Taq DNA polymerase (Roche; Mannheim, Germany) and MilliQ water to a final reaction volume of 20 μl. A two-step PCR amplification program was used and consisted of an initial denaturation step of 5 min at 95°C followed by 10 amplification cycles of denaturation at 95°C for 48 s, annealing at 63°C for 48 s, followed by extension at 72°C for 48 s. These cycles were repeated a further 25 times with the annealing temperature dropped to 61°C. A final extension of 10 min at 72°C was performed and the reactions were held at 4°C. A decreased annealing temperature combination of 59 to 57°C was used to amplify exons 17 and 44. Annealing at 61 to 59°C was used for exons 38 and 39 and 66 to 64°C for exons 10, 15, 23, and 24 to eliminate the amplification of nonspecific products. Protein residue numbering was according to NP_005493.2.
Mutant ABCA1-green fluorescent protein (GFP) expression vectors under control of the heterologous cytomegalovirus (CMV) promoter were created by PCR site-directed mutagenesis (primers are provided in supplementary Table II) of a wild-type pCIneo-ABCA1-GFP cDNA vector (kindly provided by Professor Christiane Albrecht, Institute of Biochemistry and Molecular Medicine, University of Bern). One mutation was induced per round of mutagenesis using high fidelity, nonstrand displacing, Pfu DNA polymerase (Promega; Madison, WI). PCR reactions contained 1× Pfu Buffer, 200 μM dNTPs, 0.5 μmol each of the forward and reverse mutagenic primers, 75 ng template plasmid DNA, 0.4 U Pfu polymerase, and MilliQ water to make up a 20 μL reaction volume. Cycling parameters consisted of initial denaturation at 98°C for 2 min followed by 35 cycles of denaturation at 98°C for 40 s, annealing at 55°C for 1 min, and extension at 72°C for 15 s per kb of template DNA. A final extension at 72°C for 10 min was performed. The mutation position and the entire cDNA insert for all vectors were verified by sequencing.
Cell culture
Primary fibroblast cultures were established from 2 mm punch biopsies (20) and were cultured in Advanced DMEM. HEK293 cells were obtained from the American Type Tissue Collection (Manassas, VA) and maintained in DMEM. Both media were supplemented with 10% FBS (Bio International; Auckland, New Zealand), 2 mM L-glutamine, 0.25 μg/ml amphotericin B, 100 U/ ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified environment with 5% CO2. Unless otherwise stated, all cell culture reagents were purchased from Invitrogen (Carlsbad, CA). A 100 mM stock of 4-PBA (Tocris Bioscience; Ellisville, MO) was made using sterile water and diluted to a 10 mM working stock in serum-free culture medium. Culture medium was replaced with 4-PBA supplemented serum-free medium for 24 h prior to the experiment endpoint. ApoA-I protein used for functional experiments was isolated from normolipidemic human plasma according to the method of Brace et al. (21), and was presented to cells in serum-free medium, supplemented with 10 mM 4-PBA when appropriate.
Cellular transfection protocol
Twenty four hours after seeding in 12-well culture plates, HEK293 cells were transiently transfected with the ABCA1-GFP cDNA expression vectors or the ABCA1 promoter vector using FuGENE 6 (Roche) transfection reagent at a ratio of 3:1 reagent (μL) to DNA (μg). Serum-free DMEM was mixed with FuGENE 6 and incubated at room temperature for five minutes. The plasmid DNA was added, mixed well, and incubated for a further 20 min at room temperature. Culture medium was replaced with fresh medium (containing [3H]cholesterol when required) and transfection mix was added to the cells in a drop-wise manner.
Cholesterol efflux assays
Cholesterol efflux assays were performed using primary fibroblast cells and transfected HEK293 cells as described previously (20). Twenty four hours after cells were seeded, the culture medium was replaced with fresh medium containing 0.5 μCi/well [3H]cholesterol (Amersham Biosciences; Piscataway, NJ). HEK293 cells were transiently transfected with 1 μg ABCA1-GFP cDNA expression vector per well immediately following the addition of the [3H]cholesterol. Following a 48 h incubation, to allow for cellular incorportation of the labeled cholesterol and expression of the transfected cDNA in HEK293 cells, the cells were equilibrated in serum-free medium for 12 h and were then incubated in serum-free medium containing 10 μg/ml apoA-I protein. After 8 (fibroblast) or 12 (HEK293 cells) h incubation the medium was collected and centrifuged at 3,000 g for 5 min to pellet cellular debris and the cellular lysates were harvested from the culture plate by addition of 0.1 M NaOH. The tritium decay over 5 min was determined for both the media and cellular fractions using Optiphase Hisafe II scintillation fluid (Perkin Elmer; Boston, MA). Cholesterol efflux was calculated as the proportion of total radioactivity present in the medium fraction. Nonspecific efflux in the absence of apoA-I was determined and subtracted from each experimental measurement.
Confocal microscopy
The cellular localization of ABCA1-GFP proteins was examined using confocal microscopy (19). Transfected HEK293 cells were cultured on poly-l-ornithine coated glass coverslips and 48 h following transfection the cells were incubated in serum-free medium overnight. The following day, cells were incubated with fresh medium containing 10 μg/ml apoA-I for 6 h. Cells were fixed using 4% formaldehyde and stained with 5 μg/ml AlexaFluor594 Wheat Germ agglutinin (Invitrogen) to label cell membranes and mounted on glass slides. Images were obtained using a Zeiss LSM 510 confocal microscope with the Argon (488 nm) and HeNe (633 nm) lasers and were analyzed using the Zeiss LSM Image Examiner software. The Pearson's correlation coefficient between the GFP and AlexaFluor594 signal was calculated from images using the Colocalisation Finder plugin of the ImageJ software.
