Abstract
Background
The YABBY (YAB) family of transcription factors participate in a diverse range of processes that include leaf and floral patterning, organ growth, and the control of shoot apical meristem organisation and activity. How these disparate functions are regulated is not clear, but based on interactions with the LEUNIG-class of co-repressors, it has been proposed that YABs act as transcriptional repressors. In the light of recent work showing that DNA-binding proteins associated with the yeast co-repressor TUP1 can also function as activators, we have examined the transcriptional activity of the YABs.
Results
Of the four Arabidopsis YABs tested in yeast, only FILAMENTOUS FLOWER (FIL) activated reporter gene expression. Similar analysis with Antirrhinum YABs identified the FIL ortholog GRAMINIFOLIA as an activator. Plant-based transactivation assays not only confirmed the potential of FIL to activate transcription, but also extended this property to the FIL paralog YABBY3 (YAB3). Subsequent transcriptomic analysis of lines expressing a steroid-inducible FIL protein revealed groups of genes that responded either positively or negatively to YAB induction. Included in the positively regulated group of genes were the polarity regulators KANADI1 (KAN1), AUXIN RESPONSE FACTOR 4 (ARF4) and ASYMMETRIC LEAVES1 (AS1). We also show that modifying FIL to function as an obligate repressor causes strong yab loss-of-function phenotypes.
Conclusions
Collectively these data show that FIL functions as a transcriptional activator in plants and that this activity is involved in leaf patterning. Interestingly, our study also supports the idea that FIL can act as a repressor, as transcriptomic analysis identified negatively regulated FIL-response genes. To reconcile these observations, we propose that YABs are bifunctional transcription factors that participate in both positive and negative regulation. These findings fit a model of leaf development in which adaxial/abaxial patterning is maintained by a regulatory network consisting of positive feedback loops.
Keywords: YABBYs, Arabidopsis thaliana, Leaf patterning and development, Adaxial-abaxial polarity, Lateral organ formation
Background
The YABBY (YAB) family of transcription factors regulates various aspects of vegetative and floral development in flowering plants. First identified in Arabidopsis, YABs have a characteristic structure that includes an N-terminal zinc finger domain and a C-terminal YABBY domain containing a helix-loop-helix motif similar to that found in the high mobility group (HMG) of proteins [1,2]. Analysis of the zinc finger domain has shown that it mediates homo- and heterodimerization between the YABs, as well as interactions with other proteins [3,4]. In contrast, the YABBY domain is associated with non-specific DNA-binding [5]. Phylogenetic analysis distinguishes five sub-families of YABs in the angiosperms, represented by the FILAMENTOUS FLOWER/YABBY3 (FIL/YAB3), YAB2, YAB5, CRABS CLAW (CRC) and INNER NO OUTER (INO) clades. In eudicots such as Arabidopsis, the so-called vegetative YABs - FIL, YAB3, YAB2 and YAB5 - are expressed in the abaxial domain of developing leaf and floral organ primordia [2,6], whereas CRC is restricted to the developing carpel and nectaries, and INO is expressed specifically in the outer integument of the ovule [7,8]. Characterisation of the vegetative YABs through mutant and gain-of-function analyses has shown that they regulate cell identity in developing organs and thus play an important role in establishing organ polarity and subsequent lamina growth [1,2,4,6,9]. This regulation is complex, however, as loss of FIL and YAB3 activities is associated with the partial adaxialisation of organs [2], whereas combined loss of FIL, YAB3 and YAB5 results in organ abaxialisation [4]. Similar observations have been made in Antirrhinum, where mutations in the FIL/YAB3 ortholog, GRAM, are associated with a loss of abaxial cell identity, whereas when combined with mutations in the YAB5 ortholog, PROLONGATA (PROL), they result in a loss of adaxial cell identity [10]. YABs fit into a highly complex and redundant network of transcription factors and small RNAs that promote organ polarity (for example see [11]). Factors that promote adaxial identity include the AS2, ARP and the class III HOMEODOMAIN-LEUCINE ZIPPER (C3 HD-ZIP) transcription factors, as well as the trans-acting small interfering RNAs generated by the miR390-TAS3-RDR6 pathway [12-14]. In contrast, the KANADI (KAN1, KAN2 and KAN3) and AUXIN RESPONSE FACTOR (ETTIN (ETT)/ARF3 and ARF4) classes of transcription factors promote abaxial identity, together with the microRNAs miR165/166[15-18]. The precise position of YABs within these networks is not certain, but based on yab loss-of-function phenotypes it has been proposed that YABs integrate adaxial-abaxial patterning with a program of lamina growth [6].
In addition to promoting lamina growth, vegetative YABs also prevent shoot apical meristem (SAM) regulators from being expressed in the developing leaf primordia. In the absence of vegetative YAB activity, KNOX and WUSCHEL (WUS) expression is detected in leaves, where it is associated with the formation of SAM-like structures [6,19]. YABs also play a significant role in regulating SAM activity, as the inflorescence meristem of fil mutants is noticeably enlarged and displays aberrant phyllotaxy [20,21]. In addition, these meristem defects are associated with laterally expanded expression of the meristem regulators WUS and CLAVATA3 (CLV3) [20]. Major disruptions in SAM size and organisation, culminating in SAM arrest, are also observed in higher order Arabidopsis and Antirrhinum yab mutants [4,6,10]. This aspect of YAB function is apparently conserved in angiosperms as a recent study has shown that TONGARI-BOUSHI1, a FIL-like YAB from rice, controls floret meristem activity [22]. As YAB accumulation in both Arabidopsis and Antirrhinum is limited to the abaxial domain of developing lateral organs and floral primordia [20,23], their SAM-promoting activity presumably reflects non-cell-autonomous activity [10,20].
Recent studies have begun to address how YABs function at a molecular level. For instance, physical and genetic interactions between the vegetative YABs and the co-repressors LEUNIG (LUG) and the closely related LEUNIG_HOMOLOG (LUH) suggest that YABs act in repressive complexes [4]. LUG and LUH share structural and functional similarity with TUP1, Groucho (Gro) and TLE co-repressors that are present in yeast, Drosophila and vertebrates [24]. Given this similarity, it is likely that LUG and LUH display a similar range of interactions with DNA-binding factors as that described for fungal and animal co-repressors. In this regard it is interesting to note that in yeast, some DNA-binding proteins are capable of functioning as activators when not associated with a repressor complex [25,26]. Several recent studies have also identified plant transcription factors with dual activities, behaving as either activators or repressors on different sets of target genes [27-29] or in different tissues [30]. This raises the intriguing possibility that some DNA-binding proteins associated with Gro-like co-repressors may also possess bifunctional activity. In this study, we test this possibility by examining the regulatory properties of the vegetative YABs. We show, using a combination of transient transactivation assays and genome-wide transcriptomic analysis, that FIL and possibly YAB3 function as activators during vegetative development. Consistent with the biological importance of activation activity, fusing the repressive motif SRDX to FIL results in a dominant negative phenotype when expressed in plants. Based on transcriptomic analysis, several polarity regulators were identified as FIL targets. Significantly, two of these target genes, KAN1 and ARF4 are thought to lie upstream of the YABs. We therefore propose a model in which YAB proteins maintain leaf polarity by establishing a positively reinforcing feedback loop following the emergence of adaxial/abaxial patterning.
Results
FIL and FIL-like YABs function as activators in yeast
We previously used the yeast two-hybrid assay to examine physical interactions between the YABs and various components of the LUG co-repressor complex. In testing YAB constructs for autoactivation, we discovered that yeast expressing the GAL4 DNA binding domain (BD) fused to FIL produced a noticeable colour change after 4 h growth on media containing X-α-Gal ([4]; Figure 1). Interestingly, this property is not shared with the closely related YAB3 or with other vegetatively expressed YABs (YAB2 and YAB5) (Figure 1). To determine whether activation is a conserved feature of the FIL subgroup of YABs, we examined whether the Antirrhinum FIL ortholog, GRAM, behaves as an activator in yeast. For comparison, we also tested the YAB5 ortholog PROL and the YAB2 ortholog AmYAB2. When grown on media containing X-α-Gal, yeast lines expressing BD:GRAM produced a colour change after 16 h, whereas no colour change was detected with either BD:PROL or BD:AmYAB2. While the behaviour of YAB proteins in yeast does not necessarily reflect their activity in the plant, these results nonetheless raised the intriguing possibility that the FIL subgroup of YABs function as activators.
Figure 1.
