Abstract
Background
The naturally fluorescent compound quinine has long been used to treat malaria infections. Although some evidence suggests that quinine acts in the parasite food vacuole, the mechanism of action of quinine has not yet been resolved. The Plasmodium falciparum multidrug resistance (pfmdr1) gene encodes a food vacuolar membrane transporter and has been linked with parasite resistance to quinine. The effect of multiple pfmdr1 copies on the subcellular localization of quinine was explored.
Methods
Fluorescence microscopy was used to evaluate the subcellular localization of quinine in parasites containing different pfmdr1 copy numbers to determine if copy number of the gene affects drug localization. The acidotropic dye LysoTracker Red was used to label the parasite food vacuole. Time-lapse images were taken to determine quinine localization over time following quinine exposure.
Results
Regardless of pfmdr1 copy number, quinine overlapped with haemozoin but did not colocalize with LysoTracker Red, which labeled the acidic parasite food vacuole.
Conclusions
Quinine localizes to a non-acidic compartment within the food vacuole possibly haemozoin. Pfmdr1 copy number does not affect quinine subcellular localization.
Keywords: Quinine, Plasmodium falciparum, Pfmdr1, Copy number
Background
Quinine (QN) was isolated from the bark of the Peruvian Cinchona tree in the mid-1800s [1] and quickly became the treatment of choice for intermittent fever worldwide [2]. Although efficacious, QN use as an anti-malarial requires a relatively long treatment regimen and has substantial side effects. As a result, artemisinin-based combination therapy (ACT) has now been implemented as first-line treatment regimens due to drug efficacy and better patient tolerance. However, QN remains an important treatment option for severe malaria infections [3-5]. Despite the longevity of its use as an anti-malarial, the mechanism of action of QN has not yet been fully resolved.
Early investigations into the anti-malarial activity of QN demonstrated the ability to inhibit chloroquine-induced clumping of haemozoin in the parasite food vacuole [6]. Subsequent evidence found that, like chloroquine, QN interferes with haem crystallization [7,8], indicating that QN acts in the food vacuole. In recent years, parasite resistance to QN has been reported in Africa and Southeast Asia, prompting investigations into resistance mechanisms with a focus on the food vacuole [4,9-13]. Decreased QN sensitivity has been strongly linked to a protein on the food vacuolar membrane, encoded by the Plasmodium falciparum multidrug resistance (pfmdr1) gene [10-12,14-16]. Whereas some studies have found that pfmdr1 mutations exert a significant effect on in vitro QN sensitivity [10,13,14], other studies have found no association between pfmdr1 mutations and QN sensitivity [17-19]. Elevated pfmdr1 gene copy number, however, has been linked to reduced parasite susceptibility to QN in both in vitro and clinical studies [12,16,18-20]. Thus, pfmdr1 has an effect on quinine activity.
Quinine is an aromatic alkaloid that naturally fluoresces under ultraviolet (UV) light. Because of its relatively constant fluorescence quantum yield, QN is commonly used in photochemistry as a fluorescence standard [21]. Surprisingly, the inherent fluorescence of QN has not previously been exploited to investigate its mechanism of action. In this study, the fluorescent properties of QN were exploited to evaluate subcellular localization of the drug by fluorescence microscopy in parasites containing different pfmdr1 copy numbers in order to determine if copy number of the gene affects drug localization.
Methods
Parasite cultivation
Plasmodium falciparum cultures of two clonal parasite strains (3D7 and Dd2) with different pfmdr1 copy numbers and quinine (QN) sensitivity were used in this study. The 3D7 strain has one pfmdr1 copy and is sensitive to QN. The Dd2 strain has four pfmdr1 copies [22] and is resistant to QN. Cultures were maintained in candle jars at 37°C under the Trager and Jensen method for malaria parasite culturing [23]. Media was changed every 24 hours. Type O+ serum at 10% and red blood cells (Research Blood Components, LLC: Boston, MA, USA) at 2% haematocrit in malaria culture media (MCM) [24] were used in culturing and experimental conditions.
