Abstract
Proteasome is the main intracellular organelle involved in the proteolytic degradation of abnormal, misfolded, damaged or oxidized proteins 1, 2. Maintenance of proteasome activity was implicated in many key cellular processes, like cell's stress response 3, cell cycle regulation and cellular differentiation 4 or in immune system response 5. The dysfunction of the ubiquitin-proteasome system has been related to the development of tumors and neurodegenerative diseases 4, 6. Additionally, a decrease in proteasome activity was found as a feature of cellular senescence and organismal aging 7, 8, 9, 10. Here, we present a method to measure ubiquitin-proteasome activity in living cells using a GFP-dgn fusion protein. To be able to monitor ubiquitin-proteasome activity in living primary cells, complementary DNA constructs coding for a green fluorescent protein (GFP)–dgn fusion protein (GFP–dgn, unstable) and a variant carrying a frameshift mutation (GFP–dgnFS, stable 11) are inserted in lentiviral expression vectors. We prefer this technique over traditional transfection techniques because it guarantees a very high transfection efficiency independent of the cell type or the age of the donor. The difference between fluorescence displayed by the GFP–dgnFS (stable) protein and the destabilized protein (GFP-dgn) in the absence or presence of proteasome inhibitor can be used to estimate ubiquitin-proteasome activity in each particular cell strain. These differences can be monitored by epifluorescence microscopy or can be measured by flow cytometry.
Keywords: Cellular Biology, Issue 69, Molecular Biology, Medicine, Biomedical Engineering, Virology, proteasome activity, lentiviral particles, GFP-dgn, GFP-dgnFS, GFP, human diploid fibroblasts, flow cytometry, plasmid, vector
Protocol
1. Plasmid Construction
Order custom oligo-nucleotides encoding for dgn (ACKNWFSSLSHFVIHL11) and for dgnFS (HARTGSLACPTSSSICE) and ligate it into the pEGFP-C1 vector to obtain the fusion of the GFP with dgn/dgnFS (Figure 1).
Amplify the coding sequence for GFP-dgn and GFP-dgnFS by PCR according to the protocol of the pENTR Directional TOPO Cloning Kit and continue with the pLenti6/V5 Directional TOPO Cloning Kit (Figure 6).
2. Virus Production
The day before transfection (Day 1) seed out HEK 293FT cells in a T75 flasks so that they will be 90-95% confluent on the day of transfection.
On the day of transfection dilute 36 μl of lipofectamine 2000 Reagent in 1.5 ml of DMEM without serum and antibiotics in a 15 ml falcon. Use another 15 ml falcon and dilute 3 μg pLenti6/V5 carrying the complementary DNA construct for either GFP-dgn or GFP-dgnFS, 2.5 μg envelope encoding plasmid pMD2.G and 7.5 μg vector backbone psPAX2 in 1.5 ml of DMEM without serum and antibiotics. After 5 min the diluted DNA is combined with the diluted lipofectamine 2000 Reagent.
Incubate the mixture for 20 min at room temperature to allow the DNA-lipofectamine complexes to form.
Remove the medium of HEK 293FT cells, replace it carefully by 7 ml of medium (without antibiotics) and add the DNA-lipofectamine mixture to the flask and rock it back and forth for mixing (Day 2). Incubate the flask overnight at 37 °C in a humidified 5% CO2 incubator.
Change medium the next day (10 ml medium without antibiotics; Day 3).
After 48 hr (Day 5) harvest supernatant, centrifuge at 300 x g for 5 min at room temperature and filter the supernatant through a 0.45 μm PVDF filter.
Culture medium - DMEM (HEK 293FT)
DMEM 10% FBS 0.1 mM MEM NEAA 6 mM L-glutamine 1 mM MEM Sodium Pyruvate 1% Pen-Strep (optional) 500 μg/ml Geneticin (optional)
3. Virus Concentration by Polyethylene Glycol (PEG) Precipitation
Add one volume Polyethylene glycol solution (50 mM Polyethylene glycol, 41 mM NaCl, autoclave, pH =7.2; PEG) to four volumes of supernatant and incubate for 2 hr at 4 °C. Carefully mix it every 20-30 min by inverting.
Spin down at 1500 x g for 30 min at 4 °C. A white pellet should be visible.