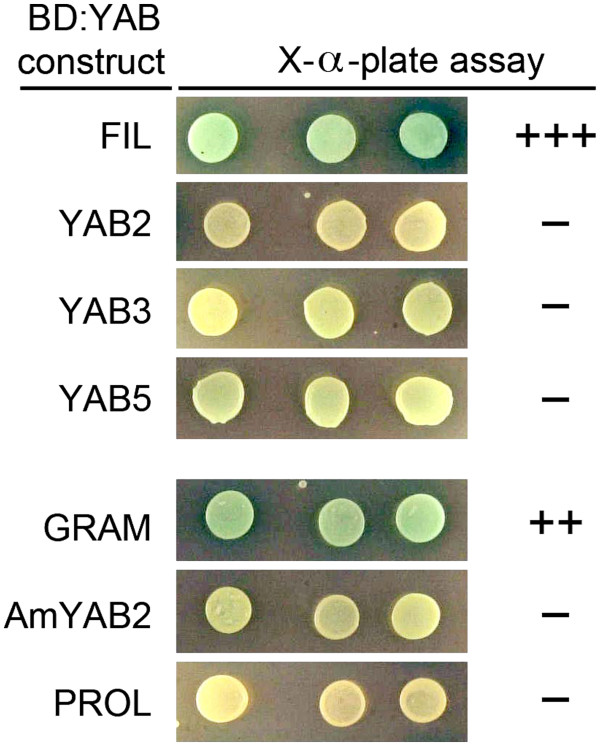
YAB transcriptional activity in yeast. Three independently transformed yeast strains expressing Arabidopsis YABs (FIL, YAB2, YAB3, YAB5) or Antirrhinum YABs (GRAM, AmYAB2, PROL) fused to the GAL4 DNA-binding domain (BD) were assayed for MEL1 reporter activity using an X-α-gal plate assay. Colour change after 4 h (+++), after 16 h (++), or no colour change after 24 h (−) are shown.
In planta assays indicate that FIL/YAB3 can function as transcriptional activators
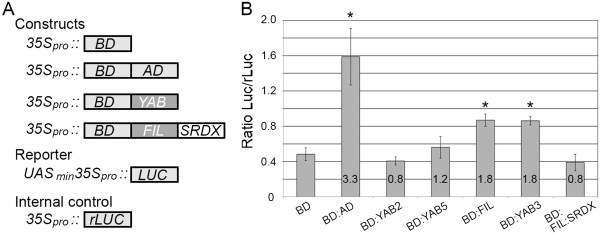
To further investigate the properties of the Arabidopsis YABs, we used an in vivo plant transcription assay to determine whether the vegetatively expressed YABs (FIL, YAB2, YAB3, YAB5) function as activators. YABs were translationally fused to GAL4 BD (Figure 2A) and introduced into Arabidopsis leaves along with the luciferase reporter UAS::LUC. Following transfection, proteins were extracted from leaf tissue and assayed for luciferase activity. We first established a baseline level of luciferase activity by assaying extracts from leaves transfected with 35Spro::BD, and then found that a construct expressing BD fused to the Gal4 activation domain (AD; 35S::BD:AD) produced a 3.3 fold increase in luciferase activity. While BD:YAB2 and BD:YAB5 constructs produced no more than baseline luciferase activity, BD:FIL and BD:YAB3 both activated the UAS::LUC reporter, resulting in a 1.8-fold increase in luciferase activity (Figure 2B). These results not only confirm that FIL functions as an activator in plants, but extend this function to the closely related YAB3 protein.
Figure 2.
Transcriptional activities of vegetatively expressed YAB proteins. (A) Outline of constructs used for transactivation assays. AD, GAL4 activation domain; BD, GAL4-DNA binding domain; YAB, vegetatively expressed YABs (FIL, YAB2, YAB3, YAB5); SRDX, repressive domain (see text for details); UAS, BD binding site; LUC, Firefly Luciferase; rLUC, Renilla Luciferase. (B) YAB transcriptional activation assays with Gal4 BD and BD-AD used as negative and positive control respectively. rLuc was used as an internal control to determine the relative bioluminescence for each sample (ratio Luc/rLuc). Numbers in the shaded boxes indicate fold activation as calculated by dividing total Luc activity of samples by the baseline values arising from the 35Spro::BD construct. Asterisks indicate significant differences (Student's t-test; p < 0.05) and error bars indicate SEM.
Genome-wide transcript profiling following FIL activation
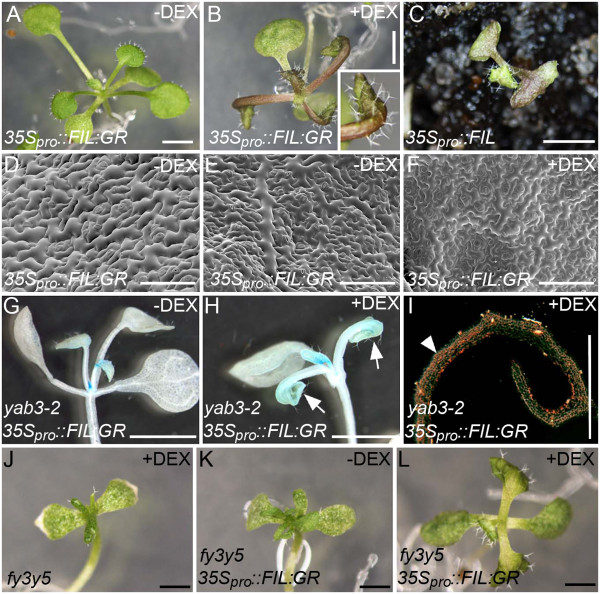
To identify genes that are positively regulated by FIL during shoot development, we opted to assay genome-wide changes in the Arabidopsis transcriptome following rapid induction of FIL activity. Lines expressing an inducible form of FIL (35Spro::FIL:GR) were generated by fusing FIL to the ligand-binding domain of the rat glucocorticoid receptor (GR; [31]). In the absence of the synthetic hormone dexamethasone (DEX), these lines were phenotypically normal, but when grown on media with DEX, or sprayed with DEX, they produced downward curling epinastic leaves that accumulated high levels of anthocyanin (Figure 3A,B). These characteristics are similar to those reported for 35Spro::FIL lines displaying an intermediate over-expression phenotype (Figure 3C; [2]). Moreover, induction of FIL activity was also associated with leaf abaxialisation, as the adaxial epidermal pavement cells of DEX-treated 35Spro::FIL:GR leaves had an abaxial morphology, appearing smaller and more irregularly shaped than cells of mock treated leaves (Figure 3D-F). Abaxialisation was subsequently confirmed when GUS activity derived from the yab3-2 allele, a gene trap that produces abaxially-restricted GUS activity [19], was detected in the adaxial domain of leaves of DEX-grown yab3-2/35Spro::FIL:GR plants (compare Figure 3G-I). Activation of FIL:GR also completely suppressed the formation of narrow or needle-like leaves of the yab triple mutants when activated in this background (Figure 3J-L).
Figure 3.
Vegetative phenotypes associated with steroid-induced constitutive activation of FIL. (A,B) A 35Spro::FIL:GR plant grown on media without DEX (A) or with DEX (B). Inset shows close-up view of an epinastic leaf. (C) 35Spro::FIL plants displaying an intermediate phenotype. (D-F) Scanning electron micrographs showing the adaxial (D, F) or abaxial (E) surface of mature leaves of 35Spro::FIL:GR plants grown on media without DEX (D,E) or with DEX (F). (G,H) Histochemical staining for YAB3:GUS activity in yab3-2/35Spro::FIL:GR plants grown in the absence of DEX (G) or with DEX (H). Arrows indicate prolonged GUS activity in the first true leaves to emerge following germination. (I) Section through a histochemically stained leaf shown in (H) viewed by dark field optics. Arrowheads indicate adaxial accumulation of GUS activity. (J-L) Twenty one day-old fil yab3 yab5 (abbreviated as fy3y5) triple mutants (J) and fy3y5/35Spro::FIL:GR (K,L) lines grown on media with DEX (J, L) or without DEX (K) under short days. Scale bar: 1 mm in (A-C,G,H, J-L), 100 μm in (D-F, I).
Having shown that the FIL:GR fusion is biologically indistinguishable from FIL, we assessed transcriptional changes in the shoots of ten-day-old plants following activation of FIL:GR. As plants were grown under short-day conditions, shoot tissue consisted of mature cotyledons, hypocotyl, and approximately four fully emerged leaves at various stages of development. These included leaves undergoing leaf blade expansion (first true leaves), as well as leaves that had only just emerged from the shoot. Although not visible, initiating organ primordia were also part of these samples. RNA was extracted from this tissue following exposure to DEX or a mock treatment, and used to probe Affymetrix ATH1 genome microarrays. Each treatment consisted of four biological replicates with expression being sampled at 4 h and 8 h post-DEX induction. Following normalization of the expression data and comparisons between mock and DEX treated samples, differentially expressed genes were ranked according to their fold change in expression. We considered a gene differentially expressed if there was a 1.5 fold or greater change in expression following DEX treatment, and if this expression difference was associated with a 0.05 or smaller q-value (see Additional file 1 for a list of differentially expressed genes). The 4 h dataset included 252 up-regulated genes and 251 down-regulated genes, while the 8 h dataset had 166 up-regulated and 184 down-regulated genes. Of these, 219 genes (107 up-regulated/112 down-regulated) were present in both datasets and thus represent a group of genes that consistently respond to FIL activation over an 8 h time period (Group A; Additional file 2). A second group of genes were differentially expressed at 8 h but were not present in the 4 h dataset (Group B; Additional file 2). Because Group A represents genes with robust and consistent responses to FIL activation, we primarily focused on this group.