Imaging of parasites using fluorescence microscopy
Live asynchronous cultures at 5-8% parasitaemia with 1μM of QN were imaged over an 11-hour period using fluorescence microscopy. Parasitized RBC preparations were plated onto 35mm glass bottom dishes (MatTek, Corp.: Ashland, MA, USA) coated with Poly-L-lysine. A coverslip was carefully applied, and the excess fluid gently squeezed out. The coverslip was sealed with warmed Vaseline to prevent the sample from drying out. Time-lapse imaging of multiple parasites was carried out with a Prior Scientific (Prior Scientific: Rockland, MA, USA) motorized x-y stage. An incubator box maintained the microscope work area including the objective at 37°C. Because P. falciparum grows well at 3% oxygen but will tolerate levels as low as 0.5% [25], air was in the incubator box, and the samples were sealed during imaging as stated above. LysoTracker Red DND-99 (Invitrogen: Carlsbad, CA, USA) (50nM) was used to stain the food vacuole and DRAQ5 (Biostatus Limited: Leicestershire, UK) (1μM) was used to stain DNA [26]. Fluorescence images were acquired using an Olympus FV1000 inverted IX81 microscope with a 63x 1.42 NA oil immersion objective at the UNC-Chapel Hill Michael Hooker Microscopy Core Facility. Serial sections with step sizes of 0.20 or 0.25μm were gathered using sequential scanning at 405nm (QN), 559nm (LysoTracker Red), and 635nm (DRAQ5) excitation, and 435±25nm, 595±25nm and 705±50nm emission filters for QN, LysoTracker Red, and DRAQ5 respectively. Confocal pinhole was set to maximum (800μm, ~8 Airy units). Laser transmitted light differential interference contrast (DIC) images were collected at an optimum z-level separately. Image stacks were deconvolved using the iterative blind deconvolution method with Volocity Image Analysis software, version 5.52 (Perkin Elmer: Waltham, MA, USA). Occasionally, Tetraspec 0.1μm beads (Molecular Probes, OR) were added to the sample and used for deconvolution with measured point spread functions. Three-dimensional (3-D) opacity volume rendering and 3-D co-localization analysis was carried out with Volocity.
Results
Quinine localizes to the parasite food vacuole
Two-dimensional (2-D) fluorescence and DIC images of both 3D7 (one pfmdr1 copy) and Dd2 (four pfmdr1 copies) parasites showed that quinine (QN) co-localizes with haemozoin, which suggests localization in the parasite food vacuole (Figure 1). Similar results were found using a Leica SP2 microscope with a 351nm excitation laser (data not shown). The co-localization was highly repeatable and seen in four fields in at least 10 experiments. Parasites were also imaged at earlier time points following the addition of QN (beginning at one hour of exposure through 11 hours). Quinine fluorescence was found in the food vacuole as early as one hour of exposure and remained there for the duration of the experiment (Figure 2). This effect appears to be independent of pfmdr1 copy number as experiments in both strains rendered similar results. The same pattern was seen regardless of whether the infected red cells were maintained on the heated microscope stage or in a candle jar, indicating that photobleaching was not a factor. The parasites did not significantly progress through the intraerythrocytic cell cycle during this 11-hour period consistent with a previously observed maturation delay in the intraerythrocytic cell cycle following QN exposure at IC50 and IC99 concentrations for 12 hours [27]. Thus, the apparent halt in cell cycle progression observed in the current study is likely due to the previously reported QN-induced maturation delay phenomenon.
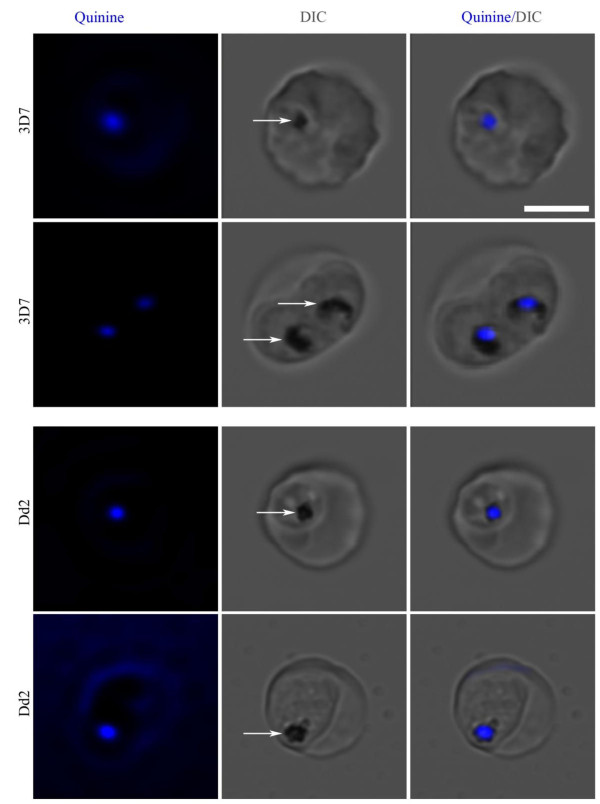
Figure 1.