Protein isolation and Western blot analysis
Western blots of cell lysates were performed as described previously (19). Cells were grown to confluence and lysed in RIPA buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS] supplemented with complete mini protease inhibitors (Roche). Lysates were separated by SDS-PAGE using 7.5% gels and transferred to nitrocellulose membrane. Membranes were probed with a polyclonal antibody to actin (A5060: Sigma; St. Louis, MO) and either a monoclonal antibody to ABCA1 (ab18180: AbCam; Cambridge, UK) or a polyclonal antibody to GFP (ab290, AbCam). Quantification of band intensity was performed using ImageQuant TL software (Amersham Biosciences).
RNA isolation and quantitative RT-PCR
Total RNA was isolated from cells cultured to confluence using the RNeasy mini kit (Qiagen, Hilden, Germany). Isolated RNA was treated with DNase I (Qiagen) and 1 μg was reverse transcribed to cDNA using Transcriptor Reverse Transcriptase (Roche) according to the manufacturer's instructions. Quantitative RT-PCR was performed using a LightCycler® 480 (Roche) with the SYBR Green Master Mix kit (Roche). Primer sequences are provided in supplementary Table III. Reactions were set up in 96-well plates and contained 1 μM forward and reverse primer, 5 μL SYBR Green I Master (Roche), 2 μL diluted cDNA and sterile water to a final volume of 10 μL. Amplification of the template was performed using an initial denaturation step of 95°C for 5 min followed by 40 to 50 cycles of denaturation at 95°C, annealing at 61°C for 5 s, and extension at 72°C for 8 s. There was a single data acquisition point at the end of each extension. The final step of each run was a melt curve analysis with continuous data acquisition and consisted of denaturing at 95°C for 30 s, cooling to 65°C, and heating with a ramp rate of 0.11°C per second to 95°C. ABCA1 mRNA expression relative to GAPDH was calculated from in-run standard curves.
Luciferase reporter gene assays
The transcriptional response of the native ABCA1 promoter to 4-PBA was examined using HEK293 cells transfected with luciferase reporter constructs. To create an ABCA1 promoter reporter vector, a 715 bp region (position -714 to +1) of the wild-type ABCA1 promoter was amplified and cloned into the pCR2.1 entry vector (Invitrogen). A 700 bp region of the promoter (position −699 to +1) was then subcloned from the pCR2.1 vector into the pGL4.10 reporter vector containing firefly luciferase (Promega; Madison, WI) using primers containing EcoRV and HindIII restriction sites (primers are provided in supplementary Table IV). Cells were transiently cotransfected with 0.5 μg of the ABCA1-promoter pGL4.10 vector, and 5 ng of the phRL-SV40 vector, designed to constitutively expression renilla luciferase. Growth medium was replaced 24 h following transfection with fresh medium, containing 4-PBA when required. After a further 24 h, cell lysates were harvested and assayed for renilla and firefly luciferase activity using the Dual Luciferase® Reporter Assay System (Promega). Firefly luciferase measurements were normalized to renilla measurements and expressed relative to untreated ABCA1 promoter activity as 1.0.
Data analysis
Data are expressed as mean + SEM unless otherwise stated. Differences of means were assessed using the Student's t-test or one-way ANOVA with Bonferroni posthoc comparisons.
RESULTS
ABCA1 mutants with impaired localization have reduced cholesterol efflux function
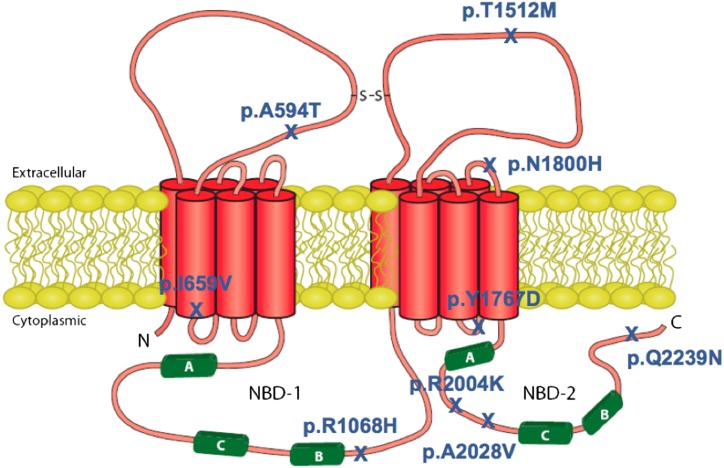
We identified three novel ABCA1 variants, p.A594T, p.Y1767D, and p.Q2239N, in heterozygote form in three individuals with HDL-C levels of 0.61, 0.17, and 0.37 mmol/L, respectively. The individual heterozygote for p.Y1767D was also heterozygote for the p.N1800H ABCA1 mutation. The position of all ABCA1 protein variants investigated within this study is shown in Fig. 1.
Fig. 1.
Schematic of the ABCA1 protein showing the position of the nine mutants studied.