Aspirate the supernatant and centrifuge again at 1500 x g for 5 min at 4 °C. Aspirate the remaining PEG solution.
Resuspend the pellet in medium or phosphate-buffered saline (PBS) by pipetting up and down and vigorously vortex for 20 to 30 seconds. As a guideline use 500 μl for one T75 flask and aliquot it to 100 μl. Store the virus at -80 °C.
4. Titering of Virus
Seed out 5×104 U2-OS cells to each well of a 6-well plate the day before transfection.
On the day of transfection remove the culture medium and replace it by 1 ml of DMEM containing 8 μg/ml polybrene (hexadimethrine bromide). Dilute the concentrated virus supernatant 1:10 and 1:100 and add 1 μl each to one well. For higher virus concentrations use 1 μl, 5 μl and 10 μl of concentrated virus supernatant for the remaining wells. One well is left as a positive control.
The following day replace the medium by 2 ml of DMEM.
Start with the selection the next day. Apply 10 μg/ml Blasticidin on each well. Replace the antibiotic containing medium every second day.
Approximately 6-7 days after starting the selection the untransduced cells are dead. For the staining wash the cells three times with PBS and cover the cells with crystal violet (10 mg/l crystal violet in 20% ethanol) and incubate for 5-10 min at room temperature. Aspirate crystal violet (can be reused a few times) and rinse the wells twice with ddH2O and dry the plate.
Count the colonies and multiply with 1,000 and the corresponding dilution (Figure 7). This procedure allows to calculate transfection units (TU) of the virus / ml.
Culture medium - DMEM (HFF-2/U2-OS)
DMEM 10% FBS 6mM L-glutamine 1% Pen-Strep
5. Transduction of Human Diploid Fibroblasts (This procedure can be used for any cell type.)
Seed out 5×104 human diploid fibroblasts (HDFs) to 6-well plates the day before transduction.
Use a multiplicity of infection of two together with 8 μg/ml polybrene as transduction enhancer in a total of one milliliter.
Change medium the next day.
When the cells are at 70-80 % of confluency start the selection with 10 μg/ml blasticidin. After the selection is completed (no more untransfected cells die) the expansion of the cells can start and the cells are ready for experiments. Chronic selection with blasticidin allows to generate a stable cell line overexpressing GFP-dgn or GFP-dgnFS fusion proteins, ready to measure proteasome activity. This procedure guarantees a very high transfection efficiency independent of the cell type.
6. Measurement by Flow Cytometry
Seed out 1×105 cells (carrying either GFP-dgn or GFP-dgnFS) the day before the experiment.
Treat control cells 3 hr prior the measurement with the proteasome inhibitor, e.g. 100 μg/ml LLnL.
Wash twice with PBS and harvest the cells using 0.5 ml of trypsin. To stop the trypsinisation use 4.5 ml of culture medium and transfer the cells to a 15 ml falcon.
Spin the cells at 300 x g for 5 min at room temperature.
Aspirate the medium, resuspend the cells in PBS and spin down again at 300 x g for 5 min at 37 °C.
Aspirate the PBS, resuspend in 400 μl of FACS buffer (10 mM Hepes, 140 mM NaCl, 2.5 mM CaCl2, pH=7.4) and transfer the cells into FACS tubes.
Keep the cells on ice and measure samples by flow cytometry. The cells should not be stored longer than 30 min at 4 °C.
7. Representative Results
The GFP-dgn fusion protein carries a sequence which is targeted to the proteasome and therefore the protein is immediately degraded; it corresponds to the decrease in GFP fluorescence signal. The frameshift mutant (GFP-dgnFS) carries a mutated version of this sequence and is not degraded by proteasome; it leads to the higher green fluorescence. For these reasons, young HDFs with expected high proteasome activity and transduced with GFP-dgn show a low (6% positive cells) fluorescence signal in both flow cytometry measurement and in epifluorescence (Figure 2A and B, Figure 3). The same HDFs transduced with GFP-dgnFS display 39.7% of positive cells. The treatment of the cells with proteasome inhibitor LLnL (N-acetyl-L-leucyl-L-leucyl-L-norleucinal) raised the signal to maximum of 62.9% in both, GFP-dgn and GFP-dgnFS cells (Figure 2A and B).
