Abstract
Background
The NPC1 gene encodes a protein involved in intracellular lipid trafficking; its second endosomal loop (loop 2) is a receptor for filoviruses. A polymorphism (His215Arg) in NPC1 was associated with obesity in Europeans. Adaptations to diet and pathogens represented powerful selective forces; thus, we analyzed the evolutionary history of the gene and exploited this information for the identification of variants/residues of functional importance in human disease.
Methods
We performed phylogenetic analysis, population genetic tests, and genotype-phenotype analysis in a population from Saudi Arabia.
Results
Maximum-likelihood ratio tests indicated the action of positive selection on loop 2 and identified three residues as selection targets; these were confirmed by an independent random effects likelihood (REL) analysis. No selection signature was detected in present-day human populations, but analysis of nonsynonymous polymorphisms showed that a variant (Ile642Met, rs1788799) in the sterol sensing domain affects a highly conserved position. This variant and the previously described His215Arg polymorphism were tested for association with obesity and type 2 diabetes (T2D) in a cohort from Saudi Arabia. Whereas no association with obesity was detected, 642Met allele was found to predispose to T2D. A significant interaction was noted with sex (P = 0.041), and stratification on the basis of gender indicated that the association is driven by men (P = 0.0021, OR = 1.5). Notably, two NPC1 haplotypes were also associated with T2D in men (rs1805081-rs1788799, His-Met: P = 0.0012, OR = 1.54; His-Ile: P = 0.0004, OR = 0.63).
Conclusions
Our data indicate a sex-specific effect of NPC1 variants on T2D risk and describe putative binding sites for filoviruses entry.
Keywords: NPC1, filovirus, natural selection, type 2 diabetes
Background
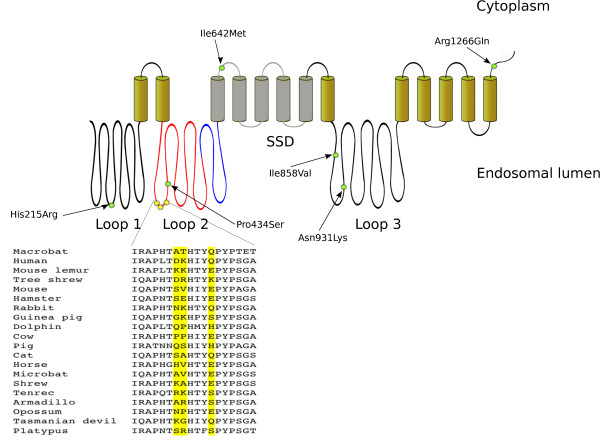
The NPC1 gene encodes a large multi-domain protein involved in the intracellular trafficking of sterols. Mutations in the gene are responsible for a rare and fatal lipid storage disorder, Niemann-Pick disease type C. The product of NPC1 resides in the limiting membrane of late endosomes and lysosomes where it facilitates lipid transport to various cellular compartments (reviewed in [1]). The protein displays 13 transmembrane domains, and three large loops are present in the lumen of the endosome (Figure 1) [2]. Interaction with lipid substrates is mediated by the most N-terminal luminal loop (loop 1) and by a sterol-sensing domain (SSD) which comprises five central transmembrane regions [2] (Figure 1). Recent works showed that the subcellular localization of NPC1 has been exploited by viruses of the Filoviridae family for host invasion [3-5]. Thus, viruses such as Ebola and Marburg require NPC1 protein expression for productive infection and the second luminal domain of NPC1 binds directly and specifically to the GP1 viral glycoprotein [3]. Consistently, primary fibroblasts from human Niemann-Pick type C1 disease patients are resistant to infection by filoviruses [4].
Figure 1.
Schematic representation of the NPC1 protein (not to scale). Cylinders represent the transmembrane regions; the SSD domain is depicted in grey. Luminal loop 2 is represented in blue and red to account for the recombination breakpoint. The position of the three positively selected sites in loop 2 is shown (yellow circles) with the alignment of a few representative mammalian species. The position of nonsynonymous polymorphisms with a minor allele frequency higher than 1% is represented by the green circles. SSD, sterol sensing domain
Mice lacking Npc1 function display a phenotype recapitulating Niemann-Pick disease type C [6], whereas haploinsufficiency for the gene results in weight gain and insulin resistance [7,8]. In fact, Npc1+/- mice display increased adiposity and adipocyte hypertrophy; these animals also show dyslipidemia and higher plasma glucose levels compared to their wild-type litter mates. In line with this evidence, a nonsynonymous polymorphism (rs1805081, His215Arg) in the human NPC1 gene has recently been associated with severe and early onset obesity in European populations [9]. A subsequent study confirmed the predisposing role of rs1805081 to obesity and increased body mass index (BMI) in Europeans, but found no association between the variant and type 2 diabetes (T2D) or fasting plasma lipid levels [10]. Conversely, the effect on obesity risk and higher BMI of the NPC1 SNP in Asian populations is still controversial [11,12]. The molecular mechanisms underlying the association between genetic variation in NPC1 and metabolic phenotypes remain to be clarified. However, analysis of Npc1 mutant mice revealed that these animals are characterized by increased liver accumulation of triacylglycerol [7], higher hepatic expression of caveolin-1 [13], a protein involved in liver lipid metabolism [14], and of sterol regulatory element-binding proteins (SREBPs) [15]. These observations suggest that mutations or polymorphisms in NPC1 result in alteration of hepatic lipid homeostasis eventually leading to weight gain and insulin resistance.
Adaptations to diet and to pathogen exposure are thought to have represented a powerful driving force throughout the evolutionary history of mammals [16]. Thus, we performed a phylogenetic analysis of NPC1 genes in mammals and a population genetics study of diversity in human populations. We identified three residues that have been targets of positive selection, possibly mediated by filovirus-exerted selective pressure. No selection signature was detected in present-day human populations, but analysis of nonsynonymous polymorphisms identified a variant (Ile642Met) in the SSD domain that affects a highly conserved position. This variant and NPC1 haplotypes were found to modulate the risk of T2D (but not BMI or obesity) in a population from Saudi Arabia.
Methods
Evolutionary analysis
Most mammalian NPC1 sequences were retrieved from the Ensembl website [17]. The sequence of baboon was obtained though blast search in the National Center for Biotechnology Information (NCBI) Trace Archive against Papio hamadryas whole genome sequence. NPC1 coding sequences for Cricetulus griseus and Mustela putorius (C-terminal portion only) were retrieved from the NCBI nucleotide database (NM_001246687.1 and JP014452, respectively).