Quinine localizes with haemozoin in the parasite food vacuole. Asynchronous parasites of PQuinine localizes with haemozoin in the parasite food vacuole. Asynchronous parasites of P. falciparum strains 3D7 (one pfmdr1 copy) and Dd2 (four copies) were incubated with QN to determine its localization. The scale bar represents 5μm. White arrows point to haemozoin crystals in the trophozoite food vacuole. QN (blue) overlapped with haemozoin in both strains and thus localized to the food vacuole.
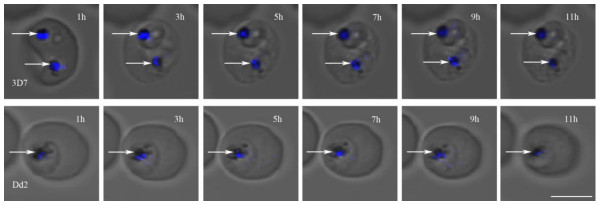
Figure 2.
Quinine localization over time in P. falciparum. Asynchronous parasites were incubated with quinine (blue) to determine subcellular localization of the drug over an 11-hour period. White arrows point to haemozoin crystals in the food vacuole of trophozoites. Quinine overlapped with haemozoin crystals at all time points assessed. The scale bar represents 5μm.
To further characterize subcellular localization of QN, co-localization of QN with either LysoTracker Red, which localizes to acidic compartments, or DRAQ5, which localizes to the nucleus, was investigated using 3-D reconstruction of z-stack fluorescence images. As expected, no co-localization with DRAQ5 was seen. However, QN was found to localize in a separate but adjacent compartment from that in which LysoTracker Red was localized (Figure 3). This 3-D imaging method does not allow for haemozoin to be visualized. The pattern of co-localization was the same for 3D7 and Dd2 parasites, despite their differences in pfmdr1 copy number. These observations suggest that QN localizes to a distinct, non-acidic compartment within the parasite food vacuole, possibly overlapping with haemozoin in parasites with both low and high pfmdr1 copy number.
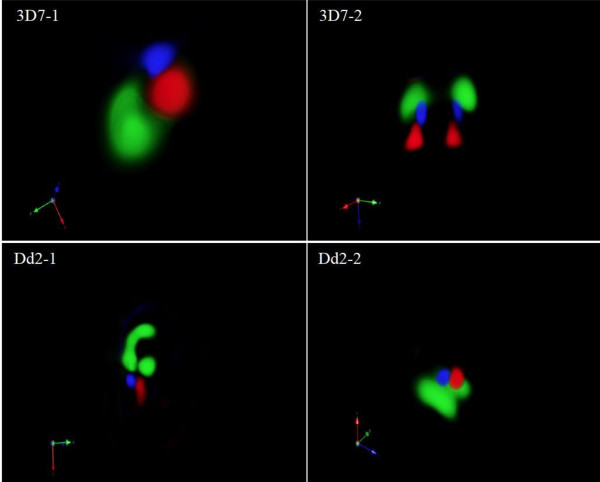
Figure 3.
3-D reconstruction of P. falciparum parasites. Z-stack images of each parasite shown in Figure 1 were used for 3-D volume rendering and colocalization analysis. No QN colocalization with LysoTracker Red was observed in either the 3D7 (left) or the Dd2 (right) strain. Because LysoTracker Red is highly specific for acidic organelles, QN appears to be in a separate and non-acidic compartment of the food vacuole. QN is shown in blue, LysoTracker Red in red, and the DNA stain DRAQ5 in green.
Discussion
Although QN was the first therapeutic compound used to treat malaria infection [1], its mechanism of action has never been fully resolved [6-8]. Some evidence suggests that parasite resistance to QN is associated with mutations and/or elevated copy number of the pfmdr1 gene, which encodes for a transporter protein found in the membrane of the parasite food vacuole [9,10,12-14,16,18,20]. Here, the natural fluorescent properties of QN were exploited to obtain insight into the mechanism of action of the drug. Although knowledge of QN’s fluorescent properties has been around since the late-1800s [28], this is the first study to employ the QN’s fluorescence for imaging in the malaria parasite.
Fluorescence microscopy was employed to image QN subcellular localization in two P. falciparum strains that contained different pfmdr1 copy numbers. Quinine consistently overlapped with the haemozoin crystals in both strains when evaluated by 2-D microscopy (Figure 1). However, upon 3-D reconstruction of serial z-stack images, QN was found to reside in a distinct compartment, which is contiguous to, but separate from, the compartment stained by LysoTracker Red. The lack of co-localization with the acidotropic dye suggests that QN resides in a non-acidic compartment within the food vacuole, possibly the same one occupied by haemozoin. This would be consistent with previous reports that quinolone compounds including QN interact with haemozoin crystals directly or with enzymes involved in the haemozoin crystallization process, as previously reported [7,8,29-33]. In summary, these findings suggest that QN is localized in a non-acidic compartment in the food vacuole, possibly that which contains haemozoin.