The close match between the number of group A genes showing reduced expression and the number showing elevated expression following DEX treatment suggested that FIL functions as both an activator and a repressor. To determine whether these two functions are associated with different biological processes, we used gene ontology analysis to identify terms that are enriched in the up- and down-regulated gene sets. Down-regulated genes were enriched for GO terms associated with growth processes (corrected p-value = 0.088) and responses to auxin (corrected p-value = 0.01), whereas up-regulated genes were enriched for terms associated with metabolic processes (auxin biosynthesis; p-value = 0.0466), responses to stimuli including abiotic stress (osmotic), biotic agents (fungal, bacterial) and chemicals (ethylene, chitin), as well as multi-cellular organismal processes (p = 0.0864) involved in lateral root formation. Based on this analysis, it appears likely that FIL regulates distinct processes by functioning as an activator in some cases and as a repressor in others.
Previous work has established that YABs promote organ polarity, and ectopic FIL expression is associated with the partial abaxialisation of leaves (this study; [1,2,4,6]). Consistent with this function, we found that two well-defined abaxial-promoting regulators, KAN1 and ARF4, are elevated ~2-fold following FIL:GR activation (Table 1; Additional file 1). Surprisingly, ETT/ARF3, a gene that functions redundantly with ARF4 in promoting abaxial cell fate [18], and other KAN family members associated with abaxial patterning (KAN2, KAN3[32,33]) were not found in either Group A or B. This suggests that FIL activates some, but not all cell polarity regulators.
Table 1.
Group A FIL-response genesselected for further analysis
|
Identifiers |
|
Fold change detected by microarray |
Ratio |
Fold change detected byqRT-PCR |
|
|---|---|---|---|---|---|
| AGI | Name and Symbol | 4ha | 8ha | 8h/4h | 4ha,b |
| AT5G04340 |
C2H2 zinc-finger transcription factor (CZF2) |
2.72 |
13.05 |
4.80 |
1.96 |
| AT2G38470 |
WRKY family transcription factor (WRKY33) |
5.61 |
15.11 |
2.69 |
3.09 |
| AT1G56650 |
MYB75/PAP1 |
9.78 |
22.20 |
2.27 |
3.35 |
| AT1G06160 |
Ethylene response factor (ERF59) |
8.66 |
19.05 |
2.20 |
8.79 |
| AT3G23550 |
MATE efflux family protein |
14.24 |
20.02 |
1.41 |
22.73 |
| AT5G60450 |
AUXIN RESPONSE FACTOR 4 (ARF4) |
3.64 |
4.14 |
1.14 |
2.74 |
| AT4G36410 |
Ubiquitin-conjugating enzyme 17 (UBC17) |
4.11 |
4.69 |
1.14 |
3.13 |
| AT5G16560 |
KANADI1 (KAN1) |
2.96 |
3.35 |
1.13 |
3.33 |
| AT3G45640 |
Mitogen-activating protein kinase 3 (MPK3) |
2.55 |
2.80 |
1.10 |
2.24 |
| AT3G62150 |
ATP-BINDING CASSETTE B21 |
5.35 |
5.45 |
1.02 |
3.15 |
| AT2G28350 |
AUXIN RESPONSE FACTOR 10 (ARF10) |
1.86 |
1.89 |
1.02 |
2.12 |
| AT1G56010 |
NAC1 |
3.57 |
3.16 |
0.88 |
1.08c |
| AT5G22580 |
Stress responsive A/B Barrel Domain |
3.44 |
2.81 |
0.82 |
2.5 |
| AT1G55200 |
Protein kinase |
4.13 |
3.26 |
0.79 |
2.28 |
| AT3G13672 |
Seven in absentia-like protein |
4.06 |
2.94 |
0.72 |
3.97 |
| AT3G13690 |
Protein kinase |
3.17 |
2.09 |
0.66 |
2.30 |
| AT5G07580 |
Ethylene response factor (ERF5) |
3.87 |
2.29 |
0.59 |
2.93 |
| AT2G37630 |
ASYMMETRIC LEAVES1 (AS1) |
-d |
1.70 |
- |
2.01 |
| AT3G15570 |
NON-PHOTOTROPIC3 (NPH3) |
−1.89 |
−3.49 |
1.84 |
−1.80 |
| AT3G23050 |
IAA7 |
−1.74 |
−2.04 |
1.17 |
−1.82 |
| AT2G40610 |
Expansin (Exp8) |
−3.28 |
−3.76 |
1.15 |
−2.97 |
| AT5G05690 |
Cytochrome P450 (CPD) |
−2.21 |
−1.69 |
0.76 |
−1.50 |
| AT1G20190 | Expansin (Exp11) | −6.17 | −4.57 | 0.74 | −3.71 |
aLength of DEX treatment. bStatistically significant fold change (p<0.05) was calculated using a Student's t-test; cFC is not statistically significant, dGroup B gene.
Validation of microarray data
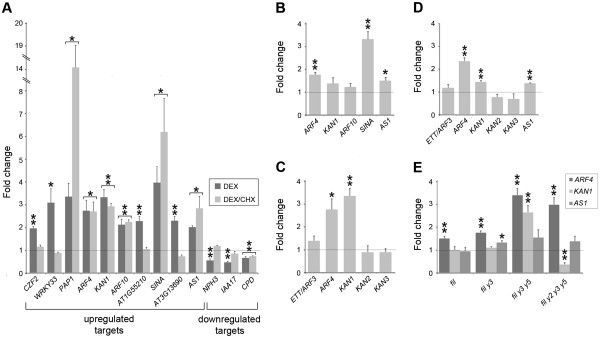
To verify our microarray data, expression of selected Group A genes in 35Spro::FIL:GR shoot tissue exposed to DEX or a mock DEX treatment for 4 h was assessed by quantitative RT-PCR. Included in this group were genes associated with polarity regulation (KAN1, ARF4, AS1) as well as genes associated with auxin (ARF10, IAA7, NPH3), ethylene responses (ERF5, ERF59) and anthocyanin regulation (MYB75/PAP1). With the exception of NAC1, all of the genes displayed a statistically significant change in expression following DEX application, which corroborated the microarray data (Table 1, Figure 4A). As microarray analysis was performed 4 h after DEX treatment, it is likely that many of the differentially expressed genes are direct downstream targets of FIL. This was tested by examining responsiveness of selected FIL-target genes to DEX induction in the presence of the translational inhibitor cycloheximide (CHX). Using an inducible transactivation system, where GUS activity is controlled by a DEX-inducible synthetic transcription factor GR-LhG4 (abbreviated to 35SproI> > GUS; [34]), we showed that combined exposure to CHX and DEX was sufficient to suppress GUS activity (see Additional file 3). Having established the effectiveness of CHX treatments, we next examined the transcriptional responses of twelve Group A genes (nine in the up-regulated class and three in the down-regulated class) and one up-regulated Group B gene following combined DEX/CHX exposure. Seven of these genes showed a statistically significant change in expression following these treatments and were thus considered direct FIL targets (Figure 4A).
Figure 4.
FIL-response genes that areimmediate targets of FIL. (A) Fold change in expression of FIL-response genes in ten-day-old 35Spro::FIL:GR seedlings following a 4 h DEX or DEX/CHX treatment. Brackets indicate genes that display significant transcriptional responses to both treatments and hence mark direct targets of FIL. (B) Response of selected positively regulated FIL-response genes in 10 day-old FILpro::FIL:GR seedlings following a 4 h DEX treatment. (C,D) Induction of abaxial polarity regulators in 10 day-old 35Spro::FIL:GR (C) or 35Spro::YAB3:GR (D) seedlings following a 4 h DEX treatment. (E) Expression of KAN1, ARF4 and AS1 in fil single and yab double, triple and quadruple mutants. Expression in a minimum of three biological replicates was determined using quantitative real-time RT-PCR and normalized first to a housekeeping gene and then to mock treatment controls. Asterisks mark significant differences determined by a Student’s t-test (one asterisk, 0.01<p<0.05; two asterisks, 0.001<p<0.005) and error bars are SEM. The grey line marks the expression level expected if there is no response to treatment.
A complicating factor in our analysis is that FIL:GR expression is not confined to the abaxial domain, and thus transcriptional changes induced by FIL activation may not reflect the normal behaviour of the endogenous protein. To address this, we generated a FILpro::FIL:GR line in which inducible FIL activity is limited to the abaxial domain of developing leaves. Growing this line on media with DEX did not result in a YAB over-expression phenotype, although some leaf epinasty was apparent (see Additional file 4). We next used qRT-PCR to assess the transcriptional response of five positively regulated FIL-response genes in the FILpro::FIL:GR line following a 4 h DEX treatment. All five genes responded positively to induction, although statistically significant elevation was only apparent for three of these: ARF4, SINA and AS1 (Figure 4B). Despite the limited nature of this survey, the observed similarity in transcriptional response between the 35Spro::FIL:GR and FILpro::FIL:GR lines suggests that many of the FIL-response genes identified in our study are indeed immediate targets of FIL.