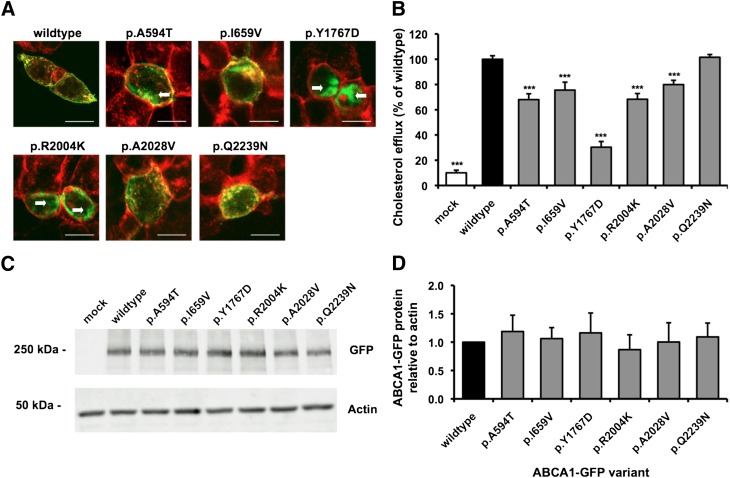
We first characterized the three novel ABCA1 mutants and three previously identified but uncharacterized mutants (p.I659V, p.R2004K, and p.A2028V) in HEK293 cells, which lack the endogenous ABCA1 protein (see supplementary Fig. I). Investigation of the six uncharacterized mutations in transfected HEK293 cells showed the p.A594T, p.I659V, p.Y1767D, p.R2004K, and p.A2028V mutants to have various degrees of mislocalization (Fig. 2A). Mislocation was indicated by areas of ABCA1-GFP (green) signal inside the cell as opposed to a yellow signal around the plasma membrane indicating normal localization of ABCA1-GFP to the plasma membrane (red). Areas of mislocalized ABCA1-GFP are indicated by arrows in Fig. 2A with the p.Y1767D and R2004K mutants being the most affected. The mislocalization of mutant ABCA1s was associated with a reduced cholesterol efflux function compared with wild-type-GFP ABCA1 (Fig. 2B) with the p.Y1767D mutant being the most affected (30.3% the efflux of wild-type). The p.Q2239N mutation did not affect plasma localization or cholesterol efflux function. A similar amount of radioactive cholesterol was incorporated into mutant and wild-type cells and the nonspecfic efflux did not differ signficantly between ABCA1 genotypes.
Fig. 2.
Mislocalization of ABCA1 to the plasma membrane correlates with reduced cholesterol efflux function in transfected HEK293 cells. A: Confocal microscopy of HEK293 cells expressing ABCA1-GFP (green), counterstained with AlexaFluor594 WGA membrane stain (red); regions of yellow indicate colocalization of ABCA1-GFP with membrane. Scale bars represent 10 μm and arrows indicate areas of substantial intracellular GFP signal. B: Cholesterol efflux from transfected cells to 10 μg/ml apoA-I protein. C: Western blot and (D) quantification for ABCA1-GFP protein level. Data represent mean + SEM of at least three experiments performed in quadruplicate for efflux and duplicate for protein quantification. ***p < 0.001 compared with wild-type or to all other values when assessed by ANOVA (D).
The ABCA1-GFP protein level in transfected HEK293 cells was assessed by Western blotting using an anti-GFP antibody (Fig. 2C). Quantification of the Western blot images showed the ABCA1-GFP signal to be comparable across all genotypes (as assessed using one-way ANOVA, Fig. 2D), which indicates all ABCA1-GFP proteins were expressed to a similar level.
4-PBA rescues mutant ABCA1 localization and improves cholesterol efflux function in transfected HEK293 cells
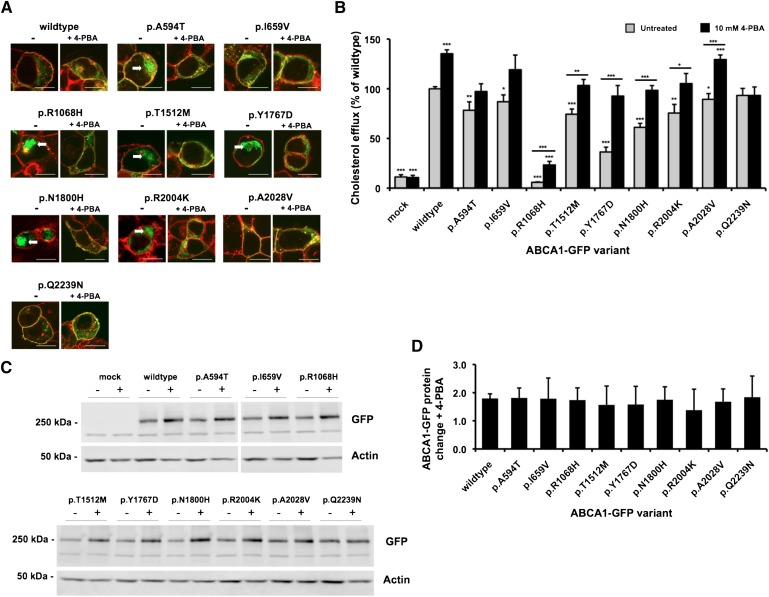
4-PBA treatment was applied to the six uncharacterized ABCA1 mutants as well as to three mutants that we have previously shown to have reduced cholesterol efflux function, p.R1068H (19), p.T1512M (20), and p.N1800H (20, Fig. 1). Treatment of transfected HEK293 cells with 4-PBA improved plasma membrane localization for all of the mislocated mutants (Fig. 3A). This was shown by a visible reduction in the intracellular GFP green signal (indicated by arrows in the most affected mutants) and a corresponding increase in yellow signal around the plasma membrane, indicating colocalization of ABCA1-GFP to the plasma membrane (red signal). Quantification of the colocalization of the ABCA1-GFP signal with plasma membrane signal showed a significantly reduced colocalization in most of the mutant cell lines compared with wild-type in the untreated state (supplementary Fig. II). Treatment with 4-PBA induced a significant increase in colocalization for the p.A594T, p.R1068H, p.T1512M, p.Y1767D, p.N1800H, and p.R2004K mutants. Treatment with 4-PBA did not affect the colocalization of the wild-type ABCA1-GFP protein (supplementary Fig. II).