DNA alignment was performed using the The RevTrans 2.0 utility [18], which uses the peptide sequence alignment [see Additional file 1, Figure S1] as a scaffold for constructing the corresponding DNA multiple alignment. This latter was checked and edited by hand to remove alignment uncertainties. The alignment was used for Genetic Algorithm Recombination Detection (GARD) [19] analysis through the DataMonkey [20]. Similarly, the evolutionary selection distance (ESD), random effects likelihood (REL) and branch-site REL analyses were performed using DataMonkey [20]. For phylogenetic analysis by maximum likelihood (PAML) analyses we used multiple alignments of NPC1 sub-regions and trees generated by maximum-likelihood using the program DnaML (PHYLIP Package). To detect selection, Nssite models that allow (M8) or disallow (M7 and M8a) a class of codons to evolve with dN/dS >1 were fitted to the data using both the F61 (Table 1) and the F3X4 [see Additional file 1, Table S1] codon frequency models. Sites under selection for the M8 model were identified using Bayes empirical Bayes (BEB) analysis using a significance cutoff of 0.90 [21,22].
Table 1.
Likelihood ratio test statistics for models of variable selective pressure among sites (F61 model of codon frequency).
| Region/selection model(number of codons) | d.f. | -2ΔLnL | P value | % of sites(average dN/dS) | Selected sites |
|---|---|---|---|---|---|
| Loop1 (269) | |||||
| M7 versus M8 | 2 | 10.04 | 0.0066 | 1.3% (1.37) | 182 (0.99, n.s.) |
| M8a versus M8 | 1 | 4.87 | 0.027 | - | - |
| Loop 2 N-term (251) | |||||
| M7 versus M8 | 2 | 25.86 | <0.0001 | 1.3% (1.78) | 416 (0.99, 0.99),417 (0.98, 1), 421 (0.91, 0.99) |
| M8a versus M8 | 1 | 10.98 | 0.0009 | - | |
| Loop 2 C-term (81) | |||||
| M7 versus M8 | 2 | <0.01 | >0.99 | - | - |
| M8a versus M8 | 1 | 1.96 | 0.16 | - | |
| SSD (164) | |||||
| M7 versus M8 | 2 | 5.70 | 0.058 | - | - |
| M8a versus M8 | 1 | 0.02 | >0.88 | - | - |
| Loop 3 (248) | |||||
| M7 versus M8 | 2 | 0.004 | >0.99 | - | - |
| M8a versus M8 | 1 | 1.52 | 0.21 | - | - |
M7 is a null model that assumes that 0<ω<1 is beta distributed among sites; M8a assumes 0<ω≤1; these two models are compared to M8, which includes an extra category of sites with ω>1 (positive selection). d.f., degree of freedom; 2ΔLnL, twice the difference of the natural logs of the maximum likelihood of the models being compared; P value, P value of rejecting the neutral models (M7 or M8a) in favor of the positive selection model (M8); % of sites (average dN/dS), estimated percentage of sites evolving under positive selection by M8 (dN/dS for these codons); selected sites, the position refers to the entire NPC1 human sequence, P values obtained from BEB and REL analyses are shown. BEB, Bayes empirical Bayes; REL, random effects likelihood; SSD, sterol sensing domain.
Population genetic analyses
Data from the Pilot 1 phase of the 1000 Genomes Project were retrieved online [23]. Low-coverage SNP genotypes were organized in a MySQL database. A set of programs was developed to retrieve genotypes from the database and to analyze them according to selected regions/populations. These programs were developed in C++ using the GeCo++ [24] and the libsequence [25] libraries. Genotype information was obtained for NPC1 and for 2,000 randomly selected RefSeq genes.
Sliding window analysis was performed on overlapping 5 kb windows moving with a step of 500 bp. For each window we calculated θW, π, and FST and these values were used to obtain the empirical distributions and to calculate percentiles. Values for the integrated haplotype_score (iHS) for HapMap Phase II SNPs were derived from a previous work [26].
Patients and controls
All subjects recruited in the study are part of the Biomarker Screening in Riyadh Project (RIYADH COHORT), a capital-wide epidemiologic study that has so far enrolled more than 17,000 Saudis from different Primary Health Care Centers. Demographic and medical information is recorded for all individuals participating in the program. DNA samples have been collected from more than 1,600 of these individuals. These individuals were selected to represent case-control cohorts for T2D. Subjects with medical complications (coronary artery disease, nephropathy, and end stage renal disease or liver disease) were excluded and a similar percentage of men and women were enrolled among T2D patients and controls. After discarding samples with poor DNA quality, 1,468 subjects were included in the study (644 T2D, 52% women; 824 controls, 54% women). Diagnosis of T2D was based on the World Health Organization proposed cut-off (fasting plasma glucose > or = 7.0 mmol/L or 126 mg/dl) as previously described [27].Written consent was obtained from all participants, and ethical approval was granted by the Ethics Committee of the College of Science Research Center, King Saud University, Riyadh, Kingdom of Saudi Arabia (KSA).
Anthropometry and DNA extraction
After an overnight fast, subjects underwent anthropometry and blood withdrawal. Anthropometry included measurement of height (to the nearest 0.5 cm) and weight (to the nearest 0.1 kg); BMI was calculated as kg/m2. According to the World Health Organization (WHO) criteria, individuals were classified as obese if their BMI was > 30 kg/m2. Whole blood was collected in ethylenediaminetetraacetic acid (EDTA)-containing tubes; genomic DNA was isolated using the blood genomic prep minispin kit (GE Healthcare, Milano, Italy). Genotyping and statistical analysisThe two NPC1 SNPs were genotyped by allelic discrimination real-time PCR, using predesigned TaqMan probe assays (Applied Biosystems, Foster City, CA, USA). Reactions were performed using TaqMan Genotyping Master Mix in an ABI 9700 analyzer (Applied Biosystems). Genotyping rate was >0.97 for both variants. In the text and tables, the allelic status of the two variants is shown with reference to the transcript orientation with the ancestral allele reported first. Genetic association was investigated by multiple linear or logistic regression (as appropriate) using genotypes/haplotypes as the independent predictor variables with sex and age as covariates; BMI was added as a covariate when addressing the association between T2D and NPC1 variants; T2D was accounted for when addressing the effect of SNPs/haplotypes on obesity and BMI. Before carrying out parametric statistical procedures, total cholesterol and triglyceride levels were logarithmically transformed to ensure a more normal distribution. Analyses were performed using PLINK [28].