To confirm that members of the KAN family (KAN2, KAN3) and the ARF4-related gene ETT/ARF3 did not respond to FIL activation, we used qRT-PCR to assay their expression in the 35Spro::FIL:GR line following a 4 h DEX treatment. Consistent with the microarray experiment, this analysis failed to detect significant changes in the expression of these genes (Figure 4C). We next considered whether the FIL paralog YAB3 regulated ARF4, KAN1 and AS1. To test this possibility, we generated an inducible YAB3 line (35Spro::YAB3:GR) and showed that growth on media containing DEX induced a YAB over-expression phenotype (see Additional file 4). qRT-PCR revealed significant elevation of ARF4, KAN1 and AS1 following DEX treatments, but not of ETT/ARF3, KAN2 or KAN3 (Figure 4D). This analysis not only indicates that FIL and YAB3 regulate the same target genes, which is consistent with the observed redundancy between these close paralogs [2], but that there is also a high degree of specificity in this regulation, as related family members do not respond to YAB induction.
Analysis of FIL-regulated polarity regulators in mutant lines
Our data indicated that FIL, and possibly other vegetative YABs, regulate ARF4, KAN1 and AS1 during vegetative development. To test this possibility further, we analysed the expression of these genes in shoot tissue derived from 28-day-old fil mutants, as well as from mutant combinations containing a progressively reduced complement of active vegetative YAB genes (fil yab3, fil yab3 yab5 and fil yab2 yab3 yab5 mutants). A complicating factor in this analysis is the observed abaxialisation of yab triple and quadruple mutants [4], which would be expected to cause elevated expression of abaxial polarity regulators. Consistent with this, we found that both ARF4 and KAN1 expression was elevated in triple yab mutants, with ARF4 expression remaining high in the quadruple mutants (Figure 4E). Unlike ARF4, KAN1 expression was significantly reduced in quadruple mutants, suggesting that KAN1 is regulated directly and positively by the YABs. AS1 expression remained unchanged in the multiple yab mutant lines (Figure 4E), with the exception of fil yab3 mutants, in which it showed a slight increase. While this analysis clearly demonstrates the predominant role of YABs in regulating KAN1, regulation of ARF4 and AS1 presumably involves other factors that function redundantly with the YABs.
Our expression studies suggested the possibility that ectopic expression of the polarity regulators KAN1 and ARF4 following FIL:GR activation may be the cause of the polarity defects observed in the 35Spro::FIL:GR line when continuously exposed to DEX. To test this, kan1 and arf4 mutations were introduced sequentially or together into the 35Spro::FIL:GR background and their phenotype assessed following continuous exposure to DEX. Lines receiving DEX treatment displayed the same leaf epinastic and adaxialisation phenotype as that of 35Spro::FIL:GR lines (see Additional file 5). This suggests that the abaxialisation phenotype associated with constitutive YAB activity is the result of multiple factors being ectopically expressed during leaf development.
Expression of FIL:SRDX induces yab loss-of-function phenotypes
The ERF-associated amphiphilic repressor motif (EAR/SRDX; [35]) has been used to convert positively-acting transcription factors into dominant repressors. Thus, to address the importance of transcriptional activation for YAB function, we investigated the effects of constitutive FIL:SRDX expression in plants. Before performing these experiments, we confirmed that the SRDX motif suppressed FIL activation by showing that a FIL:SRDX fusion was incapable of activating the luciferase reporter in plant-based transactivation assays (Figure 2B).
To compare the effects of constitutive FIL:SRDX expression to that of FIL, plants were transformed with either a 35Spro::FIL:SRDX or 35Spro::FIL construct. While fifty-five 35Spro::FIL primary transformants were recovered following Basta treatment, the same selection yielded only a single 35Spro::FIL:SRDX transformant. This plant had a wild type appearance, suggesting that the FIL:SRDX transgene was unlikely to be active in this line. Given the failure to isolate significant numbers of transgenic plants, we assumed that embryonic or early seedling activity of the FIL:SRDX transgene was likely to be associated with lethality. To circumvent this problem, we used the GR:LhG4/pOP transactivation system to drive post-embryonic expression of FIL transgenes (35Spro::GR:LhG4/pOP::FIL; abbreviated to 35SproI> > FIL). Growth of 35SproI> > FIL lines in the presence of DEX produced the same epinastic cotyledon phenotype as 35S::FIL:GR lines exposed to DEX (Figure 5A,B compare with Figure 3B), although subsequent leaf development exhibited weaker phenotypes, even when the DEX concentration was doubled (data not shown). In contrast, the early stage of 35SproI> > FIL:SRDX growth on media with DEX was characterised by the development of narrow, upwardly curling, hyponastic cotyledons and leaves (Figure 5C,D). The fact that this phenotype differs from the one associated with constitutive FIL expression is consistent with FIL:SRDX no longer functioning as an activator.
Figure 5.
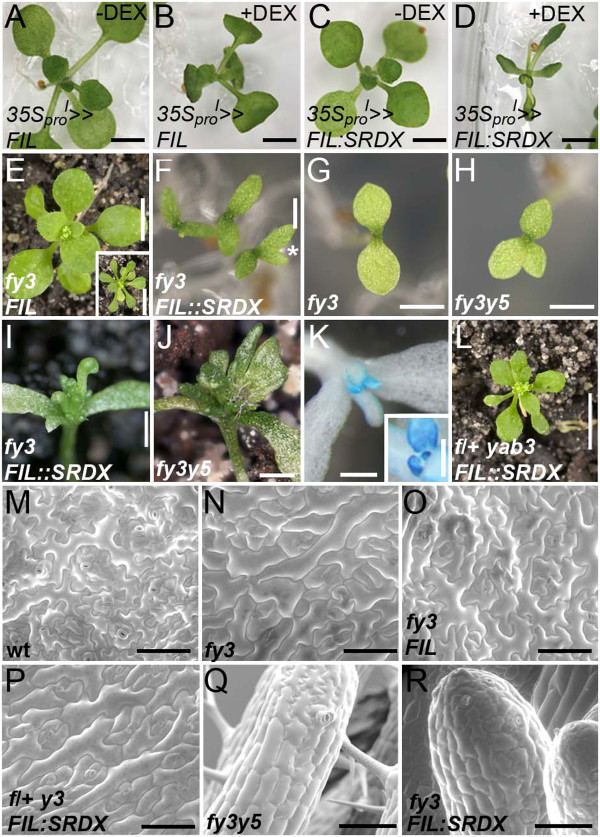
Phenotypes induced by adominant negative FIL:SRDX construct. (A,B) 35SproI>>FIL plants grown on media without DEX (A) or with DEX (B). (C,D) 35SproI>>FIL:SRDX plants grown on media without DEX (C) or with DEX (D). (E) A fil yab3/FILpro::FIL plant displaying full complementation. Inset: fil yab3 double mutant plant. (F) fil yab3/FILpro::FIL:SRDX seedlings showing narrow cotyledons that are sometimes bifurcated (asterisk). Cotyledons of fil yab3 (G) and fil yab3 yab5 mutant seedlings (H). (I) fil yab3/FILpro::FIL:SRDX plant with needle-like leaves. (J) fil yab3 yab5 triple mutant plant with narrow and needle-like leaves. (K) Histochemical staining for YAB3:GUS activity in a fil yab3/FILpro::FIL:SRDX plant. YAB3 promoter activity is detected throughout young radial leaves. Inset: fil yab3/FILpro::FIL stained for GUS activity. (L) A fil/+ yab3/FILpro::FIL:SRDX plant with a fil yab3 mutant leaf phenotype. (M-R) Scanning electron micrograph showing the abaxial epidermis of wildtype (M), fil yab3 (N), fil yab3 FILpro::FIL (O) and fil/+ yab3/FILpro::FIL:SRDX (P) leaves. Note that the larger cell morphology in (N,P) is due to leaf adaxialisation. (Q,R) SEM showing epidermal cell morphology of fil yab3 yab5 needle leaves (Q) and those of fil yab3/FILpro::FIL:SRDX plants (R). Scale bars are 5 mm in (E,L) and inset in (A); 2 mm for (A-D, F-H); 1 mm for (I,J); 200 μm for (K) and inset in (K); 100 μm in (M-R).