Fig. 3.
The chemical chaperone 4-PBA restores membrane localization and rescues cholesterol efflux function of ABCA1 mutants. Transfected HEK293 cells were treated with 10 mM 4-PBA for 24 h. A: Confocal microscopy of cells expressing ABCA1-GFP (green) counterstained with AlexaFluor594 WGA membrane stain (red); regions of yellow indicate colocalization of ABCA1-GFP with membrane. Scale bars represent 10 μm and arrows indicate areas of substantial intracellular GFP signal. B: Cholesterol efflux from ABCA1-GFP transfected cells to 10 μg/ml apoA-I. C: Western blot and (D) quantification of foldchange in ABCA1-GFP protein level with 4-PBA treatment. Data represent mean + SEM of two experiments performed in quadruplicate for efflux and duplicate for Western blot protein quantification. *p < 0.05, **p < 0.01, ***p < 0.001 compared with untreated wild-type value (symbols directly above error bars) and for treated versus untreated values of the same genotype (symbols above lines). For (D), the comparison was to all other values, as assessed by ANOVA.
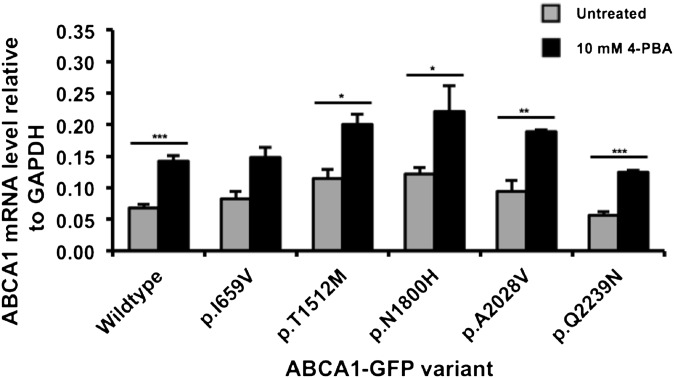
Upon 4-PBA treatment, efflux function was significantly increased relative to the untreated level for the p.R1068H, p.T1512M, p.Y1767D, p.N1800H, p.R2004K, and p.A2028V mutants (Fig. 3B). There was no change in cholesterol efflux function for the mock transfected cells, confirming that the 4-PBA treatment was specifically acting on the ABCA1-GFP proteins. Two of the three most severely mislocated mutants, p.Y1767D, and p.N1800H, showed a restored efflux function that was equivalent to wild-type untreated cells. However, the cholesterol efflux of the p.R1068H mutant was unable to be restored to wild-type levels. Interestingly, the cholesterol efflux of the wild-type ABCA1-GFP transfected cells was also significantly increased after 4-PBA treatment (Fig. 3B). Treatment of transfected HEK293 cells with 4-PBA increased ABCA1-GFP protein level by a similar fold increase (1.4- fold to 1.8-fold) across all genotypes including wild-type (Fig. 3C, D). Furthermore, 4-PBA increased the level of wild-type and mutant ABCA1-GFP mRNA (1.8-fold to 2.1-fold) in transfected cells (Fig. 4). These results indicate that 4-PBA promotes a similar transcriptional upregulation of the heterologous promoter within the wild-type and mutant ABCA1-GFP vectors that results in an increased ABCA1 protein level.
Fig. 4.
4-PBA increases ABCA1-GFP mRNA level in transfected HEK293 cells. HEK293 cells transfected with wild-type ABCA1-GFP were treated with 10 mM 4-PBA for 24 h, total mRNA was harvested, reverse transcribed, and ABCA1 mRNA level examined by quantitative RT-PCR relative to GAPDH. Data represents mean + SEM for two experiments performed in duplicate. *p < 0.05, **p < 0.01, ***p < 0.001 compared with the untreated condition.
4-PBA improves mutant ABCA1 efflux function independent of protein level in primary fibroblasts
We assessed the effect of 4-PBA treatment in the context of the native ABCA1 promoter using available p.R1068H, p.N1800H, and wild-type primary fibroblast cell lines (19, 20). Fibroblasts were obtained from two p.R1068H heterozygote carriers (RH1 and RH2), two p.N1800H heterozygote carriers (NH1 and NH2), and two ABCA1 wild-type subjects (WT1 and WT2). The mutant carriers RH1, RH2, and NH1 were confirmed to carry no other ABCA1 mutations on either their mutant or wild-type alleles. We had previously determined that subject NH2 was also heterozygous for the p.C978fsX988 mutation, which encodes a truncated protein that is functionally null and in this individual was present on a seperate allele to the p.N1800H mutation (20).
