Abstract
A-kinase anchoring proteins (AKAPs) are signaling scaffolds that contribute to various aspects of cAMP signaling. They do this by tethering protein kinase-A to specific subcellular sites, thereby focusing its activity toward relevant substrates. Recently the structural basis for these protein–protein interactions has been elucidated by x-ray crystallography. Recent reports have identified AKAPs that bind to adenylyl cyclases to regulate cAMP synthesis and that sequester phosphodiesterases to break down this second messenger locally. Another emerging aspect of AKAP function is their role in integrating cAMP signaling with other signaling pathways. For example, molecular and genetic approaches have been used to show that the neuronal anchoring protein WAVE1 integrates signaling from PKA and Cdk5 to regulate actin polymerization and cytoskeletal events.
Signaling scaffolds
Over the past twenty years, a hallmark achievement in cell biology has been the elucidation of the fundamental role that protein–protein interactions play in cellular signaling. Indeed, the recent large-scale genomics and proteomics projects have shown that after a certain point the evolution of complex metazoans is driven not by the creation of entirely new genes but rather by the combinatorial shuffling of modular protein–protein interaction domains [1,2]. Among different signaling pathways, this shuffling of modular domains drives the creation of new connectivities and regulatory networks [2]. Prime examples of this strategy are the numerous scaffolding and adaptor proteins that function in the assembly of multi-protein signaling complexes [3,4]. These signaling scaffolds serve as platforms for the integration and simultaneous dissemination of multiple signals. By sequestering a signaling enzyme to a specific subcellular environment, these proteins ensure that upon activation the enzyme is near its relevant targets. Thus scaffolds contribute to the spatiotemporal resolution of cellular signaling and are a key means by which a common signaling pathway can serve many different functions.
One family of scaffolding proteins are the A-kinase anchoring proteins (AKAPs), which anchor protein kinase A (PKA) to specific subcellular locations [5,6]. AKAPs are a well-studied family of signaling scaffolds and because of the range of their interactions serve as a good model for these systems. As PKA is the primary effector of the second messenger 3′5′-cyclic-adenosinemonophosphate (cAMP), AKAPs play an important role in the targeting and regulation of PKA-mediated phosphorylation events. An equally important role of AKAPs is their capacity to form multi-protein complexes that integrate cAMP signaling with other pathways and signaling events. In this review we focus on recent advances in the study of AKAPs. In terms of AKAP function, our discussion of these advances is divided into three main areas: structural analysis of the AKAP/PKA interaction, the role of AKAPs in the spatiotemporal dynamics of cAMP signaling, and the ability of AKAPs to integrate signals from multiple pathways.
The AKAP/PKA complex
AKAPs make up a structurally diverse protein family with >50 members. Functionally, these proteins share three common features: first, they contain a PKA-anchoring domain; second, they bind other signaling enzymes to form multi-protein complexes; and third, they target these signaling complexes to specific subcellular sites through various targeting motifs, like lipid modifications and protein–protein interaction domains [5]. PKA is a broad spectrum Ser/Thr kinase that can phosphorylate a range of proteins. Anchoring of PKA by AKAPs confines PKA activity to a relevant subset of potential substrates. The activity of PKA is also regulated by its two regulatory subunits, which form a dimer that binds to the two catalytic subunits. Activation of PKA occurs through the binding of cAMP to the regulatory dimer, causing dissociation of the catalytic subunits. The two main PKA subtypes are defined by the identity of their regulatory subunits, RI and RII. The majority of known AKAPs bind specifically to the RII holoenzyme. However, several dual specificity AKAPs, which bind to both PKA subtypes, have been identified. These include the dual-function anchoring proteins D-AKAP1 and D-AKAP2.
Early biochemical studies mapped the PKA-anchoring domain of AKAPs to a 14–18-residue amphipathic helix [7]. Subsequent structural work using NMR showed that this helix bound to a hydrophobic groove in the docking and dimerization (D/D) domain of the RII dimer [8,9]. Recently, two x-ray crystal structures of complexes between an AKAP amphipathic helix and the RII D/D domain were published. Gold et al. used an engineered helix termed AKAPis [10••] that optimally binds to RII [11], while Kinderman et al. used a peptide corresponding to the amphipathic helix of D-AKAP2 [12••]. Both structures confirm much of the previous structural work using NMR, showing that the hydrophobic face of the AKAP helix docks to a hydrophobic groove formed by a four-helix crossing bundle in the RII D/D domain (Figure 1a). However, the crystal structures show that upon binding the AKAP helix creates an asymmetric complex, in which the N-terminal loop of only one of the RII protomers wraps around to make van der Waals contact with side-chains of the AKAP helix. This adds an element of flexibility to the otherwise rigid docking platform of the RII D/D domain and may explain how RII is able to recognize a diverse array of AKAP amphipathic helices.