In contrast to previous experiments using SRDX fusions [36,37], the presence of this motif was not sufficient to induce a strong loss-of-function phenotype in 35SproI> > FIL:SRDX lines. This could be due to levels of FIL:SRDX not being sufficiently high to interfere with the activity of the native FIL/YAB3 proteins. To address this, we introduced a FILpro::FIL:SRDX construct into yab3 mutants that were heterozygous for the fil mutation (fil/+ yab3) and analysed the phenotype of transgenic fil yab3 mutants in the subsequent generation. As a control for these experiments, we first showed that FILpro::FIL complemented the fil yab3 mutant phenotype (n = 19; Figure 5E). None of the FILpro::FIL:SRDX lines complemented the fil yab3 phenotype, instead, the majority (n = 19/30) had cotyledons that were noticeably smaller than those of fil yab3 mutants and, like fil yab3 yab5, displayed frequent bifurcations (Figure 5F-H). Leaves arising in over half of the fil yab3/FILpro::FIL:SRDX lines (n = 13/19) were similar in appearance to fil yab3 yab5 mutant leaves in being radial and extremely short (Figure 5I, J; [4]). Presence of YAB3:GUS activity throughout the developing leaves of these lines indicated that these organs were partially or fully abaxialised (Figure 5K). In lines where this abaxialisation phenotype was most prevalent (n = 6/13), transgenic fil/+ yab3 sibs also had a fil yab3 leaf phenotype (Figure 5L), which is consistent with FIL:SRDX acting dominantly to suppress native FIL activity.
Analysis of leaf epidermal cell morphology confirmed that FILpro::FIL complemented the polarity defects of fil yab3 mutants, as abaxial cells displayed a wildtype morphology (Figure 5M-O). In contrast, fil/+ yab3/FILpro::FIL:SRDX cells were larger and less irregular than wildtype abaxial cells and similar in appearance to the partially adaxialised cells of fil yab3 mutants (Figure 5N,P). Indeed, SEM analysis of fil yab3/FILpro::FIL:SRDX needle-like leaves revealed a mixture of oblong and jigsaw-shaped cells; exactly the same cellular morphology observed in fil yab3 yab5 triple mutant leaves (Figure 5Q,R; [4]). In summary, these experiments show that inverting the transcriptional activity of FIL through fusions to SRDX leads to a strong yab loss-of-function phenotype when expressed in plants lacking native FIL/YAB3 activity. A plausible explanation for this phenotype is that the FIL:SRDX protein functions dominantly by preventing YAB2 and YAB5 from activating their targets. This experiment therefore provides the first direct evidence that YABs function as activators during the early stages of leaf development.
Discussion
Based on earlier work showing physical and genetic interactions with members of the plant Gro-like family of transcriptional co-repressors, we proposed that YABs function as transcriptional repressors [4,23]. However, genetic analysis indicates that YABs perform their functions independently of these co-repressors. For instance, the conspicuous loss of adaxial cell identity characteristic of yab triple or quadruple mutants is not seen in lines lacking co-repressor activity, nor when single or double yab mutants are combined with mutations in the LUG and LUH co-repressors [10,23]. In this study, we investigated the regulatory activity of the vegetative YABs and found that, in yeast-based transcriptional and in plant transactivational assays, FIL functions as an activator. This property is likely shared with the closely related YAB3 protein, but seemingly not with YAB2 and YAB5. Analysis of the ovule specific YAB, INO, indicates that this factor also functions as an activator, although in this case only autoregulation was investigated [8,38]. This conclusion was based on the observation that abundance of INO transcripts and activity of an INO promoter:reporter construct are reduced in strong ino mutants.
Direct evidence for FIL functioning as an activator also comes from our microarray analysis, where changes in gene expression were assessed 4 h and 8 h following DEX-induced activation. Of the response genes identified, approximately half were under positive regulation, whereas the remainder were under negative regulation. Subsequent qRT-PCR analysis incorporating CHX treatments confirmed that half of the genes tested were indeed likely to be direct FIL-targets. An increasingly common way to establish whether a transcription factor functions as an activator is to convert it into a repressor through a fusion to the SRDX motif, and then show that this chimeric protein produces a dominant negative phenotype when expressed in planta. While FIL:SRDX expression in wildtype plants did not generate yab loss-of-function leaf phenotypes, prominent leaf patterning and growth defects were observed in a fil yab3 mutant background. This suggests that native FIL and YAB3 proteins suppress the repressive activity of FIL:SRDX, perhaps through competition for target binding sites, as FIL and YAB3 are both expressed at high levels in the abaxial domain of developing leaves. In the absence of competing FIL/YAB3, FIL:SRDX constitutively represses genes normally activated by the remaining vegetative YABs. This implies that all four vegetative YABs function as activators during the early stages of leaf development. While this is consistent with the observed functional redundancy between members of the YAB family [2,4,6], our analysis did not reveal activation activity associated with either YAB2 or YAB5 (Figures 1 and 2). This may reflect the relative insensitivity of our yeast and transactivation assays or, alternatively, a requirement for YAB-interacting proteins that function as activators.
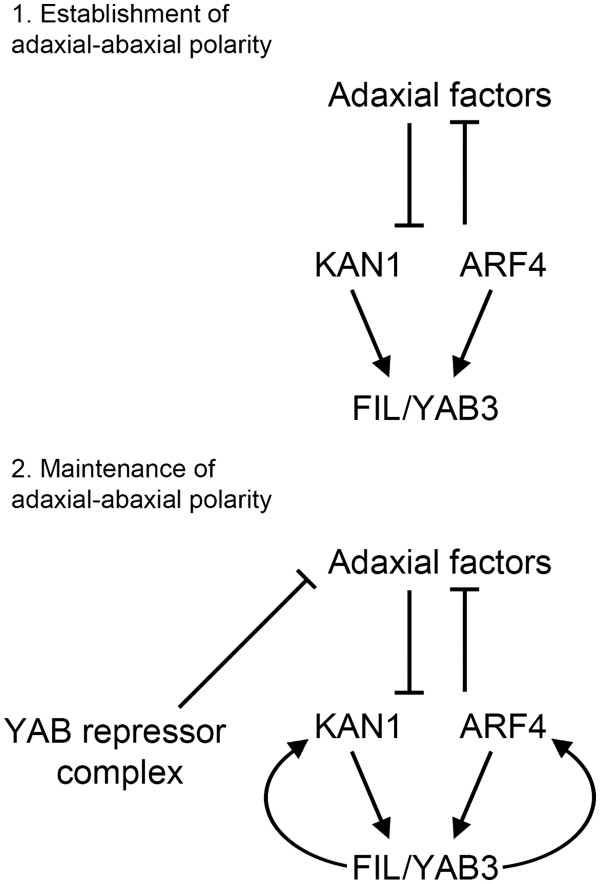
Transcription factors that act as both activators and repressors have been reported in animals [39] and several examples are also known in plants. For instance, the meristem-regulator WUSCHEL activates AGAMOUS (AG) expression in the floral meristem [40], but represses the A-type Arabidopsis Response Regulator7 (ARR7) in the shoot apical meristem [41]. Extensive functional characterisation has confirmed the dual nature of WUS, as well as showing that this activity applies to many more genes [29,42]. Bifunctional activity is also a feature of the senescence-related WRKY53 factor, which acts as an activator or repressor according to sequences surrounding the WRKY-binding motif (W-box) present in the promoters of target genes [27]. On the basis of our results, we propose a model in which YABs are bifunctional, acting either as activators or, when associated with LUG and LUH, as repressors (Figure 6).
Figure 6.
Model for YAB functionduring the early stagesof leaf development. Adaxial-abaxial patterning is established during the early stages of leaf development and is closely associated with the onset of FIL/YAB3 expression. FIL/YAB3 maintain KAN1 and ARF4 expression through direct positive regulation, which in turn establishes a positive feedback loop. As well as acting as positive regulators, YABs associate with transcriptional co-repressors, forming a repressive complex that potentially targets adaxial-promoting factors. It is likely that this regulatory network is confined to the early stages of leaf development, as at later stages FIL/YAB3 expression is present at high levels in the margins of the growing lamina, but KAN1 expression is not readily detectable.
A novel aspect of our study is the finding that the abaxial polarity regulators KAN1 and ARF4 are likely to be immediate FIL targets. This is supported by the observed increase in KAN1 and ARF4 expression in 35Spro::FIL:GR plants following combined DEX/CHX treatment. Furthermore, finding reduced KAN1 expression in yab quadruple mutants indicates that KAN1 regulation is a feature common to the other vegetative YABs. In contrast to KAN1, ARF4 expression was elevated in all yab mutant backgrounds examined. This finding was surprising, given that FIL acts positively on ARF4. Previous studies of yab mutants have established that in addition to leaf patterning defects, these lines lack stipules, display retarded growth and have defects in meristem activity [4,6]. yab mutants also display defects in auxin-regulated processes, such as vascular patterning and formation of the leaf margin, and have reduced activity of the auxin-responsive reporter DR5[6]. Given this, it is possible that increased ARF4 expression observed in yab mutants is a consequence of pleiotropy, and that this obscures the role played by YABs in regulating ARF4.