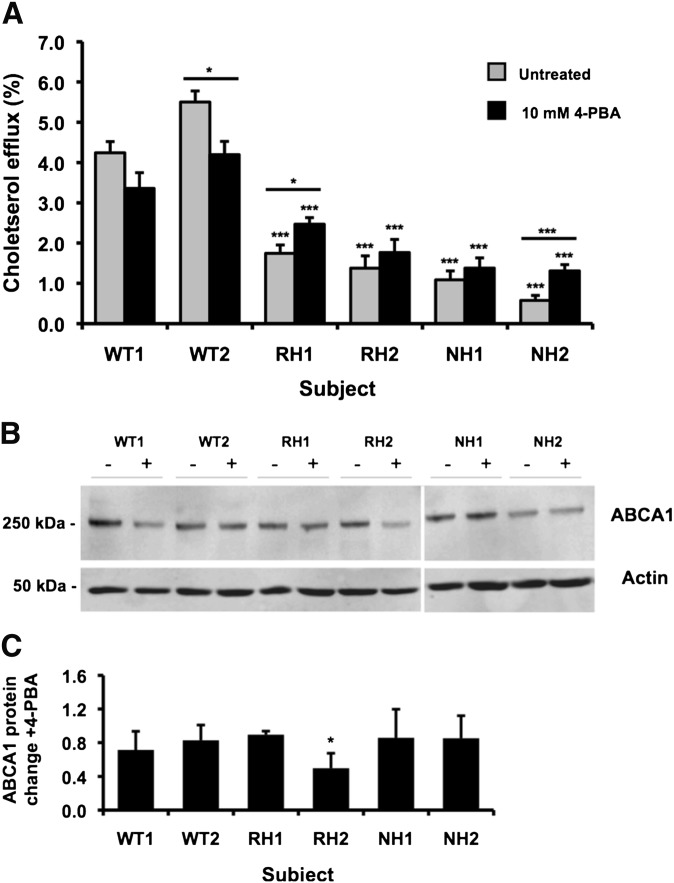
The cholesterol efflux function of the mutant fibroblast cell lines was lower than the efflux from wild-type cells (Fig. 5A) as previously shown (19, 20). After treatment with 4-PBA, the cholesterol efflux function of wild-type ABCA1 fibroblast cells was reduced (significantly in the case of WT2). In contrast, there was a clear trend for increased efflux in the mutants fibroblast cells with statistically significant increases for the p.R1068H heterozygote RH1 (p < 0.05) and the p.N1800H heterozygote NH2 (p < 0.001). Despite this improvement in efflux function in the mutant cell lines, the efflux remained 34.6–65.3% that of untreated wild-type cells (p < 0.001, Fig. 5A). It was noted that the increase in cholesterol efflux promoted by 4-PBA in the p.N1800H fibroblasts was not as striking as that seen in HEK293 cells transfected with this mutant where 4-PBA promoted cholesterol efflux back up to untreated wild-type levels.
Fig. 5.
Treatment with 4-PBA increases the function of mutant ABCA1 primary fibroblast cells and decreases protein expression. Fibroblast cultures established from wild-type (WT1, WT2), p.R1068H carriers (RH1, RH2) and p.N1800H carriers (NH1, NH2) were treated with 10 mM 4-PBA for 24 h. A: Cholesterol efflux from fibroblast cells to 10 μg/ml apoA-I. B: Western blot and (C) quantification of foldchange for ABCA1 protein level with 4-PBA treatment. Data represent the mean + SEM of two experiments performed in quadruplicate for efflux and in duplicate for Western blot protein quantification. *p < 0.05, ***p < 0.001 compared with untreated values of both wild-type subjects (symbols directly above error bars) and for treated versus untreated values for each subject (symbols above lines) or to all other values when assessed by ANOVA (C).
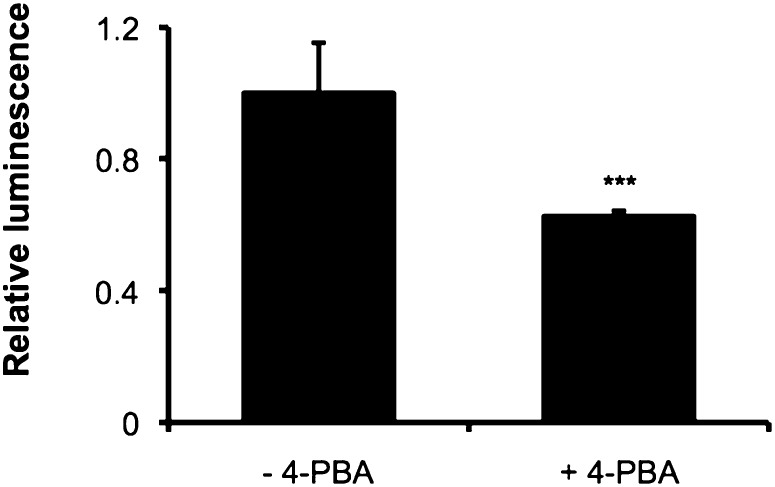
In contrast to transfected HEK293 cells, the ABCA1 protein level was decreased for all wild-type and mutant fibroblast cell lines following 4-PBA treatment (Fig. 5B). There was a similar decrease in protein level for most cells lines with a greater decrease for RH2 cells (p < 0.05), as assessed using one-way ANOVA (Fig. 5C). Further investigation using luciferase reporter assays showed 4-PBA to decrease the transcriptional activity of the native ABCA1 promoter (Fig. 6), indicating the lower protein level in treated fibroblasts to be the result of transcriptional downregulation of ABCA1 by 4-PBA.