Results
Evolutionary analysis of NPC1 mammalian genes
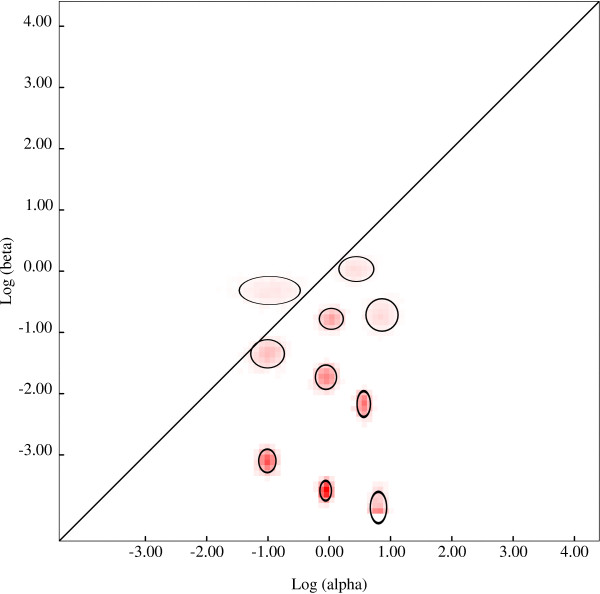
To analyze the evolutionary history of NPC1 in mammals we retrieved coding sequence information for 41 species from public databases (see methods). Alignment of these sequences revealed that NPC1 evolved under purifying selection, as the average non-synonymous substitution rate (dN) was generally much lower than the rate for synonymous substitutions (dS) (average dN/dS = 0.12). Nonetheless, natural selection might act on a few sites within a gene that is otherwise strongly constrained. Before testing this possibility, we screened the NPC1 alignment for evidence of recombination using a recently developed algorithm (GARD) [19]; this analysis uncovered the presence of one single recombination breakpoint at nucleotide position 1619 (ΔAICc = 53.7), falling within luminal loop 2 (Figure 1). After taking this information into account, we analyzed the evolutionary fingerprint of NPC1 by applying the ESD method [29], which uses the site-by-site probability distribution of synonymous and nonsynonymous substitution rates to partition sites into selective classes. ESD estimated 10 substitution rate classes (Figure 2), one of which showing dN/dS (ω) >1, indicative of positive selection. Specifically, the estimated average ω for this class was 1.98 with an estimated percentage of sites of 2% (95% IC: 0.1 to 0.3). We next applied maximum-likelihood analyses implemented in the PAML package [30,31] to single NPC1 domains. Specifically, we separately analyzed luminal loops 1 and 3, as well as the SSD domain; luminal loop 2 was divided into two halves to account for the recombination breakpoint. Results indicated that a model allowing sites to evolve with ω >1 (M8) had significantly better fit to the data than models assuming no positive selection (M7 and M8a) for the N-terminal portion of loop 2 (Table 1, and Additional file 1, Table S1). Some evidence of positive selection was also evident for loop 1. No selection signature was detected for the remaining NPC1 regions. Three sites in the N-terminal portion of loop 2 were found to have a high posterior probability of being under positive selection according to BEB analysis (P >0.90) (Table 1 Figure 1) [21,22]. These three sites were confirmed by an independent REL analysis that allows variation of dS among sites [32] (Table 1). BEB analysis also identified one site in luminal loop 1, which was not confirmed by REL analysis. Finally, we verified whether any lineage shows evidence of episodic positive selection by applying a branch-site REL analysis [33]. Results indicated that a proportion of sites has evolved under episodic diversifying selection in the gorilla and baboon lineages, although the proportion of sites evolving with ω >1 was very low (about 1%) in both lineages. Thus, the branch-site REL test should be interpreted with caution, as sequencing errors in the reference sequences of these two primates might be partially responsible for these results [see Additional file 1, Figure S2].
Figure 2.
Evolutionary fingerprint of NPC1. The estimate of the distribution of synonymous and nonsynonymous substitution rates is plotted on a log-log scale. The ellipses reflect a Gaussian-approximated variance in each individual rate estimate, and colored pixels show the density of the posterior sample of the distribution for a given rate. The diagonal line represents the neutral evolution expectation (ω = 1), points above the line correspond to positive selection (ω >1), and points below the line to purifying selection (ω <1)
Population genetics in humans
The human NPC1 gene spans about 55 kb on chromosome 18. To gain insight into its evolutionary history in human populations, we exploited sequencing data from the 1000 Genomes Pilot Project [34], which generated low-coverage whole genome sequencing data of 179 individuals with different ancestry (Yoruba from Nigeria, Europeans and Asians). Nucleotide diversity for the entire NPC1 gene region was calculated using θW, an estimate of the expected per site heterozygosity [35] and π, the average number of pair-wise sequence nucleotide differences between haplotypes [36]. As a comparison, the same indexes were obtained for 2,000 randomly selected human genes. Both θW and π for NPC1 ranged from the 29th to the 40th percentiles in the distribution of values calculated for the 2,000 reference genes in the three populations (not shown). In order to address the possibility of local selection affecting NPC1 sub-regions, we performed a sliding window analysis of θW, π, and Yoruba/European/Asian population genetic differentiation (FST) [37] along the gene. Again, we applied the same procedure to 2,000 randomly selected human genes, allowing calculation of the 2.5th and 97.5th percentiles to be used as reference cutoffs. No region in NPC1 displayed nucleotide diversity outside the calculated cutoffs [see Additional file 1, Figure S3]. As for FST, a peak was evident in the middle of the gene, but it did not exceed the 97.5th percentile [see Additional file 1, Figure S4]. Analysis of iHS [26] for variants within the peak revealed no absolute value higher than 2 (data not shown). Overall, these analyses suggest that NPC1 is neutrally evolving in humans or that selection signatures are too weak to be detected using these approaches.