The observation that FIL regulates KAN1 and ARF4 is also surprising given that previous studies have placed the YABs downstream of such regulators. For instance, FIL expression occurs after the establishment of adaxial-abaxial patterning in developing lateral organ primordia [43], which itself depends on antagonistic interactions between the C3 HD-ZIPs and KAN genes [15,32]. Thus, KAN activity is likely to precede that of the YABs, a hierarchy that is further supported by the observed reduction of FIL/YAB3 expression in developing leaves of kan double and triple mutants [15,33]. Similarly, ARF4 and the related ETT/ARF3 gene are thought to function upstream of the YABs [44]. Our findings can be reconciled with YABs functioning downstream of these abaxial regulators if it is assumed that FIL functions after the establishment of adaxial-abaxial patterning. Such a scenario is attractive, as activating KAN1 and ARF4 following organ patterning would generate a positive feedback loop that maintains abaxial identity (see Figure 6). An important test of this model is to determine whether expression of KANs and C3 HD-ZIPs is initially polarized in the organ primordia of yab quadruple mutants. This could be addressed using live cell imaging to monitor the dynamics of KAN and C3 HD-ZIP expression during the early stages of lateral organ development. Another aspect of our model is the role played by the LUG-YAB regulatory complex in the early patterning of the leaf. While our microarray analysis did not identify any known adaxial regulators that are negatively regulated by FIL, previous studies have shown that lug mutants enhance the adaxialisation phenotype of fil yab3 mutant leaves [4]. We therefore propose that the LUG-YAB regulatory complex is involved in the repression of adaxial regulators, although whether this is mediated by direct or indirect regulation remains to be determined. What is currently missing from this model is an understanding of the temporal-spatial distribution of each of these transcriptional activities. For instance, can repression and activation occur in the same cell, or are they spatially separated activities? Determining the expression pattern of positively and negatively regulated YAB-target genes within the developing leaf may help resolve these issues. Additionally, establishing the precise distribution of the LUG-YAB complex within cells of the developing leaf will also provide insight into the likely transcriptional activities of vegetative YABs. While technically challenging, this could be achieved using in planta BRET measurements [45].
The activation of KAN1, ARF4 and AS1 by FIL and YAB3 has recently been integrated into a computational-derived model of sepal patterning [11]. With the exception of AS1, the inclusion of these regulatory relationships produced a coherent model with other known patterning pathways. The inconsistency with AS1 regulation concerns the apparent restriction of AS1 expression to the adaxial domain of the developing sepal where FIL presumably does not act. However, closer inspection of AS1 expression domain in sepals revealed that this overlapped with FIL during the early stages of organ formation. Thus, FIL regulation of AS1 could also be incorporated into this model, if it is assumed that there are also adaxial factors that promote AS1 expression and abaxial factors that repress AS1 expression during the later stages of sepal development [11].
Auxin distribution is affected in yab loss-of-function mutants, leading to disruptions in the marginal region of developing leaves, as well as the formation of multiple cotyledons during embryogenesis [6]. How YABs regulate auxin distribution in the developing leaf blade is not well understood, but may reflect roles in auxin biosynthesis, movement or signalling. Our study confirms the link between the YABs and auxin responses, as many of the genes responding to FIL activation have GO terms associated with either auxin responses or biosynthesis. In addition we show that ARF4 and ARF10 are positively regulated by FIL and may thus influence the sensitivity of leaf cells to auxin. It is, however, unlikely that the auxin-associated defects of yab mutants are due to reduced activity of ARF4, as ARF4 expression was elevated in triple and quadruple yab mutants, presumably as a result of abaxialisation that is associated with the loss of vegetative YAB activity. Instead, we propose that YABs mediate auxin signalling either through direct association with ARFs, or more likely as part of a regulatory complex. Supporting this possibility is evidence that some ARFs interact with components of the LUG regulatory complex ([46] and our unpublished data) and hence may be indirectly associated with the YABs that are also part of these complexes.
Conclusion
This study has shown that FIL is a bifunctional transcription factor that acts either as an activator or as a repressor. Our finding that the conversion of FIL into an obligate repressor resulted in yab loss-of-function phenotypes when expressed in plants provides direct evidence that activation is required for leaf patterning. Consistent with this finding, FIL is likely to function as a direct activator of the abaxial-promoting factors KAN1 and ARF4, as well is adaxial regulator AS1. On the basis of these results, it is proposed that FIL generates positive feedback loops that maintain adaxial-abaxial polarity following initial polarization of the leaf primordium. This study has therefore provided important new insights into YAB function, as well as laying the foundations for future studies aimed at understanding these enigmatic transcription factors.
Methods
Plant material and growth conditions
Arabidopsis thaliana wildtype plants were either Columbia (Col) or Landsberg erecta (Ler). All transgenic plants were generated using an Agrobacterium floral dipping method [47] and transgenic lines identified by BASTA treatment. The steroid inducible FIL construct 35Spro::FIL:GR, 35Spro::FIL and 35Spro::FIL:SRDX were introduced into Col plants. pOP::FIL and pOP::FIL:SRDX constructs were introduced into the inducible 35Spro::GR:LhG4 driver line [34] that was kindly provided by Miltos Tsiantis (University of Oxford, UK). FILpro::FIL or FILpro::FIL:SRDX were introduced into yab3-2 mutant plants segregating for fil-8, and transgenic fil-8 yab3-2 plants were identified by PCR genotyping in either the T1 population or T2 populations. As a result of segregation, fil-8 was absent in some transgenic T1 plants. These lines were backcrossed to fil-8 yab3-2 double mutants and fil-8 yab3-2 progeny identified in subsequent populations.
The steroid-inducible FIL transgene was introgressed into various yab mutant backgrounds, which have been described previously and are in either a Ler or mixed Ler/Col er background [4,19]. To minimize the effects of er and background variation, we characterized a minimum of 10 F2 or F3 plants that were wildtype for ERECTA. Presence of yab mutant alleles and the transgene was confirmed by PCR genotyping. The 35Spro::FIL:GR transgenic line was also crossed to kan1-12 and arf4-3 mutants [16,48] that were kindly provided by Scott Poethig (University of Pennsylvania). Homozygous mutant F2 lines were identified by PCR genotyping. Details of all PCR genotyping are available upon request.
Plants were either grown on soil or on 0.5x Murashige and Skoog media, in a growth room at 18°C or growth cabinet kept at 21°C, under lights for 8 h (short days) or 16 h (long days). For DEX induction, plants were grown on media containing 10 μM DEX/0.1% DMSO or media containing 0.1% DMSO for mock treatment. Alternatively, DEX-induction was performed on soil grown plants. In this case, DEX treatment involved spraying plants with a 20 μM DEX solution containing 0.2% ethanol and 0.05% Silwet L-77 every 2–3 days, or with a solution of 0.2% ethanol and 0.05% Silwet L-77 for mock treatments. In addition to spraying, plants receiving a DEX treatment were watered with a 20 μM DEX/0.2% ethanol solution.
Constructs
To generate the 35Spro::FIL:GR construct, the FIL coding sequence was PCR-amplified from wildtype inflorescence cDNA using FIL-F3 and FIL-R1.1 primers (see Additional file 6) and a high fidelity Taq polymerase (Kod HiFi; Novagen). Presence of XbaI and BamHI sites in these primers allowed the product to be cloned into the equivalent sites of pBIΔGR, a binary vector containing the ligand-binding domain of the rat glucocorticoid receptor (GR) downstream of the cauliflower mosaic virus 35S promoter (kindly provided by Robert Sablowski, John Innes Centre). As a result, the GR domain was fused in frame with the FIL C-terminus. 35Spro::FIL was generated by amplifying the FIL coding sequence with primers cFIL-KpnI and Fil R-X (see Additional file 6) and cloning this product into the SmaI and XbaI sites located between the 35S promoter and 3’ UTR of the octopine synthase gene of vector pART7 [49]. 35Spro::FIL:SRDX was generated by amplifying FIL coding sequence with cFIL-KpnI and FILR1.1 primers and cloning the product into KpnI and BamHI sites of BJ36-SRDX, a shuttle vector containing the 12 amino acid synthetic ERF-associated amphiphilic repression motif (SRDX; [35]). This placed the SRDX domain in frame with the C-terminal end of the FIL coding sequence. The FIL:SRDX fusion was then moved into the KpnI and XbaI sites of pART7. The FILpro::FIL construct was made by placing the 3.8 kb FIL promoter [50], amplified from Col genomic DNA with pFIL-Xho and pFIL-KpnI (see Additional file 6), into XhoI and KpnI sites upstream of the FIL ORF in the shuttle vector BJ36. A similar approach was also used to generate the FILpro::FIL:SRDX construct.
FIL-containing cassettes from pART7 and BJ36 were excised with NotI and introduced into the NotI site of binary vector pMLBART [49]. Binaries were then introduced into Agrobacterium tumefaciens (GV3101) by electroporation.