Fig. 6.
4-PBA decreases transcriptional activity of the native ABCA1 promoter. Wild-type ABCA1 promoter activity in transfected HEK293 cells, as measured by dual luciferase reporter assay. Data represents mean + SEM for two experiments performed in triplicate. ***p < 0.001 compared with the untreated condition.
DISCUSSION
The ABCA1 protein functions as a tetramer in the plasma membrane (22) where it facilitates the efflux of cholesterol onto apoA-I to form HDL. The ABCA1 protein is very intolerant to structural change and many ABCA1 mutants exhibit misfolding and intracellular retention (1, 3, 4), leading to a lack of functional protein at the plasma membrane and associated low HDL-C levels. We hypothesized that restoring localization of mutant ABCA1 proteins would improve their function. We show that the chemical chaperone 4-PBA can restore localization of mutant ABCA1 proteins and enhance their cholesterol efflux function. Our study is the first application of a chemical chaperone treatment to ABCA1 and our results highlight that correct localization of ABCA1 is a major factor determining its function.
Our analysis in transfected HEK293 cells using nine different ABCA1 missense mutants with varying degrees of mislocalization showed that mislocalization coincided with a loss in cholesterol efflux function in some of the mutants. Treatment of the mutant cell lines with 4-PBA ameliorated intracellular retention and improved plasma membrane localization for all mislocated mutants. Most importantly, the correction of localization restored efflux function to the level of untreated wild-type transfected cells for the majority of mutants. The magnitude by which 4-PBA rescued function was mutant-dependent as restored localization did not always result in a restoration of function, as was the case for the p.R1068H mutant. Interestingly, despite no alteration to membrane localization, efflux function of the wild-type protein was also increased by 4-PBA. This is most likely due to the increase in total wild-type ABCA1 protein level that was induced by 4-PBA in HEK293 cells.
For two of the most dramatically affected mutants, p.R1068H and p.N1800H, a functional improvement with 4-PBA treatment was also confirmed ex vivo using primary fibroblast cells. Although 4-PBA also increased the cholesterol efflux function of ABCA1 mutants in primary fibroblast, the magnitude of the effect was much less than that seen in the HEK293 system. The lesser effect in fibroblasts may relate to the reduced expression of ABCA1 promoted by 4-PBA, which, for wild-type ABCA1, is associated with a reduced efflux. This would be expected to dampen the magnitude of functional improvements in heterozygous mutant fibroblasts harboring a wild-type allele. This is supported by the results seen in the NH2 versus NH1 fibroblasts. The NH2 fibroblasts, which harbor a p.N1800H allele and a null allele, show a significant improvement in function, yet the NH1 fibroblasts, which harbor a p.N1800H allele and a wild-type allele, show no significant functional improvement. Hence, for fibroblasts only harboring mutant ABCA1 alleles, the increase in function promoted by 4-PBA reflects the increase in mutant allele function whereas in fibroblasts harboring a mutant and wild-type ABCA1 allele, the increase in function promoted by 4-PBA reflects the increase in mutant allele function combined with the decrease in wild-type allele function.
Improved positioning of ABCA1 at the plasma membrane has been shown previously for a p.Q597R mutant under conditions of induced ER stress; however, in that instance, no improvement in efflux activity was seen (23). This is in contrast with our results where the majority of the mutants showed significant improvements in function, including the p.A594T mutant, which is of a similar location to p.Q597R. This discrepancy could be due to the mechanism by which plasma membrane localization was restored by the two different treatments. Plasma membrane localization of the p.Q597R protein was promoted with thapsigargin, an inducer of ER stress, whereas 4-PBA is known to alleviate ER stress (14) by acting as a chaperone itself (13) as well as inducing the expression of endogenous chaperone proteins (15). The exact nature of the mutation could also be important. Indeed, our data shows that the specific location of the mutation is an important determinant of the capacity for functional rescue. For example, the p.Y1767D mutant showed a much enhanced localization and dramatic increase in cholesterol efflux after 4-PBA treatment whereas the efflux function for the p.R1068H mutant remained low despite showing a similar enhancement in localization. The p.R1068H mutant, located in the first ATP binding domain, has been shown to be defective in oligomerisation (19) and it is likely that although localization was improved for this mutant with 4-PBA treatment, the oligomerisation and therefore efflux function remained defective.
As well as restoring localization, 4-PBA treatment had the effect of increasing ABCA1 mRNA and protein expression in transfected HEK293 cells. This effect is most likely due to its histone deacetylase inhibitor action. The ABCA1 expression vectors used here are driven by a heterologous CMV promoter and histone deacetylase inhibitors are known to exert a transcriptional effect on the CMV promoter (24). Functional improvements for other mutant proteins by 4-PBA have been attributed to increased expression (8, 10, 25), which suggests a global effect on transcription. Interestingly, we observed an opposite transcriptional effect of 4-PBA in fibroblasts harboring mutant ABCA1 proteins. 4-PBA decreased the transcriptional activity of the native ABCA1 promoter, which was associated with a decrease in ABCA1 protein level in fibroblasts. ABCA1 is highly regulated by many factors at the transcriptional level and one possibile mechanism for the decrease in ABCA1 promoter activity promoted by 4-PBA is that it could reduce the expression or activity of transcriptional regulators. As a fatty acid, 4-PBA may regulate transcription in a manner similar to unsaturated fatty acids that supress ABCA1 transcription through posttranslational antagonism of liver X receptor α (26).