Association of NPC1 SNPs with obesity and T2D
To shed light on the distribution of polymorphisms segregating in NPC1 we again exploited the 1000 Genomes Project data [34] by selecting nonsynonymous variants that have been detected in the gene with a minor allele frequency higher than 1%. Six variants were identified; only two of them were located in domains possibly affecting sterol homeostasis: rs1805081 (His215Arg), located in loop 1 and previously associated with obesity in Europeans [9], and rs1788799 (Ile642Met), located in the SSD (Figure 1). Analysis of the mammalian NPC1 alignment indicated that codon 215 is relatively variable, whereas position 642 is conserved (Ile) in all species [see Additional file 1, Figure S1]. We analyzed the role of these two SNPs in predisposing to obesity and weight gain by recruiting a population consisting of 1,468 subjects (820 obese individuals and 648 non-obese controls) from Saudi Arabia (Table 2). The two polymorphisms displayed limited linkage disequilibrium (LD) in our study population (D' = 0.93, r2 = 0.080) and both complied with Hardy-Weinberg equilibrium. Minor allele frequencies for rs1788799 (G, 642Met) and rs1805081 (G, 215Arg) in this cohort amounted to 0.41 and 0.12, respectively. Association of these SNPs with obesity was assessed by fitting a logistic regression model using age, sex, and absence/presence of T2D as covariates. Results indicated that neither SNP associates with obesity (Table 3). Similarly, no association between NPC1 variants and BMI was detected (Table 3).
Table 2.
Characteristics of the Saudi cohort.
| Obese | Non-obese | |||
|---|---|---|---|---|
| T2D | No T2D | T2D | No T2D | |
| Sample size | 441 | 379 | 203 | 445 |
| Females (%) | 261 (59) | 233 (61) | 73 (36) | 216 (48.5) |
| Males (%) | 180 (41) | 145 (39) | 130 (64) | 229 (51.5) |
| Age ± s.d. (years) | 53.07 ± 10.61 | 42.79 ± 10.15 | 55.54 ± 16.19 | 38.56 ± 13.90 |
| BMI ± s.d., kg/m2 | 35.62 ± 6.83 | 34.67 ± 3.92 | 24.25 ± 3.31 | 22.74 ± 3.3 |
T2D, type 2 diabetes.
Table 3.
Association analysis of NPC1 polymorphisms with obesity, BMI, and T2D.
| Trait | Minor allele | Whole sample | Males | Females | |||
|---|---|---|---|---|---|---|---|
| P valuea | OR (95% CI)b | P valuea | OR (95% CI)b | P valuea | OR (95% CI)b | ||
| Obesity | |||||||
| rs1788799 (C/G) | G (Met) | >0.5 | 1.05 (0.87 to 1.18) | >0.5 | 1.02 (0.81 to 1.27) | >0.5 | 1.02 (0.83 to 1.27) |
| rs1805081 (A/G) | G (Arg) | >0.5 | 0.83 (0.78 to 1.25) | >0.5 | 1.06 (0.76 to 1.47) | >0.5 | 0.93 (0.67 to 1.30 |
| BMI | P valuea | BETAc | P valuea | BETAc | P valuea | BETAc | |
| rs1788799 (C/G) | G (Met) | 0.462 | 0.328 | >0.5 | 0.490 | >0.5 | 0.231 |
| rs1805081 (A/G) | G (Arg) | >0.5 | -0.411 | >0.5 | -0.343 | >0.5 | -0.450 |
| T2D | P valuea | OR (95% CI)b | P valuea | OR (95% CI)b | P valuea | OR (95% CI)b | |
| rs1788799 (C/G) | G (Met) | 0.027 | 1.24 (1.05 to 1.48) | 0.004 | 1.50 (1.15 to 1.95) | >0.5 | 1.07 (0.85 to 1.36) |
| rs1805081 (A/G) | G (Arg) | >0.5 | 1.06 (0.82 to 1.39) | >0.5 | 1.11 (0.76 to .60) | >0.5 | 1.04 (0.71 to 1.52) |
aP values were calculated using logistic or linear regression (as appropriate) with an additive model; Bonferroni-corrected P values are reported; nominally significant P values are in bold; bodds ratio and 95% confidence intervals refer to the minor allele. cregression coefficient. BMI, body mass index; OR, odds ratio; T2D, type 2 diabetes.
We next evaluated the role of rs1805081 and rs1788799 in predisposing to T2D; to this aim all subjects were analyzed by fitting a logistic regression using age, sex, and BMI as covariates. No effect of rs1805081 on T2D susceptibility was observed; conversely, a significant association between rs1788799 and T2D was detected (for the minor allele 642Met, P = 0.0137, odds ratio (OR) = 1.24) (Table 3). A significant interaction was also noted between allelic status at this variant and sex (Pinteraction = 0.041); stratification of the population on the basis of gender indicated that the association between rs1788799 and T2D is driven by male subjects (Table 3). Thus, we next analyzed the effect of NPC1 haplotypes on susceptibility to diabetes. After correcting for age, sex and BMI, two haplotypes were found to be associated with T2D with an opposite effect. Specifically, AC and AG (rs1805081-rs1788799, 215His-642Ile and 215His-642Met) haplotypes were observed to protect and predispose to the disease, respectively (Table 4). Again, the association could only be detected in men and occurred in both obese and non-obese individuals (Table 4).
Table 4.
Association analysis of NPC1 haplotypes with T2D.
| Trait | Haplotype (rs1805081 -rs1788799, residues) |
Whole sample | Males | Females | |||
|---|---|---|---|---|---|---|---|
| Freq.a | P value (OR) | Freq.a | P value (OR) | Freq.a | P value (OR) | ||
| T2D (all) | |||||||
| GC (Arg-Ile) | 0.12 | 0.451 (1.11) | 0.12 | 0.495 (1.14) | 0.12 | 0.653 (1.09) | |
| AG (His-Met) | 0.41 | 0.009 (1.26) | 0.40 | 0.0012 (1.54) | 0.41 | 0.545 (1.08) | |
| AC (His-Ile) | 0.47 | 0.003 (0.77) | 0.48 | 0.0004 (0.63) | 0.47 | 0.425 (0.91) | |
| T2D (obese) | |||||||
| GC (Arg-Ile) | 0.12 | 0.577 (1.10) | 0.12 | 0.498 (1.20) | 0.12 | 0.780 (1.07) | |
| AG (His-Met) | 0.42 | 0.230 (1.14) | 0.42 | 0.013 (1.44) | 0.41 | 0.745 (0.95) | |
| AC (His-Ile) | 0.46 | 0.142 (0.85) | 0.46 | 0.004 (0.60) | 0.47 | 0.813 (1.03) | |
| T2D (non-obese) | |||||||
| GC (Arg-Ile) | 0.11 | 0.77 (1.07) | 0.11 | 0.957 (1.02) | 0.12 | 0.666 (1.19) | |
| AG (His-Met) | 0.40 | 0.0125 (1.46) | 0.40 | 0.033 (1.52) | 0.41 | 0.158 (1.40) | |
| AC (His-Ile) | 0.49 | 0.009 (0.67) | 0.49 | 0.042 (0.69) | 0.47 | 0.088 (0.65) | |
ahaplotype frequency. Significant P values are in bold. Freq., frequency; T2D, type 2 diabetes.