This study underscores the importance of utilizing the 3-D reconstruction software in imaging studies, since the localization of QN into this novel compartment would not have been detected otherwise. A recent study revealed that QN-haem adducts exhibit fluorescence at least seven-fold greater than QN alone [34]. Thus, it is possible that QN exists in other areas of the food vacuole but cannot be visualized due to the fluorescence intensity of the QN-haem adducts present in these parasites.
Although there was no apparent difference in localization in strains containing different pfmdr1 copy numbers, the possibility that the pfmdr1 gene has a role cannot be ruled out. Single nucleotide polymorphisms in pfmdr1 have also been associated with decreased sensitivity to QN [10,13,14]. Because the protein encoded by pfmdr1 is a membrane transporter that pumps solutes into the food vacuole, it is possible that mutations within the pfmdr1 gene could affect the transporter function of the protein by altering the conformation or function of the transporter protein.
In summary, this study is novel because it is the first to exploit quinine fluorescence to study the intracellular distribution of the drug. Here, QN was shown to enter a distinct, non-acidic compartment inside the parasite food vacuole. These results are important because they provide visual support for the hypothesis that QN interferes with haemozoin production, which could guide future studies to investigate a possible interaction between QN and enzymes involved in the haemozoin formation process.
Conclusions
Quinine localizes to a non-acidic compartment within the food vacuole and pfmdr1 copy number does not affect QN subcellular localization. These results support previous findings that QN acts in the parasite food vacuole, perhaps interfering with haemozoin formation. This is the first study to provide evidence for QN localization in a sub-compartment of the food vacuole.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EBB participated in the design of the study, carried out all of the fluorescence microscopy experiments and data analysis, and drafted the manuscript. MC participated in the acquisition and analysis of microscopy data. SRM conceived of the study and participated in its design. All authors read and approved the final manuscript.
Contributor Information
Elaine B Bohórquez, Email: elaine_brooks@med.unc.edu.
Michael Chua, Email: michael_chua@med.unc.edu.
Steven R Meshnick, Email: meshnick@email.unc.edu.
Acknowledgements
Funding for this project was provided by NIH grant 1-R21-AI076785-01A1.
References
- Bruce-Chwatt LJ. Three hundred and fifty years of the peruvian fever bark. Br Med J (Clin Res Ed) 1988;296:1486–1487. doi: 10.1136/bmj.296.6635.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeka A, Achan J, D'Alessandro U, Talisuna AO. Quinine monotherapy for treating uncomplicated malaria in the era of artemisinin-based combination therapy: an appropriate public health policy? Lancet Infect Dis. 2009;9:448–452. doi: 10.1016/S1473-3099(09)70109-4. [DOI] [PubMed] [Google Scholar]
- WHO. World malaria report. World Health Organization, Geneva; 2010. [Google Scholar]
- Okombo J, Ohuma E, Picot S, Nzila A. Update on genetic markers of quinine resistance in Plasmodium falciparum. Mol Biochem Parasitol. 2011;177:77–82. doi: 10.1016/j.molbiopara.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Kyu HH, Fernandez E. Artemisinin derivatives versus quinine for cerebral malaria in african children: a systematic review. Bull World Health Organ. 2009;87:896–904. doi: 10.2471/BLT.08.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W. Antimalarial drugs and their actions. Postgrad Med J. 1973;49:573–583. doi: 10.1136/pgmj.49.574.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater AF, Cerami A. Inhibition by chloroquine of a novel haem polymerase enzyme activity in malaria trophozoites. Nature. 1992;355:167–169. doi: 10.1038/355167a0. [DOI] [PubMed] [Google Scholar]
- Egan TJ, Ross DC, Adams PA. Quinoline anti-malarial drugs inhibit spontaneous formation of beta-haematin (malaria pigment) FEBS Lett. 1994;352:54–57. doi: 10.1016/0014-5793(94)00921-X. [DOI] [PubMed] [Google Scholar]
- Zalis MG, Pang L, Silveira MS, Milhous WK, Wirth DF. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am J Trop Med Hyg. 1998;58:630–637. doi: 10.4269/ajtmh.1998.58.630. [DOI] [PubMed] [Google Scholar]
- Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 Mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in plasmodium falciparum. Mol Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- Nkrumah LJ, Riegelhaupt PM, Moura P, Johnson DJ, Patel J, Hayton K, Ferdig MT, Wellems TE, Akabas MH, Fidock DA. Probing the multifactorial basis of plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium-proton exchanger PfNHE. Mol Biochem Parasitol. 2009;165:122–131. doi: 10.1016/j.molbiopara.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 Modulates sensitivity and resistance to multiple antimalarials in plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, Subramanian G, Aravind L, Cooper RA, Wootton JC, Xiong M, Su XZ. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Cowman AF, Galatis D, Thompson JK. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci U S A. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaijaroenkul W, Wisedpanichkij R, Na-Bangchang K. Monitoring of in vitro susceptibilities and molecular markers of resistance of plasmodium falciparum isolates from thai-myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 2010;113:190–194. doi: 10.1016/j.actatropica.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant plasmodium falciparum malaria. Clin Infect Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price RN, Uhlemann AC, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. Mefloquine resistance in plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Kawamoto F, Miller RS, Meshnick SR. Resistance to antimalarials in southeast asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–2423. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Nair S, Qin H, Singlam S, Brockman A, Paiphun L, Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AN. Quinine sulfate as a fluorescence quantum yield standard. Photochem Photobiol. 1969;9:439–444. doi: 10.1111/j.1751-1097.1969.tb07311.x. [DOI] [PubMed] [Google Scholar]
- Lim P, Alker AP, Khim N, Shah NK, Incardona S, Doung S, Yi P, Bouth DM, Bouchier C, Puijalon OM, Meshnick SR, Wongsrichanalai C, Fandeur T, Le Bras J, Ringwald P, Ariey F. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Cultivation of malarial parasites. Nature. 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- Methods in malaria research. Malaria Research and Reference Reagent Resource Center (MR4)/American Type Culture Collection (ATCC), Manassas, VA; 2008. [Google Scholar]
- Scheibel LW, Adler A, Trager W. Tetraethylthiuram disulfide (antabuse) inhibits the human malaria parasite plasmodium falciparum. Proc Natl Acad Sci U S A. 1979;76:5303–5307. doi: 10.1073/pnas.76.10.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purfield AE, Tidwell RR, Meshnick SR. The diamidine DB75 targets the nucleus of plasmodium falciparum. Malar J. 2009;8:104. doi: 10.1186/1475-2875-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga MI, Ferreira PE, Schmidt BA, Ribacke U, Bjorkman A, Tichopad A, Gil JP. Antimalarial exposure delays plasmodium falciparum intra-erythrocytic cycle and drives drug transporter genes expression. PLoS One. 2010;5:e12408. doi: 10.1371/journal.pone.0012408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach F. The historical aspects of sunscreens. J Photochem Photobiol B. 2001;64:99–104. doi: 10.1016/S1011-1344(01)00202-0. [DOI] [PubMed] [Google Scholar]
- de Villiers KA, Gildenhuys J, le Roex T. Iron(III) protoporphyrin IX complexes of the antimalarial cinchona alkaloids quinine and quinidine. ACS Chem Biol. 2012;7:666–671. doi: 10.1021/cb200528z. [DOI] [PubMed] [Google Scholar]
- Pisciotta JM, Coppens I, Tripathi AK, Scholl PF, Shuman J, Bajad S, Shulaev V, Sullivan DJ Jr. The role of neutral lipid nanospheres in Plasmodium falciparum haem crystallization. Biochem J. 2007;402:197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leed A, DuBay K, Ursos LM, Sears D, De Dios AC, Roepe PD. Solution structures of antimalarial drug-haem complexes. Biochemistry. 2002;41:10245–10255. doi: 10.1021/bi020195i. [DOI] [PubMed] [Google Scholar]
- Egan TJ, Ncokazi KK. Quinoline antimalarials decrease the rate of beta-hematin formation. J Inorg Biochem. 2005;99:1532–1539. doi: 10.1016/j.jinorgbio.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Correa Soares JB, Menezes D, Vannier-Santos MA, Ferreira-Pereira A, Almeida GT, Venancio TM, Verjovski-Almeida S, Zishiri VK, Kuter D, Hunter R, Egan TJ, Oliveira MF. Interference with haemozoin formation represents an important mechanism of schistosomicidal action of antimalarial quinoline methanols. PLoS Negl Trop Dis. 2009;3:e477. doi: 10.1371/journal.pntd.0000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alumasa JN, Gorka AP, Casabianca LB, Comstock E, de Dios AC, Roepe PD. The hydroxyl functionality and a rigid proximal N are required for forming a novel non-covalent quinine-haem complex. J Inorg Biochem. 2011;105:467–475. doi: 10.1016/j.jinorgbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]