The improvement in cholesterol efflux of ABCA1 mutants in fibroblasts despite the reduction in ABCA1 protein expression indicates that ABCA1 function and expression can be uncoupled. This uncoupling is not seen in wild-type fibroblasts where a decreased protein level is associated with a decreased efflux function. We are not the first to observe an uncoupling effect of a drug on ABCA1 function and expression. The cholesterol lowering drug probucol increases ABCA1 protein level by inhibiting its proteosomal degradation (27) but simultaneously inhibits efflux function by inhibiting translocation to the plasma membrane (28). As such, probucol provides an inverse example to 4-PBA.
This study provides the first evidence that the activity of mutant ABCA1s can be enhanced by the chemical chaperone 4-PBA through restoration of cellular localization. 4-PBA is already approved for clinical use and our data suggests that it could provide an unexplored avenue for the development of HDL-C raising therapies to reduce CVD risk. The direct application of 4-PBA as a therapeutic agent, however, requires caution and more investigation. The effect of 4-PBA on ABCA1 expression and function in other cell types, particularly hepatic and macrophage cells, should be investigated because both are important cell types in determining HDL-C levels. In addition, the exact mechanism for how 4-PBA restores cellular localization requires further investigation.
Supplementary Material
Acknowledgments
The authors thank the p.R1068H and p.N1800H family members for their participation in this study. We are grateful to Professor Christiane Albrecht for kindly providing the pCIneo-ABCA1-GFP expression vector.
Footnotes
Abbreviations:
- 4-PBA
- sodium 4-phenylbutyrate
- CFTR
- cystic fibrosis transmembrane regulator
- CMV
- cytomegalovirus
- CVD
- cardiovascular disease
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- HDL-C
- HDL cholesterol
This work was supported by the Health Research Council of New Zealand and University of Otago postgraduate scholarships (B.S and R.J.S).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and four tables.
REFERENCES
- 1.Candini C., Schimmel A. W., Peter J., Bochem A. E., Holleboom A. G., Vergeer M., Dullaart R. P., Dallinga-Thie G. M., Hovingh G. K., Khoo K. L., et al. 2010. Identification and characterization of novel loss of function mutations in ATP-binding cassette transporter A1 in patients with low plasma high-density lipoprotein cholesterol. Atherosclerosis. 213: 492–498 [DOI] [PubMed] [Google Scholar]
- 2.Frikke-Schmidt R., Nordestgaard B. G., Stene M. C., Sethi A. A., Remaley A. T., Schnohr P., Grande P., Tybjaerg-Hansen A. 2008. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 299: 2524–2532 [DOI] [PubMed] [Google Scholar]
- 3.Singaraja R. R., Visscher H., James E. R., Chroni A., Coutinho J. M., Brunham L. R., Kang M. H., Zannis V. I., Chimini G., Hayden M. R. 2006. Specific mutations in ABCA1 have discrete effects on ABCA1 function and lipid phenotypes both in vivo and in vitro. Circ. Res. 99: 389–397 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka A. R., Abe-Dohmae S., Ohnishi T., Aoki R., Morinaga G., Okuhira K., Ikeda Y., Kano F., Matsuo M., Kioka N., et al. 2003. Effects of mutations of ABCA1 in the first extracellular domain on subcellular trafficking and ATP binding/hydrolysis. J. Biol. Chem. 278: 8815–8819 [DOI] [PubMed] [Google Scholar]
- 5.Leandro P., Gomes C. M. 2008. Protein misfolding in conformational disorders: rescue of folding defects and chemical chaperoning. Mini Rev. Med. Chem. 8: 901–911 [DOI] [PubMed] [Google Scholar]
- 6.Iannitti T., Palmieri B. 2011. Clincal and experimental applications of sodium phenylbutyrate. Drugs R D. 11: 227–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong N., Madesh M., Gonzales L. W., Zhao M., Yu K., Ballard P. L., Shuman H. 2006. Functional and trafficking defects in ATP binding cassette A3 mutants associated with respiratory distress syndrome. J. Biol. Chem. 281: 9791–9800 [DOI] [PubMed] [Google Scholar]
- 8.Gondcaille C., Depreter M., Fourcade S., Lecca M. R., Leclercq S., Martin P. G., Pineau T., Cadepond F., ElEtr M., Bertrand N., et al. 2005. Phenylbutyrate up-regulates the adrenoleukodystrophy-related gene as a nonclassical peroxisome proliferator. J. Cell Biol. 169: 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Saux O., Fulop K., Yamaguchi Y., Ilias A., Szabo Z., Brampton C. N., Pomozi V., Huszar K., Aranyi T., Varadi A. 2011. Expression and in vivo resuce of human ABCC6 disease-causing mutants in mouse liver. PLoS ONE. 6: e24738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van den Berghe P. V., Stapelbroek J. M., Krieger E., de Bie P., van de Graaf S. F., de Groot R. E., van Beurden E., Spijker E., Houwen R. H., Berger R., et al. 2009. Reduced expression of ATP7B affected by Wilson disease-causing mutations is rescued by pharmacological folding chaperones 4-phenylbutyrate and curcumin. Hepatology. 50: 1783–1795 [DOI] [PubMed] [Google Scholar]