Finally, we evaluated the role of NPC1 haplotypes in modulating fasting plasma lipid levels. Circulating levels of total-, LDL- and HDL-cholesterol, as well as triglycerides were available for 1,443 individuals of the above described cohort. No effect of NPC1 haplotypes on total- and LDL-cholesterol was detected (Table 5). Conversely, different NPC1 haplotypes were associated, although weakly, with HDL-cholesterol and triglyceride levels both in men and women (Table 5).
Table 5.
Association analysis of NPC1 haplotypes with lipid levels.
| Trait | Haplotype (rs1805081-rs1788799, residues) |
Whole sample | Males | Females | |||
|---|---|---|---|---|---|---|---|
| BETAa | P value | BETAa | P value | BETAa | P value | ||
| HDL-cholesterol | |||||||
| GC (Arg-Ile) | -0.047 | 0.026 | -0.070 | 0.017 | -0.020 | 0.698 | |
| AG (His-Met) | -0.013 | 0.32 | 0.004 | 0.803 | -0.0278 | 0.048 | |
| AC (His-Ile) | 0.032 | 0.015 | 0.025 | 0.190 | 0.040 | 0.023 | |
| LDL-cholesterol | |||||||
| GC (Arg-Ile) | -0.068 | 0.650 | -0.140 | 0.477 | -0.034 | 0.877 | |
| AG (His-Met) | 0.149 | 0.124 | -0.125 | 0.330 | 0.200 | 0.157 | |
| AC (His-Ile) | -0.105 | 0.265 | -0.701 | 0.583 | -0.144 | 0.289 | |
| Total cholesterol | |||||||
| GC (Arg-Ile) | -0.011 | 0.408 | 0.003 | 0.872 | -0.028 | 0.166 | |
| AG (His-Met) | 0.010 | 0.224 | 0.002 | 0.873 | 0.022 | 0.097 | |
| AC (His-Ile) | -0.005 | 0.538 | -0.003 | 0.794 | -0.008 | 0.505 | |
| Triglycerides | |||||||
| GC (Arg-Ile) | 0.045 | 0.101 | 0.104 | 0.003 | -0.0313 | 0.466 | |
| AG (His-Met) | 0.024 | 0.173 | -0.014 | 0.563 | 0.071 | 0.009 | |
| AC (His-Ile) | -0.041 | 0.018 | -0.031 | 0.181 | -0.052 | 0.046 | |
aregression coefficient. Significant P values are in bold.
Discussion
During mammalian evolution genes involved in diet and immune response have been preferential targets of positive selection [16], highlighting the role of nutrient availability/preferences and pathogens as powerful selective forces. The protein product of NPC1 plays a central role in lipid metabolism, as it acts as a cholesterol transporter and its transcription is regulated by the SREBP pathway [1]. Conversely, the gene does not participate in immune response, but is exploited by members of the filovirus family as an intracellular receptor that mediates the late steps of viral invasion [3-5]. Evidence has indicated that genes directly involved in antiviral response or acting as viral receptors (for example, HAVCR1, CD4) display domains evolving under positive selection as the result of a genetic conflict with extant or extinct viral species [38-46]. Positive selection at these host genes may result from adaptation either to increase viral recognition and restriction efficiency or to avoid binding of specific viral components. Our evolutionary analysis in mammals indicated a predominant role of purifying selection in driving the evolution of NPC1 but also identified few positions that have been targeted by positive selection. Specifically, maximum-likelihood ratio tests indicated that three residues in the N-terminal portion of luminal loop 2 evolved under positive selection; these codons are located in close proximity to each other, and selection was confirmed by an independent REL analysis. PAML also identified one positively selected site in luminal loop 1, but this was not supported by REL, suggesting that it may represent a false positive, as the M8 model has been shown to be more prone than REL to false positive results when a relatively high number of sequences (species) is used for analysis [47]. These results suggest that the selective pressure responsible for positive selection in NPC1 stems from pathogens rather than from dietary changes. Indeed, a recent study has indicated that luminal loop 2 is necessary and sufficient to bind filovirus GP1 protein directly and to mediate productive infection [3]; the authors were able to map the GP1 residues involved in engaging loop 2 and determined that they are conserved among filoviruses [3]. This observation, together with evidence showing that NPC1 is required for infection of both human and rodent cells by distantly related viral species, strongly suggests that the cholesterol transporter is a necessary factor for most members of the Filoviridae family [3-5]. These pathogens display a wide host range in mammals [48] and are thought to have affected vertebrates for millions of years, as testified by the detection of filovirus-derived elements in the genome of both eutherians and marsupials [49]. Thus, we suggest that the positively selected sites we identified in luminal loop 2 evolved in response to a host-filovirus arms race and might represent relevant residues in mediating GP1 binding.
Population genetic analysis of NPC1 in humans revealed no evident signature of natural selection in loop 2 or any other gene region, although we cannot exclude that weak or geographically-restricted selective events have acted on the gene. With respect to filovirus infection, this might not be surprising as the known human pathogens Ebola and Marburg viruses are highly virulent agents that rapidly kill infected individuals, a feature that possibly limits their spreading in human populations [50] and makes them unlikely candidates to play a role as selective agents. Genetic diversity in human NPC1 has nevertheless been recently associated with metabolic dysfunction, this association being based on the central role of the gene in lipid trafficking. Specifically, the His215Arg (rs1805081) variant in luminal loop 1, which is involved in cholesterol binding, was shown to associate with obesity in populations of European descent [9,10]. It has been proposed that alleles responsible for obesity and T2D might have evolved as 'thrifty' variants in ancient populations [51,52]. In line with this hypothesis, selection signatures have been detected for a few polymorphisms associated with these conditions [53,54], although this does not seem to be the case for NPC1. Nonetheless, inspection of nonsynonymous SNPs located in the gene revealed that, in addition to the above mentioned variant in loop 1, a polymorphism (Ile642Met, rs1788799) in the SSD domain segregates at relatively high frequency in human populations and affects an isoleucine residue which is conserved in all the mammals we analyzed.