To determine possible decline in proteasome activity in aged human skin samples, dermal fibroblasts isolated from young, middle-aged and old donors were infected with GFP-dgn and GFP-dgnFS, as described above and cultivated to the same passage number before analysis by flow cytometry. In these experiments, a clear-cut increase in GFP signal between young individuals (11.2±0.88% GFP-positive cells) and middle-aged donors (20.4±2.27% GFP-positive cells, P=0.003) has been observed, indicating a decrease in proteasome activity in samples obtained from aged donors7 (Figure 4). No further decrease in proteasome activity was observed in fibroblasts isolated from oldest individuals (Figure 4). Fluorescence intensity for GFP-dgnFS was in all cases ~90% (data not shown) which indicates a high transfection efficiency for the three different age groups.
 Figure 1. The GFP-dgn and GFP-dgnFS sequence. Displayed is the 3'-end of GFP (green) the multiple cloning site of the pEGFP-CL1 vector (grey) and the dgn/dgnFS sequence (red).
Figure 1. The GFP-dgn and GFP-dgnFS sequence. Displayed is the 3'-end of GFP (green) the multiple cloning site of the pEGFP-CL1 vector (grey) and the dgn/dgnFS sequence (red).
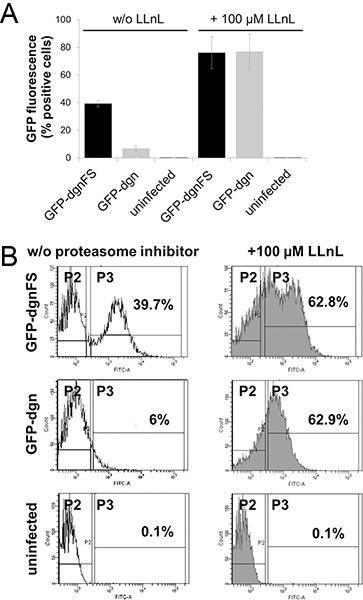 Figure 2. Flow cytometry analysis of GFP-dgn and GFP-dgnFS human diploid fibroblasts. A. Young human foreskin fibroblasts (HFF-2) were infected with lentiviral vectors carrying a green fluorescent protein (GFP)–dgn gene or a GFP–dgnFS (frameshift) construct, as indicated and analyzed for GFP fluorescence using flow cytometry (FACS Canto II, Becton Dickinson). Where indicated, cells were also treated for 3h with proteasome inhibitor N-acetyl-L-leucyl-L-leucyl-L-norleucinal (LLnL). Uninfected cells were used as a control. Experiments were performed in triplicates. B. Data are representative of three independent experiments. The numbers reflect the amount of GFP positive cells. Click here to view larger figure.
Figure 2. Flow cytometry analysis of GFP-dgn and GFP-dgnFS human diploid fibroblasts. A. Young human foreskin fibroblasts (HFF-2) were infected with lentiviral vectors carrying a green fluorescent protein (GFP)–dgn gene or a GFP–dgnFS (frameshift) construct, as indicated and analyzed for GFP fluorescence using flow cytometry (FACS Canto II, Becton Dickinson). Where indicated, cells were also treated for 3h with proteasome inhibitor N-acetyl-L-leucyl-L-leucyl-L-norleucinal (LLnL). Uninfected cells were used as a control. Experiments were performed in triplicates. B. Data are representative of three independent experiments. The numbers reflect the amount of GFP positive cells. Click here to view larger figure.
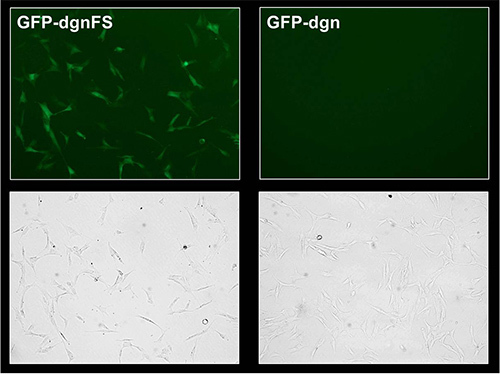 Figure 3. Fluorescence microscopy analysis of GFP-dgn and GFP-dgnFS HFF-2 cells. Young HFF-2 were treated as in Figure 2. At 9 days after infection, cells were visualized by fluorescence and phase contrast microscopy.