Yeast assays
The coding sequences of GRAM, PROL and AmYAB2 were amplified from Antirrhinum majus vegetative cDNA using a high fidelity Taq polymerase and primers that incorporate EcoRI sites (see Additional file 6). The YAB sequences were then cloned into the EcoRI site of the pGBK-T7 vector (Clontech), resulting in the coding sequence being downstream of and in frame with the Gal4 DNA-binding motif (BD). Constructs were transferred into yeast and transcriptional activity of the chimeric BD-YAB protein was assessed using X-α-Gal plate-based colorimetric assays according to published protocols.
In vivo plant transactivation assays
To generate Gal4 BD fusions, the coding sequences of FIL, FIL:SRDX, YAB2, YAB3 and YAB5 were amplified with oligonucleotides listed in Additional file 6 using a high fidelity Taq. PCR products were inserted into the SmaI site of vector pMN6 [51], downstream of the BD and 35S promoter. This resulted in BD being fused in frame to an alanine linker (AAAARS) that was incorporated into the N-terminus of the YAB coding sequence by PCR. For the transactivation experiments, pMN6 alone was used as a negative control, while a vector containing both the GAL4 activation (AD) and DNA-binding domains (AD-BD) driven by the 35S promoter (pMN7; [51]) was used as a positive control. A vector containing the Renilla luciferase (rLUC) gene under the control of the 35S promoter (pRLC) was used as the internal standard and the transactivation reporter was pGLL, a vector with the firefly LUC gene downstream of multiple Gal4 UAS fused to a minimal promoter [51]. Four micrograms of a mixture of BD-YAB construct and firefly LUC reporter (1:1 mass ratio) together with 0.5 μg internal standard were introduced into the abaxial side of mature Arabidopsis leaves using particle bombardment according to the manufacturer’s recommendation (BioRad). Bombarded tissue was left for 24 h on 0.5x MS plates under lights before extracting protein into 1x luciferase cell culture lysis reagent (Promega) with proteinase inhibitor (Roche). Activity of LUC in 20 μl of extract was then assessed following the addition of 80 μl assay buffer containing 1x luciferase cell culture reagent, 1x proteinase inhibitor, 0.1 mM EDTA, 20 mM Tricine, 27 mM MgSO4, 33 mM DTT, 270 μM CoA, 530 μM ATP and 546 nM luciferin. In a separate assay, rLUC activity was measured following the addition of coelenterazine (Promega) in a standard phosphate buffer. Luminometric assays were performed on three bombardments and were repeated three times using a Mithras LB940 Multimode Microplate Reader.
Microarray analysis
For experiments involving DEX induction, four pools of ten-day-old 35Spro::FIL:GR seedlings (grown under short days) were sprayed with a solution containing 10 μM DEX, 0.1% ethanol and 0.015% Silwet L77 (DEX treatment) or a solution containing 0.1% ethanol, 0.015% Silwet L77 (mock treatment), prior to collection at 4 h or 8 h. RNA was extracted from pools of seedlings using a Plant RNeasy Mini Kit (Qiagen). RNA integrity was confirmed by agarose gel electrophoresis before probe synthesis using 300 ng of total RNA per sample according to the AtGenExpress protocol [52]. Affymetrix ATH1 genome arrays were then hybridized and washed using a Genechip fluidics station, and scanned using a Genechip Scanner 3000. Scanned data were processed using the GeneSpring GX software package (Agilent). CEL files were analysed using the GC-Robust Multi Array average method, which incorporates background subtraction, normalization and probe summation. An unpaired Student’s t test with a Benjamin-Hochberg false discovery rate correction was applied to the data to obtain q-values (corrected p-values based on a False Discovery Rate of p = 0.05).
Quantitative real time RT-PCR
For DEX induction, four independent pools of ten-day-old transgenic plants received either a mock or a DEX treatment as outlined above. For DEX/CHX treatments, transgenic plants were first sprayed with a 10 μg/μl cycloheximide solution in 0.015% Silwet L77 and left for 1 h before spraying again with the CHX solution or with a solution containing both 10 μg/μl cycloheximide and 10 μM DEX. Plants were harvested 4 h following these treatments and RNA extracted using an RNeasy kit (Qiagen). For analysis of yab mutants, pools of 28-day-old (short-day grown) wildtype (Ler) plants and yab mutants were collected and RNA extracted. Total RNA was first treated with DNase (Ambion) before synthesizing cDNA using an oligo (dT) primer with Superscript III reverse-transcriptase (Invitrogen). PCR reactions were performed in the presence of SYBR-green on a Rotor-Gene 3200 Real-Time Cycler (Corbett Research) using ACTIN2 (ACT2; AT3g18780), TUBULIN7 (TUB7; At2g29550) or TRANSLATIONALLY CONTROLLED TUMOR PROTEIN (TCTP; At3g16640) as housekeeping controls. Expression levels were first normalized to the housekeeping controls then to the mock treatment, in the case of DEX or DEX/CHX treatments, or to wildtype plants in the case of yab mutants. Oligonucleotides used for qRT-PCR are listed in Additional file 6.
Microscopy
Imaging of the leaf surface was performed on an environmental scanning electron microscope (FEI Quanta) fitted with a Peltier cold stage operating at 2°C. Leaf samples were attached to stubs using carbon adhesive tape before viewing under an accelerating voltage of 12.5Kv and 5.3 Torr vacuum. GUS staining was performed by briefly fixing tissue in 90% acetone and then incubating tissue overnight in a 50 mM phosphate buffer containing X-Gluc and a mixture of potassium ferricyanide and ferrocyanide at 37°C. Tissue was either examined as whole mounts in 70% ethanol or embedded in Paraplast Plus, before being sectioned at 8 μm and viewed under dark-field optics.
Bioinformatics
The accession code, E-MEXP-3726, may be used to access the microarray data from the ArrayExpress website (http://www.ebi.ac.uk/arrayexpress/).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
OB conducted the microarray analyses and, together with JEL, quantified expression of FIL-response genes by qRT-PCR. LP made the yeast constructs and together with JEL performed yeast assays looking at the transcriptional activity of Antirrhinum and Arabidopsis YABs. CPS supervised the plant based transactivation and DEX/CHX qRT-PCR assays and drafted sections of the manuscript. JFG generated all transgenic plants, supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Microarray analysis of 35Spro::FIL:GR seedlings following dexamethasone treatment. This file contains lists of genes that were identified as being differentially expressed in the shoot tissue of 35Spro::FIL:GR lines exposed to DEX for 4 h or 8 h.
FIL-response genes that are differentially expressed at 4 h and 8 h following DEX treatment or at 8 h following DEX treatment. This file contains lists of genes that are have been placed into two groups. Group A genes are differentially expressed at 4 h and 8 h following DEX treatment, whereas Group B genes are differentially expressed at 8 h but not 4 h following DEX treatment. (
Histochemical staining of seedlings treated with DEX and CHX. This figure shows the effectiveness of DEX and DEX/CHX treatments on control plants. Histochemical staining for GUS activity in ten-day-old 35SproI> > GUS seedlings exposed to a mock DEX/CHX treatment (A), CHX (B), DEX (C) and DEX/CHX (D) for 9 h. Scale bars are 1 mm.
Vegetative phenotypes associated with steroid-induced activation of abaxially expressed FILand constitutively expressed YAB3. This figure shows the phenotype of FILpro::FIL:GR plants and 35Spro::YAB3:GR plants continuously exposed to DEX. (A) Fourteen-day-old FILpro::FIL:GR plants grown on soil and sprayed with DEX (see Methods). (B,C) 35Spro::YAB3:GR plant grown on media without DEX (B) or in the presence of DEX (C). Scale bars are 1 mm.
Vegetative phenotype associated with continuous FIL activation in different mutants backgrounds. This figure shows the DEX-inducible phenotype and leaf epidermal cell morphology of mutant plants harbouring the 35Spro::FIL:GR construct. (A-D) Twenty-day-old 35Spro::FIL:GR (A), 35Spro::FIL:GR/arf4 (B), 35Spro::FIL:GR/kan1 (C) and 35Spro::FIL:GR/arf4 kan1 (D), plants grown on soil and sprayed with DEX (see Methods). (E-I) SEM of the adaxial surface of a leaf taken from 35Spro::FIL:GR plants (E, F), a 35Spro::FIL:GR/arf4 plant (G), a 35Spro::FIL:GR/kan1 plant (H) or a 35Spro::FIL:GR/arf4 kan1 plant (I). Plants received a mock treatment (E) or were sprayed with DEX (F-I). Scale bars are 2 mm (A-D) and 100 μM (E-I).
Primer sequences used for qRT-PCR and cloning. This file contains a list of all the oligonucleotides used for generating constructs and conducting qRT-PCR.
Contributor Information
Oliver Bonaccorso, Email: sikma82@gmail.com.
Joanne E Lee, Email: j.lee6@student.unimelb.edu.au.
Libby Puah, Email: puah_libby@yahoo.com.sg.
Charles P Scutt, Email: charlie.scutt@ens-lyon.fr.
John F Golz, Email: jgolz@unimelb.edu.au.