We thus reasoned that this SNP might affect NPC1 function and modulate metabolic phenotypes. We tested this hypothesis in a large cohort of subjects from Saudi Arabia, a region where the prevalence of obesity and T2D is very high [55-57]. The previously described association between rs1805081 and obesity [9,10] was not replicated in the Saudi sample, although the relatively lower minor allele frequency (MAF) of the variant in this population (12%) compared to Europeans (ranging from 25% to 40%) might have limited our detection power. No effect on BMI or obesity was detected in the Saudi cohort for the Ile642Met variant either. Similarly, the role of the His215Arg variant in predisposing to obesity was not observed in a cohort of Chinese children [12], although a possible interaction between this (and other) variant and sedentary behavior has been described in a population of the same ethnicity [11]. Recently, a meta-analysis of rs1805081 on obesity risk in Europeans also revealed a weak effect of the polymorphism on body fat percentage, but not on BMI or on the odds of being obese [58]. One possibility to explain these contrasting results is that variants in NPC1 interact with environmental cues, as suggested by the Chinese study [11] and possibly with additional genetic factors. This seems to be the case for Npc1+/- mice: these animals develop increased adiposity and metabolic disturbances but the phenotype depends on both fat intake and genetic background [7,59]. These animals also present with increased fasting plasma glucose levels, glucose intolerance, and insulin resistance, indicating a T2D phenotype [7,59]. Somehow in contrast with these results, a recent study indicated that heterozygosity for a hypomorphic Npc1 mutation on the C57BL/6J 'metabolic syndrome' genetic background protects old male mice, but not females, from weight gain [60]. Overall, these observations suggest that Npc1 genetic variation interacts with diet, sex and with one or more gene(s) in modulating metabolic phenotypes.
A possible association between the two NPC1 variants and T2D was analyzed in the Saudi cohort. Overweight and obesity are strong risk factors for the development of T2D; genetic susceptibility is nevertheless believed to play a stronger role in non-obesity related T2D [61]. Thus, we verified the effect of rs1805081 and rs1788799 on diabetes susceptibility by taking BMI into account; a significant association was detected between rs1788799 and T2D, with a predisposing role for the derived 642Met allele.
Several metabolic traits are sexually dimorphic in humans and/or show sex-specific heritability linked to the autosomes [62]. Thus, it was suggested that variants with a sex-specific effect might be difficult to detect without separating the sexes or modeling for gender-based differences [62]. Testing for interaction with sex in our cohort indicated the presence of a significant effect; stratification of the population on the basis of gender revealed that the association is driven by male subjects. This was even more evident when haplotype analysis using the two coding variants was performed. Notably, two major haplotypes showed an opposite effect on T2D susceptibility in men only, and the effect was evident in both obese and non-obese individuals. An interaction between gender and genetic factors has been described for some other genes involved in T2D [63-66]; the reasons underlying these sex-specific events remain to be elucidated and might include a role for sex hormones, epistatic effects with X-linked variants, or differences in dietary habits and lifestyle between the sexes that, in turn, interact with the genetic status.
Further analyses on plasma lipid levels showed the presence of different associations with NPC1 haplotypes in men and women. Nonetheless, these effects were generally weak and should be interpreted with caution. The stronger effect was detected for triglyceride levels. Thus, in men a minor haplotype unrelated to T2D susceptibility was found to associate with higher levels, whereas in women the two major haplotypes that predispose or protect men from diabetes were found to be associated with higher and lower triglyceride levels, respectively.
Conclusions
Data reported here indicate that NPC1 has evolved adaptively in mammals and that the underlying selective pressure might be virus-driven. No selection signature was detected in present-day human populations, but analysis of nonsynonymous polymorphisms showed that a variant (Ile642Met) in the SSD domain affects a highly conserved position. This variant and haplotypes comprising Ile642Met and the previously described His215Arg polymorphism were found to modulate the risk of T2D in a population from Saudi Arabia with a sex-specific effect. Analysis of additional cohorts will be instrumental for clarifying the role of the two NPC1 variants on plasma lipid levels and T2D susceptibility. Our results indicate that haplotype analysis (as opposed to single variant association) and modeling for sex-specific effects are strongly recommended when NPC1 genetic variability is analyzed.
Abbreviations
BEB: Bayes empirical Bayes; BMI: body mass index; bp: base pair; ESD: evolutionary selection distance; GARD: Genetic Algorithm Recombination Detection; iHS: integrated haplotype score; NCBI: National Center for Biotechnology Information; OR: odds ratio; PAML: phylogenetic analysis by maximum likelihood; PCR: polymerase chain reaction; REL: random effects likelihood; SNP: single nucleotide polymorphism; SREBP: sterol regulatory element-binding proteins; SSD: sterol sensing domain; T2D: type 2 diabetes.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MS, RC, DF and UP performed the experiments and analyzed the data. MC and MS designed the study and contributed to writing the manuscript. NMA contributed to the study design and to writing the manuscript. MSA, KMA and SS are responsible for following the cohorts of patients and collecting and cataloguing the samples. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table S1: Likelihood ratio test statistics for models of variable selective pressure among sites. The table reports results of the likelihood ratio tests (M7 versus M8 and M8a versus M8) using the F3X4 codon frequency model. Figure S1: Multiple protein alignment of NPC1 mammalian genes. The figure shows the NPC1 multiple species alignment (41 species, Clustal format); positively selected sites and human nonsynonymous polymorphisms are highlighted. Figure S2: Branch-site random effects likelihood (branch-site REL) analysis of NPC1 genes. The figure shows a branch-site REL analysis of NPC1 with the width and color of each branch indicating the strength of selection. Figure S3: Sliding-window analysis of nucleotide diversity along NPC1 using the 1000 Genomes Project data. The figure shows θW and π calculated for Yoruba, Europeans and Asians in sliding windows of 5 kb moving along the NPC1 gene region. Figure S4: Sliding-window analysis of FST along NPC1 using the 1000 Genomes Project data. The figure shows FST (YRI/CEU/CHB-JPT) calculated in 5 kb windows moving along the NPC1 gene region.
Contributor Information
Nasser M Al-Daghri, Email: aldaghri2011@gmail.com.