Figure 3. Fluorescence microscopy analysis of GFP-dgn and GFP-dgnFS HFF-2 cells. Young HFF-2 were treated as in Figure 2. At 9 days after infection, cells were visualized by fluorescence and phase contrast microscopy.
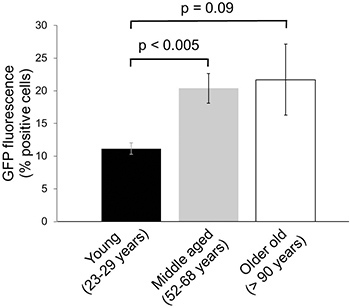 Figure 4. Changes in proteasome activity in human skin aging. The human foreskin fibroblasts from nine different donors in the indicated age groups were minimally expanded and infected with lentiviral constructs coding for GFP–dgn. At 9 days after infection, the cells were analyzed by flow cytometry. Data were obtained on three samples per age group in duplicates (± SE).
Figure 4. Changes in proteasome activity in human skin aging. The human foreskin fibroblasts from nine different donors in the indicated age groups were minimally expanded and infected with lentiviral constructs coding for GFP–dgn. At 9 days after infection, the cells were analyzed by flow cytometry. Data were obtained on three samples per age group in duplicates (± SE).
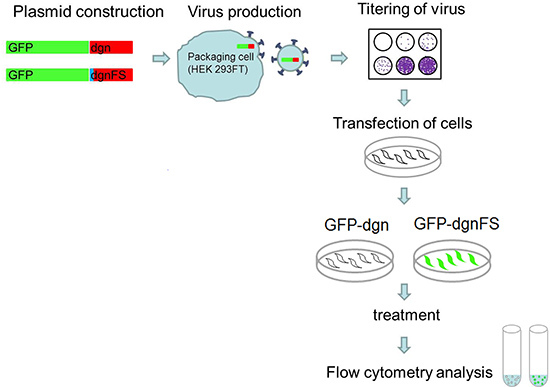 Figure 5. Experimental approach. Custom-oligo nucleotides for dgn and dgnFS are cloned into the pEGFP-C1 vector and viruses are produced for each construct using HEK 293FT cells. The titer of the viruses is determined. Cells are transduced with the virus and expanded. After the favored treatment the cells are analyzed for fluorescence signal by flow cytometry.
Figure 5. Experimental approach. Custom-oligo nucleotides for dgn and dgnFS are cloned into the pEGFP-C1 vector and viruses are produced for each construct using HEK 293FT cells. The titer of the viruses is determined. Cells are transduced with the virus and expanded. After the favored treatment the cells are analyzed for fluorescence signal by flow cytometry.
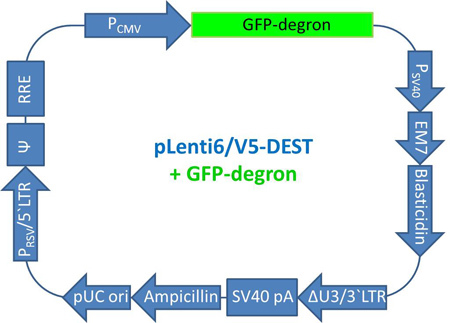 Figure 6. Map of pLenti GFP-dgn. The map of the pLenti6/V5-DEST vector including the GFP-dgn sequence is displayed. Abbreviations: PCMV (CMV promoter), GFP-dgn (sequence of GFP-dgn), PSV40 (SV40 early promoter), EM7 (EM7 promoter), Blasticidin (Blasticidin resistance gene), ΔU3/3'LTR (3'LTR with deleted U3 region), SV40 pA (SV40 polyadenylation signal), Ampicillin (Ampicillin resistance gene), pUC ori (pUC origin), PRSV/5'LTR (RSV/5'LTR hybrid promoter), Ψ (HIV-1 Ψ packaging signal), RRE (HIV-1 Rev response element).
Figure 6. Map of pLenti GFP-dgn. The map of the pLenti6/V5-DEST vector including the GFP-dgn sequence is displayed. Abbreviations: PCMV (CMV promoter), GFP-dgn (sequence of GFP-dgn), PSV40 (SV40 early promoter), EM7 (EM7 promoter), Blasticidin (Blasticidin resistance gene), ΔU3/3'LTR (3'LTR with deleted U3 region), SV40 pA (SV40 polyadenylation signal), Ampicillin (Ampicillin resistance gene), pUC ori (pUC origin), PRSV/5'LTR (RSV/5'LTR hybrid promoter), Ψ (HIV-1 Ψ packaging signal), RRE (HIV-1 Rev response element).