- 11.Zeitlin P. L., Diener-West M., Rubenstein R. C., Boyle M. P., Lee C. K., Brass-Ernst L. 2002. Evidence of CFTR function in cystic fibrosis after systemic administration of 4-phenylbutyrate. Mol. Ther. 6: 119–126 [DOI] [PubMed] [Google Scholar]
- 12.Rubenstein R. C., Egan M. E., Zeitlin P. L. 1997. In vitro pharmacologic restoration of CFTR-mediated chloride transport with sodium 4-phenylbutyrate in cystic fibrosis epithelial cells containing delta F508-CFTR. J. Clin. Invest. 100: 2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota K., Niinuma Y., Kaneko M., Okuma Y., Sugai M., Omura T., Uesugi M., Uehara T., Hosoi T., Nomura Y. 2006. Suppressive effects of 4-phenylbutyrate on the aggregation of Pael receptors and endoplasmic reticulum stress. J. Neurochem. 97: 1259–1268 [DOI] [PubMed] [Google Scholar]
- 14.Erbay E., Babaev V. R., Mayers J. R., Makowski L., Charles K. N., Snitow M. E., Fazio S., Wiest M. M., Watkins S. M., Linton M. F., et al. 2009. Reducing endoplasmic reticulum stress through a macrophage lipid chaperone alleviates atherosclerosis. Nat. Med. 15: 1383–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright J. M., Zeitlin P. L., Cebotaru L., Guggino S. E., Guggino W. B. 2004. Gene expression profile analysis of 4-phenylbutyrate treatment of IB3–1 bronchial epithelial cell line demonstrates a major influence on heat-shock proteins. Physiol. Genomics. 16: 204–211 [DOI] [PubMed] [Google Scholar]
- 16.Lea M. A., Tulsyan N. 1995. Discordant effects of butyrate analogues on erythroleukemia cell proliferation, differentiation and histone deacetylase. Anticancer Res. 15: 879–883 [PubMed] [Google Scholar]
- 17.Brusilow S. W., Finkelstein J. 1993. Restoration of nitrogen homeostasis in a man with ornithing transcarbamylase deficiency. Metabolism. 42: 1336–1339 [DOI] [PubMed] [Google Scholar]
- 18.Slatter T. L., Jones G. T., Williams M. J., van Rij A. M., McCormick S. P. 2008. Novel rare mutations and promoter haplotypes in ABCA1 contribute to low-HDL-C levels. Clin. Genet. 73: 179–184 [DOI] [PubMed] [Google Scholar]
- 19.Suetani R. J., Sorrenson B., Tyndall J. D., Williams M. J., McCormick S. P. 2011. Homology modeling and functional testing of an ABCA1 mutation causing Tangier disease. Atherosclerosis. 218: 404–410 [DOI] [PubMed] [Google Scholar]
- 20.Sorrenson B., Suetani R. J., Bickley V. M., George P. M., Williams M. J., Scott R. S., McCormick S. P. 2011. An ABCA1 truncation shows no dominant negative effect in a familial hypoalphalipoproteinemia pedigree with three ABCA1 mutations. Biochem. Biophys. Res. Commun. 409: 400–405 [DOI] [PubMed] [Google Scholar]
- 21.Brace R. J., Sorrenson B., Sviridov D., McCormick S. P. 2010. A gel-based method for purification of apolipoprotein A-I from small volumes of plasma. J. Lipid Res. 51: 3370–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trompier D., Alibert M., Davanture S., Hamon Y., Pierres M., Chimini G. 2006. Transition from dimers to higher oligomeric forms occurs during the ATPase cycle of the ABCA1 transporter. J. Biol. Chem. 281: 20283–20290 [DOI] [PubMed] [Google Scholar]
- 23.Tanaka A. R., Kano F., Ueda K., Murata M. 2008. The ABCA1 Q597R mutant undergoes trafficking from the ER upon ER stress. Biochem. Biophys. Res. Commun. 369: 1174–1178 [DOI] [PubMed] [Google Scholar]
- 24.Spenger A., Ernst W., Condreay J. P., Kost T. A., Grabherr R. 2004. Influence of promoter choice and trichostatin A treatment in expression of baculovirus delivered genes in mammalian cells. Protein Expr. Purif. 38: 17–23 [DOI] [PubMed] [Google Scholar]
- 25.Tveten K., Holla O. L., Ranheim T., Berge K. E., Leren T. P., Kulseth M. A. 2007. 4-Phenylbutyrate restores the functionality of a misfolded mutant low-density lipoprotein receptor. FEBS J. 274: 1881–1893 [DOI] [PubMed] [Google Scholar]
- 26.Uehara Y., Engel T., Li Z., Goepfert C., Rust S., Zhou X., Langer C., Schachtrup C., Wiekowski J., Lorkowski S., et al. 2002. Polyunsaturated fatty acids and acetoacetate downregulate the expression of the ATP-binding cassette transporter A1. Diabetes. 51: 2922–2928 [DOI] [PubMed] [Google Scholar]
- 27.Wu C. A., Tsujita M., Hayashi M., Yokoyama S. 2004. Probucol inactivates ABCA1 in the plasma membrane with respect to its mediation of apoliporotein binding and high density lipoprotein assembly and to its proteolytic degradation. J. Biol. Chem. 279: 30168–30174 [DOI] [PubMed] [Google Scholar]
- 28.Favari E., Zanotti I., Zimetti F., Ronda N., Bernini F., Rothblat G. H. 2004. Probucol inhibits ABCA1-mediated cellular lipid efflux. Arterioscler. Thromb. Vasc. Biol. 24: 2345–2350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.