Rachele Cagliani, Email: rachele.cagliani@bp.lnf.it.
Diego Forni, Email: diego.forni@bp.lnf.it.
Majed S Alokail, Email: msa85@yahoo.co.uk.
Uberto Pozzoli, Email: uberto.pozzoli@bp.lnf.it.
Khalid M Alkharfy, Email: alkharfy@ksu.edu.sa.
Shaun Sabico, Email: eaglescout01@yahoo.com.
Mario Clerici, Email: mario.clerici@unimi.it.
Manuela Sironi, Email: manuela.sironi@bp.lnf.it.
Acknowledgements
We wish to thank Dr Matteo Fumagalli for helpful comments about this work. The authors are grateful to Prince Mutaib Chair for Biomarkers of Osteoporosis for technical support and to the primary care physicians and nurses who recruited and collected the data on the subjects.
References
- Garver WS. Gene-diet interactions in childhood obesity. Curr Genomics. 2011;12:180–189. doi: 10.2174/138920211795677903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Ioannou YA. Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J Biol Chem. 2000;275:24367–24374. doi: 10.1074/jbc.M002184200. [DOI] [PubMed] [Google Scholar]
- Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, Ruthel G, Pfeffer SR, Dye JM, Whelan SP, Brummelkamp TR, Chandran K. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, Pentchev PG, Pavan WJ. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- Jelinek D, Millward V, Birdi A, Trouard TP, Heidenreich RA, Garver WS. Npc1 haploinsufficiency promotes weight gain and metabolic features associated with insulin resistance. Hum Mol Genet. 2011;20:312–321. doi: 10.1093/hmg/ddq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek D, Heidenreich RA, Erickson RP, Garver WS. Decreased Npc1 gene dosage in mice is associated with weight gain. Obesity (Silver Spring) 2010;18:1457–1459. doi: 10.1038/oby.2009.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyre D, Delplanque J, Chevre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proenca C, Gaget S, Korner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- Sandholt CH, Vestmar MA, Bille DS, Borglykke A, Almind K, Hansen L, Sandbaek A, Lauritzen T, Witte D, Jorgensen T, Pedersen O, Hansen T. Studies of metabolic phenotypic correlates of 15 obesity associated gene variants. PLoS One. 2011;6:e23531. doi: 10.1371/journal.pone.0023531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi B, Wang C, Wu L, Zhang M, Shen Y, Zhao X, Wang X, Mi J. Influence of physical inactivity on associations between single nucleotide polymorphisms and genetic predisposition to childhood obesity. Am J Epidemiol. 2011;173:1256–1262. doi: 10.1093/aje/kwr008. [DOI] [PubMed] [Google Scholar]
- Wu L, Xi B, Zhang M, Shen Y, Zhao X, Cheng H, Hou D, Sun D, Ott J, Wang X, Mi J. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010;59:3085–3089. doi: 10.2337/db10-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver WS, Erickson RP, Wilson JM, Colton TL, Hossain GS, Kozloski MA, Heidenreich RA. Altered expression of caveolin-1 and increased cholesterol in detergent insoluble membrane fractions from liver in mice with Niemann-Pick disease type C. Biochim Biophys Acta. 1997;1361:272–280. doi: 10.1016/S0925-4439(97)00047-1. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rojo MA, Restall C, Ferguson C, Martel N, Martin S, Bosch M, Kassan A, Leong GM, Martin SD, McGee SL, Muscat GE, Anderson RL, Enrich C, Pol A, Parton RG. Caveolin-1 orchestrates the balance between glucose and lipid-dependent energy metabolism: implications for liver regeneration. Hepatology. 2012;55:1574–1584. doi: 10.1002/hep.24810. [DOI] [PubMed] [Google Scholar]
- Garver WS, Jelinek D, Oyarzo JN, Flynn J, Zuckerman M, Krishnan K, Chung BH, Heidenreich RA. Characterization of liver disease and lipid metabolism in the Niemann-Pick C1 mouse. J Cell Biochem. 2007;101:498–516. doi: 10.1002/jcb.21200. [DOI] [PubMed] [Google Scholar]
- Kosiol C, Vinar T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six Mammalian genomes. PLoS Genet. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensembl Genome Browser. http://www.ensembl.org/index.html
- Wernersson R, Pedersen AG. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of bayes prediction of amino acid sites under positive selection. Mol Biol Evol. 2002;19:950–958. doi: 10.1093/oxfordjournals.molbev.a004152. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes. http://www.1000genomes.org/
- Cereda M, Sironi M, Cavalleri M, Pozzoli U. GeCo++: a C++ library for genomic features computation and annotation in the presence of variants. Bioinformatics. 2011;27:1313–1315. doi: 10.1093/bioinformatics/btr123. [DOI] [PubMed] [Google Scholar]
- Thornton K. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Daghri NM, Al-Attas OS, Alokail MS, Alkharfy KM, Yousef M, Sabico SL, Chrousos GP. Diabetes mellitus type 2 and other chronic non-communicable diseases in the central region, Saudi Arabia (Riyadh cohort 2): a decade of an epidemic. BMC Med. 2011;9:76. doi: 10.1186/1741-7015-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond SL, Scheffler K, Gravenor MB, Poon AF, Frost SD. Evolutionary fingerprinting of genes. Mol Biol Evol. 2010;27:520–536. doi: 10.1093/molbev/msp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Pond SK, Muse SV. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 2005;22:2375–2385. doi: 10.1093/molbev/msi232. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrell B, Fourment M, Frost SD, Delport W, Scheffler K. A random effects branch-site model for detecting episodic diversifying selection. Mol Biol Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Genetical structure of populations. Nature. 1950;166:247–249. doi: 10.1038/166247a0. [DOI] [PubMed] [Google Scholar]
- Emerman M, Malik HS. Paleovirology--modern consequences of ancient viruses. PLoS Biol. 2010;8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316:1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Satta Y, Jinnai N, Goto S, Hayasaka I, Saitou N, Guan-Jun J, Tokunaga K, Jorde LB, Emi M, Inoue I. Evidence for natural selection in the HAVCR1 gene: high degree of amino-acid variability in the mucin domain of human HAVCR1 protein. Genes Immun. 2005;6:398–406. doi: 10.1038/sj.gene.6364215. [DOI] [PubMed] [Google Scholar]