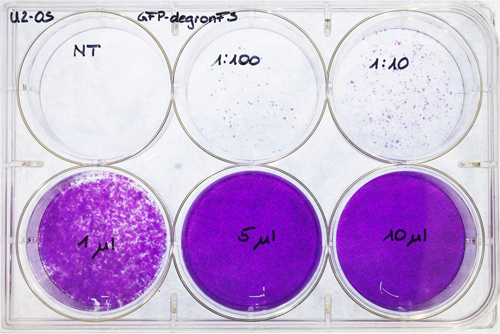 Figure 7. U2-OS titering plate. U2-OS were seeded out on a 6-well plate and transfected with different virus concentrations (dilution of 1/100 and 1/10, 1, 5 and 10 μl of concentrated viral supernatant). One well was used as untransfected control (NT). After 6-7 days, the cells were stained with crystal violet and air dried.
Figure 7. U2-OS titering plate. U2-OS were seeded out on a 6-well plate and transfected with different virus concentrations (dilution of 1/100 and 1/10, 1, 5 and 10 μl of concentrated viral supernatant). One well was used as untransfected control (NT). After 6-7 days, the cells were stained with crystal violet and air dried.
Discussion
The first publication using green fluorescent protein (GFP) as a reporter substrate for ubiquitin-proteasome activity was published in 2000 12. Since then, GFP has become a common tool to visualize cellular activities, especially the ubiquitin-proteasome process. To monitor ubiquitin-proteasome activity in vivo a transgenic mouse model with a GFP-based reporter has been introduced 13. Additional in vivo research established another transgenic mouse model with a similar degron-destabilized GFP reporter as used in this publication 14.
Unfortunately, GFP as a reporter has some imperfection which has been shown by Dantuma et al. and Bowman et al.12,15. First, destabilized GFP can accumulate without the inhibition of the proteasome due to unknown reasons. As the ubiquitin-proteasome machinery is a system whose functionality is dependent on multiple steps, a disturbance prior to the degradation can lead to the accumulation of the reporter substrate and result in false-positive effects 12. Second, GFP-based reporter substrates are due to their short half-lives sensitive to changes in transcription and translation. Any change in GFP fluorescence can therefore be a result of reduced or increased synthesis and translation 15. These problems have been addressed in other studies by modifications of GFP as follows:
Proteasome activity was found to be decreased in cellular senescence and organismal aging 7, 8, 9, 10. In the field of aging research, the focus is mainly on the removal of oxidized proteins where the proteasome plays a key role, and there is no conclusive evidence that protein ubiquitylation precedes degradation in this case 2. Previous work has also shown that the use of GFP with a mutated uncleavable ubiquitin moiety is suitable to study the complex process of protein degradation 16. This GFP-construct gives the possibility to monitor proteasome activity without the potential influence to its activity via disturbances in the ubiquitin machinery.
In this work, we used a degron-destabilized GFP-based reporter protein (GFP-dgn) to monitor ubiquitin-proteasome activity in living human diploid fibroblasts. To have an overview of the transfection efficiency, the functionality and the transcription/translation levels of the GFP-dgn reporter, several appropriate controls were included. We used young untreated GFP-dgn transfected fibroblasts to show that there is almost no green fluorescence visible and proteasome function is not affected. To validate, that the signal increases with the inhibition of the proteasome, we treated GFP-dgn transfected fibroblasts with a proteasome inhibitor to see the accumulation of GPF after inhibition. As a third control we used GFP-dgnFS. With this construct we can show the overall transfection efficiency which can differ from cell strain to cell strain and with all three controls we have an overview over the synthesis rate of the constructs within the cell.
The method presented here enables the user to measure easily and rapidly the ubiquitin-proteasome activity in living cells. A stable overexpression of the GFP reporter substrate can be achieved with different techniques 17. In this publication, transfection of the GFP-dgn cDNA into the cells is achieved by lentiviral system which provides a stable expression. Additionally, the use of this system enables high transfection efficiency independent of the cell type, cell metabolic condition or donor age 7. We have successfully used this protocol to introduce DNA to the endothelial cells in which other methods of transfection failed 18. Nevertheless, previous experiments showed that cells that are expanded longer in culture display a lower fluorescence signal than freshly transfected cells possibly due to the longer selection pressure.