Acknowledgements
The Authors would like to acknowledge the help of Sarah Sherson, Marnie Soso and Giana Romano in maintaining plant lines and Roger Curtain for help with the ESEM. We would also like to thank Chris Cobbett, Michael Hynes and members of the Golz lab for critically reading the manuscript. JEL is supported by an Elizabeth and Vernon Puzey Postgraduate Scholarship. This work was supported by an ARC Discovery Grant (JFG).
References
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum S, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Kanaya E, Watanabe K, Nakajima N, Okada K, Shimura Y. Zinc Release from the CH2C6 Zinc Finger Domain of FILAMENTOUS FLOWER Protein from Arabidopsis thaliana Induces Self-assembly. J Biol Chem. 2001;276:7383–7390. doi: 10.1074/jbc.M009491200. [DOI] [PubMed] [Google Scholar]
- Stahle MI, Kuehlich J, Staron L, von Arnim AG, Golz JF. YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell. 2009;21:3105–3118. doi: 10.1105/tpc.109.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya E, Nakajima N, Okada K. Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J Biol Chem. 2002;277:11957–11964. doi: 10.1074/jbc.M108889200. [DOI] [PubMed] [Google Scholar]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22:2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- Villanueva JM, Broadhvest J, Hauser BA, Meister RJ, Schneitz K, Gasser CS. INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules. Genes Dev. 1999;13:3160–3169. doi: 10.1101/gad.13.23.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell. 1999;99:199–209. doi: 10.1016/S0092-8674(00)81651-7. [DOI] [PubMed] [Google Scholar]
- Golz JF, Roccaro M, Kuzoff R, Hudson A. GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development. 2004;131:3661–3670. doi: 10.1242/dev.01221. [DOI] [PubMed] [Google Scholar]
- La Rota C, Chopard J, Das P, Paindavoine S, Rozier F, Farcot E, Godin C, Traas J, Monéger F. A data-driven integrative model of sepal primordium polarity in Arabidopsis. Plant Cell. 2011;23:4318–4333. doi: 10.1105/tpc.111.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Guo M, Nogueira FT, Timmermans MC. Establishing leaf polarity: the role of small RNAs and positional signals in the shoot apex. Development. 2007;134:813–823. doi: 10.1242/dev.000497. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. The Plant Journal. 2007;51:173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery JF, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran MK, Bowman JL, Sundaresan V. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell. 2002;14:2761–2770. doi: 10.1105/tpc.004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell. 2008;20:1217–1230. doi: 10.1105/tpc.107.057877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugassi N, Nakayama N, Bochnik R, Zik M. A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 2010;10:131. doi: 10.1186/1471-2229-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Ohmori Y, Yoshida A, Kawai A, Mayama-Tsuchida T, Ichikawa H, Mitsuda N, Ohme-Takagi M, Hirano HY. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell. 2012;24:80–95. doi: 10.1105/tpc.111.094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Efremova N, Golz JF, Rubiera R, Kuckenberg M, Castillo R, Tietz O, Saedler H, Schwarz-Sommer Z. Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development. 2004;131:3649–3659. doi: 10.1242/dev.01205. [DOI] [PubMed] [Google Scholar]
- Lee JE, Golz JF. Diverse roles of Groucho/Tup1 co-repressors in plant growth and development. Plant Signal Behav. 2012;7:86–92. doi: 10.4161/psb.7.1.18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman MJ, Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol Cell Biol. 2007;27:7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Reese JC. Molecular genetic analysis of the yeast repressor Rfx1/Crt1 reveals a novel two-step regulatory mechanism. Mol Cell Biol. 2005;25:7399–7411. doi: 10.1128/MCB.25.17.7399-7411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol. 2004;55:853–867. doi: 10.1007/s11103-004-2142-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamothe R, Boyle P, Dulude A, Roy V, Lezin-Doumbou C, Kaur GS, Bouarab K, Despres C, Brisson N. The transcriptional activator Pti4 is required for the recruitment of a repressosome nucleated by repressor SEBF at the potato PR-10a gene. Plant Cell. 2008;20:3136–3147. doi: 10.1105/tpc.108.061721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL Is a Bifunctional Transcription Factor That Acts as a Repressor in Stem Cell Regulation and as an Activator in Floral Patterning. Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A, Louvet R, Johansen JN, Salsac F, Morin H, Fournet F, Belcram K, Gillet F, Hofte H, Laufs P. et al. The transcription factor BELLRINGER modulates phyllotaxis by regulating the expression of a pectin methylesterase in Arabidopsis. Development. 2011;138:4733–4741. doi: 10.1242/dev.072496. [DOI] [PubMed] [Google Scholar]
- Gallois JL, Woodward C, Reddy GV, Sablowski R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development. 2002;129:3207–3217. doi: 10.1242/dev.129.13.3207. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/S0960-9822(01)00392-X. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131:2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Craft J, Samalova M, Baroux C, Townley H, Martinez A, Jepson I, Tsiantis M, Moore I. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. The Plant Journal. 2005;41:899–918. doi: 10.1111/j.1365-313X.2005.02342.x. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Heyl A, Ramireddy E, Brenner WG, Riefler M, Allemeersch J, Schmulling T. The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 2008;147:1380–1395. doi: 10.1104/pp.107.115436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell. 2007;19:473–484. doi: 10.1105/tpc.106.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister RJ, Kotow LM, Gasser CS. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development. 2002;129:4281–4289. doi: 10.1242/dev.129.18.4281. [DOI] [PubMed] [Google Scholar]
- Adkins NL, Hagerman TA, Georgel P. GAGA protein: a multi-faceted transcription factor. Biochem Cell Biol. 2006;84:559–567. doi: 10.1139/o06-062. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/S0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A. et al. Transcriptional Control of a Plant Stem Cell Niche. Developmental cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15:1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Xu X, Soutto M, Xie Q, Servick S, Subramanian C, von Arnim AG, Johnson CH. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proceedings of the National Academy Science USA. 2007;104:10264–10269. doi: 10.1073/pnas.0701987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger J, Zambryski P. The role of SEUSS in auxin response and floral organ patterning. Development. 2004;131:4697–4707. doi: 10.1242/dev.01306. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Okada K. Two discrete cis elements control the bbaxial side-specific expression of the FILAMENTOUS FLOWER gene in Arabidopsis. Plant Cell. 2003;15:2592–2602. doi: 10.1105/tpc.015214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Quail PH. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 2003;35:660–664. doi: 10.1046/j.1365-313X.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray analysis of 35Spro::FIL:GR seedlings following dexamethasone treatment. This file contains lists of genes that were identified as being differentially expressed in the shoot tissue of 35Spro::FIL:GR lines exposed to DEX for 4 h or 8 h.
FIL-response genes that are differentially expressed at 4 h and 8 h following DEX treatment or at 8 h following DEX treatment. This file contains lists of genes that are have been placed into two groups. Group A genes are differentially expressed at 4 h and 8 h following DEX treatment, whereas Group B genes are differentially expressed at 8 h but not 4 h following DEX treatment. (
Histochemical staining of seedlings treated with DEX and CHX. This figure shows the effectiveness of DEX and DEX/CHX treatments on control plants. Histochemical staining for GUS activity in ten-day-old 35SproI> > GUS seedlings exposed to a mock DEX/CHX treatment (A), CHX (B), DEX (C) and DEX/CHX (D) for 9 h. Scale bars are 1 mm.
Vegetative phenotypes associated with steroid-induced activation of abaxially expressed FILand constitutively expressed YAB3. This figure shows the phenotype of FILpro::FIL:GR plants and 35Spro::YAB3:GR plants continuously exposed to DEX. (A) Fourteen-day-old FILpro::FIL:GR plants grown on soil and sprayed with DEX (see Methods). (B,C) 35Spro::YAB3:GR plant grown on media without DEX (B) or in the presence of DEX (C). Scale bars are 1 mm.
Vegetative phenotype associated with continuous FIL activation in different mutants backgrounds. This figure shows the DEX-inducible phenotype and leaf epidermal cell morphology of mutant plants harbouring the 35Spro::FIL:GR construct. (A-D) Twenty-day-old 35Spro::FIL:GR (A), 35Spro::FIL:GR/arf4 (B), 35Spro::FIL:GR/kan1 (C) and 35Spro::FIL:GR/arf4 kan1 (D), plants grown on soil and sprayed with DEX (see Methods). (E-I) SEM of the adaxial surface of a leaf taken from 35Spro::FIL:GR plants (E, F), a 35Spro::FIL:GR/arf4 plant (G), a 35Spro::FIL:GR/kan1 plant (H) or a 35Spro::FIL:GR/arf4 kan1 plant (I). Plants received a mock treatment (E) or were sprayed with DEX (F-I). Scale bars are 2 mm (A-D) and 100 μM (E-I).
Primer sequences used for qRT-PCR and cloning. This file contains a list of all the oligonucleotides used for generating constructs and conducting qRT-PCR.