- Patel MR, Loo YM, Horner SM, Gale M Jr, Malik HS. Convergent evolution of escape from hepaciviral antagonism in primates. PLoS Biol. 2012;10:e1001282. doi: 10.1371/journal.pbio.1001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZD, Weinstock G, Gerstein M. Rapid evolution by positive Darwinian selection in T-cell antigen CD4 in primates. J Mol Evol. 2008;66:446–456. doi: 10.1007/s00239-008-9097-1. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Takada A. Filovirus tropism: cellular molecules for viral entry. Front Microbiol. 2012;3:34. doi: 10.3389/fmicb.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A. In: The Cambridge Encyclopedia of Human Evolution. Jones S, Martin R, Pilbeam D, editor. Cambridge: Cambridge University Press; 1992. People and disease; pp. 411–420. [Google Scholar]
- Neel JV. Diabetes mellitus: a "thrifty" genotype rendered detrimental by "progress"? 1962. Bull World Health Organ. 1999;77:694–703. discussion 692-693. [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo A, Hudson RR. An evolutionary framework for common diseases: the ancestral-susceptibility model. Trends Genet. 2005;21:596–601. doi: 10.1016/j.tig.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Di Rienzo A. Population genetics models of common diseases. Curr Opin Genet Dev. 2006;16:630–636. doi: 10.1016/j.gde.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Luca F, Perry GH, Di Rienzo A. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 2010;30:291–314. doi: 10.1146/annurev-nutr-080508-141048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, Singh GM, Gutierrez HR, Lu Y, Bahalim AN, Farzadfar F, Riley LM, Ezzati M. Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Body Mass Index) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377:557–567. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Othaimeen AI, Al-Nozha M, Osman AK. Obesity: an emerging problem in Saudi Arabia. Analysis of data from the National Nutrition Survey. East Mediterr Health J. 2007;13:441–448. [PubMed] [Google Scholar]
- Al-Nozha MM, Al-Maatouq MA, Al-Mazrou YY, Al-Harthi SS, Arafah MR, Khalil MZ, Khan NB, Al-Khadra A, Al-Marzouki K, Nouh MS, Abdullah M, Attas O, Al-Shahid MS, Al-Mobeireek A. Diabetes mellitus in Saudi Arabia. Saudi Med J. 2004;25:1603–1610. [PubMed] [Google Scholar]
- den Hoed M, Luan J, Langenberg C, Cooper C, Sayer AA, Jameson K, Kumari M, Kivimaki M, Hingorani AD, Grontved A, Khaw KT, Ekelund U, Wareham NJ, Loos RJ. Evaluation of common genetic variants identified by GWAS for early onset and morbid obesity in population-based samples. Int J Obes (Lond) 2012. doi: 10.1038/ijo.2012.34. [DOI] [PMC free article] [PubMed]
- Jelinek D, Heidenreich RA, Garver WS. The Niemann-Pick C1 gene interacts with a high-fat diet and modifying genes to promote weight gain. Am J Med Genet A. 2011. pp. 2317–2319. [DOI] [PubMed]
- Borbon I, Campbell E, Ke W, Erickson RP. The role of decreased levels of Niemann-Pick C1 intracellular cholesterol transport on obesity is reversed in the C57BL/6J, metabolic syndrome mouse strain: a metabolic or an inflammatory effect? J Appl Genet. 2012;53:323–330. doi: 10.1007/s13353-012-0099-8. [DOI] [PubMed] [Google Scholar]
- Matsuda A, Kuzuya T. Relationship between obesity and concordance rate for type 2 (non-insulin-dependent) diabetes mellitus among twins. Diabetes Res Clin Pract. 1994;26:137–143. doi: 10.1016/0168-8227(94)90151-1. [DOI] [PubMed] [Google Scholar]
- Weiss LA, Pan L, Abney M, Ober C. The sex-specific genetic architecture of quantitative traits in humans. Nat Genet. 2006;38:218–222. doi: 10.1038/ng1726. [DOI] [PubMed] [Google Scholar]
- Kilpelainen TO, Zillikens MC, Stancakova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, Styrkarsdottir U, Zhou Y, Smith AV, Zhao JH, Amin N, Vedantam S, Shin SY, Haritunians T, Fu M, Feitosa MF, Kumari M, Halldorsson BV, Tikkanen E, Mangino M, Hayward C, Song C, Arnold AM, Aulchenko YS, Oostra BA, Campbell H. et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Guo T, Traurig M, Mason CC, Kobes S, Perez J, Knowler WC, Bogardus C, Hanson RL, Baier LJ. SIRT1 is associated with a decrease in acute insulin secretion and a sex specific increase in risk for type 2 diabetes in Pima Indians. Mol Genet Metab. 2011;104:661–665. doi: 10.1016/j.ymgme.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy JJ, Somji A, Weiss LA, Steffy B, Vega R, Barrett-Connor E, Talavera G, Glynne R. Polymorphisms of the scavenger receptor class B member 1 are associated with insulin resistance with evidence of gene by sex interaction. J Clin Endocrinol Metab. 2009;94:1789–1796. doi: 10.1210/jc.2008-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolppanen AM, Pulkkinen L, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, Uusitupa M. Finnish Diabetes Prevention Study Group. Tenomodulin is associated with obesity and diabetes risk: the Finnish diabetes prevention study. Obesity (Silver Spring) 2007;15:1082–1088. doi: 10.1038/oby.2007.613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Likelihood ratio test statistics for models of variable selective pressure among sites. The table reports results of the likelihood ratio tests (M7 versus M8 and M8a versus M8) using the F3X4 codon frequency model. Figure S1: Multiple protein alignment of NPC1 mammalian genes. The figure shows the NPC1 multiple species alignment (41 species, Clustal format); positively selected sites and human nonsynonymous polymorphisms are highlighted. Figure S2: Branch-site random effects likelihood (branch-site REL) analysis of NPC1 genes. The figure shows a branch-site REL analysis of NPC1 with the width and color of each branch indicating the strength of selection. Figure S3: Sliding-window analysis of nucleotide diversity along NPC1 using the 1000 Genomes Project data. The figure shows θW and π calculated for Yoruba, Europeans and Asians in sliding windows of 5 kb moving along the NPC1 gene region. Figure S4: Sliding-window analysis of FST along NPC1 using the 1000 Genomes Project data. The figure shows FST (YRI/CEU/CHB-JPT) calculated in 5 kb windows moving along the NPC1 gene region.