The PEG precipitation is used to increase the titer of the transforming units per ml (TU/ml). If good titers are reached (above 5×105 TU/ml), the PEG precipitation can be omitted. An important step during PEG precipitation is to avoid vigorous suspending or pipetting which generates air bubbles. It can inactivate the virus particles and significantly decrease transfection efficiency.
When using proteasome inhibitors a priori experiments should be performed. The final concentration used and incubation time are cell type dependent and have to be determined experimentally. Previous observations on HFF-2 cells revealed that 3h incubation with LLnL should not be exceeded because of 1) the toxic effects of the inhibitor 2) too high GFP signal that can be reached, or 3) side effects of the proteasome inhibition over longer time periods.
Disclosures
No conflicts of interest declared.
Acknowledgments
This study was funded by: National Research Network on Aging (NFN S93) by the Austrian Science Foundation (FWF), European Commission Integrated Projects MiMAGE and PROTEOMAGE, Netherlands Genomics Initiative/Netherlands Organization for Scientific Research (NGI/NWO; 05040202 and 050-060-810 NCHA), the EU funded Network of Excellence Lifespan (FP6 036894), and Innovation Oriented Research Program on Genomics (SenterNovem; IGE01014 and IGE5007).
References
- Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimi. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- Stangl K, Stangl V. The ubiquitin-proteasome pathway and endothelial (dys)function. Cardiovasc. Res. 2009;85:281–290. doi: 10.1093/cvr/cvp315. [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Stoykova A. Roles of the ubiquitin-proteosome system in neurogenesis. Cell Cycle. 2010;9:3174–3180. doi: 10.4161/cc.9.16.12551. [DOI] [PubMed] [Google Scholar]
- Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118:329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel R, Greussing R, Maier AB, Declercq L, Jansen-Durr P. Functional Interplay between mitochondrial and proteasome activity in skin aging. J. Invest. Dermatol. 2010;131:594–603. doi: 10.1038/jid.2010.383. [DOI] [PubMed] [Google Scholar]
- Grillari J, Grillari-Voglauer R, Jansen-Durr P. Post-translational modification of cellular proteins by ubiquitin and ubiquitin-like molecules: role in cellular senescence and aging. Adv. Exp. Med. Biol. 2010;694:172–196. doi: 10.1007/978-1-4419-7002-2_13. [DOI] [PubMed] [Google Scholar]
- Bulteau AL, Szweda LI, Friguet B. Age-dependent declines in proteasome activity in the heart. Arch. Biochem. Biophys. 2002;397:298–304. doi: 10.1006/abbi.2001.2663. [DOI] [PubMed] [Google Scholar]
- Strucksberg KH, Tangavelou K, Schroder R, Clemen CS. Proteasomal activity in skeletal muscle: A matter of assay design, muscle type, and age. Anal. Biochem. 2009. [DOI] [PubMed]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- Lindsten K, Menendez-Benito V, Masucci MG, Dantuma NP. A transgenic mouse model of the ubiquitin/proteasome system. Nat. Biotechnol. 2003;21:897–902. doi: 10.1038/nbt851. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20:362–364. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- Bowman AB, Yoo SY, Dantuma NP, Zoghbi HY. Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 2005;14:679–691. doi: 10.1093/hmg/ddi064. [DOI] [PubMed] [Google Scholar]
- Myung J, Kim KB, Lindsten K, Dantuma NP, Crews CM. Lack of proteasome active site allostery as revealed by subunit-specific inhibitors. Mol. Cell. 2001;7:411–420. doi: 10.1016/s1097-2765(01)00188-5. [DOI] [PubMed] [Google Scholar]
- Menendez-Benito V, Heessen S, Dantuma NP. Monitoring of ubiquitin-dependent proteolysis with green fluorescent protein substrates. Methods Enzymol. 2005;399:490–511. doi: 10.1016/S0076-6879(05)99034-4. [DOI] [PubMed] [Google Scholar]
- Lener B, et al. The NADPH oxidase Nox4 restricts the replicative lifespan of human endothelial cells. Biochem. J. 2009;423:363–374. doi: 10.1042/BJ20090666. [DOI] [PMC free article] [PubMed] [Google Scholar]