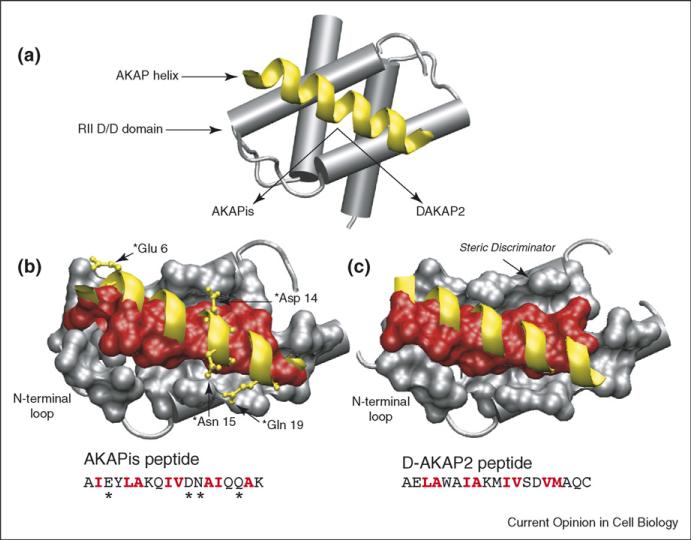
Figure 1.
Structure of the PKA/AKAP binding interface. (a) Schematic of an AKAP PKA-anchoring domain (yellow) bound to the D/D domain (residues 1–44) of RII. In the structures shown below, the helical backbones of AKAPis and DAKAP2 are depicted in yellow and residues that form the binding interface between RII and the AKAP (red) are shown in space-filling. The primary sequences of the AKAP peptides are included with interfacial residues highlighted in red. (b) Close-up of the AKAPis helix bound to the hydrophobic groove of RII. AKAPis residues that form polar interactions with RII are shown in ball and stick (yellow). (c) Close-up of the DAKAP2 PKA-anchoring domain bound to the hydrophobic groove of RII. The potential steric discriminator that contributes to the PKA-subtype specificity of AKAPs is indicated. Comparison of the two structures shows that, although they are quite similar, the RII/AKAPis binding interface comprises a greater number of residues and there are differences in the registry of the helical side-chains involved at each interface.
Although the RII/AKAP structures are quite similar to each other, they do have some interesting differences (Figure 1b,c). For example, the complex with AKAPis reveals several hydrogen bonding and electrostatic interactions between sidechains of the helix and RII [10••]. In addition, the registry of helical sidechains buried at the hydrophobic binding interface differs slightly between the two complexes. This is likely to be due to the differences in the helical peptides used for each structure. Although the presence of hydrophobic residues on the binding face of the helix is conserved among different AKAPs, the identity of those residues is not strongly conserved. This, coupled with the differences in sidechain registry seen between the two structures and the flexibility of the N-terminal interface, indicates that each AKAP may bind to RII in a slightly different manner. This in turn may explain the differences seen among AKAPs in their affinity for RII [13]. It is possible that the cell utilizes these differing affinities to control the timing and activity of signaling complexes formed by different AKAPs.
One key detail the structures begin to address is the identity of the molecular determinants that discriminate between RI and RII anchoring. Previous work using site-directed mutagenesis demonstrated that a Val-to-Trp mutation at the 13th residue of the D-AKAP2 helix abolished RII binding but not RI binding [14]. Kinderman et al. modeled this data onto the DAKAP2 crystal structure to illustrate that a bulky residue at this position would present a steric barrier to the binding of RII (Figure 1c) [12••]. The crystal structure of the uncomplexed RI D/D domain shows that there is a cavity at the corresponding position that can accommodate a larger sidechain. When combined, the information from both structures provides an ideal starting point for detailed investigation of the molecular recognition properties that drive the R-subunit-binding preferences of different AKAPs. This, as demonstrated by Gold et al., will assist in the design of PKA-anchoring inhibitors with greater affinity and PKA-subtype specificity [10••,15].
Compartmentalized cAMP signaling
A variety of imaging studies have demonstrated that cAMP levels are unevenly distributed, with dynamic pools throughout the cell [16–19,20•]. The flux of cAMP is governed by two sets of enzymes: adenyl cyclases, which are activated by Gs-proteins, synthesize cAMP from ATP; and phosphodiesterases terminate cAMP signaling by hydrolyzing it to AMP. Local cAMP gradients are at least partially generated by the tethering of PDEs to specific subcellular sites through various PDE binding proteins [17,21,22]. It is now becoming clear that in addition to anchoring PKA many AKAPs contribute to the local degradation of cAMP by colocalizing PDEs.
In Sertoli cells, PDE4D3 is targeted to the centrosome by interacting with AKAP450 [23], and in T cells PDE4A associates with several AKAPs, including AKAP95, AKAP149 and MTG16b [24]. More recently, the AKAP Gravin has been shown to target PDE4D to the plasma membrane in HEK 293 cells (Figure 2) [25]. Using a combination of RNAi and a cAMP probe based on a modified cyclic-nucleotide-gated (CNG) ion channel, these studies demonstrated that targeting of PDE4 to the plasma membrane leads to brief bursts of cAMP following stimulation by prostaglandin, whereas knockdown of Gravin by RNAi leads to sustained subplasmalemmal cAMP levels. These findings are consistent with a separate study that monitored the effects on PKA activity of a colocalized PDE [26••]. Using a fluorescent PKA activity reporter (AKAR2) that had been modified to co-anchor both PKA and a PDE [19], this study demonstrated that co-anchoring of the PDE led to brief pulses of PKA activity. In contrast, experiments using a reporter that lacked PDE anchoring showed sustained PKA activity under the same conditions. This supports the idea that, in addition to spatially restricting PKA activity, AKAP–PDE complexes also function to ensure that PKA activity is rapidly quenched by the local degradation of cAMP.
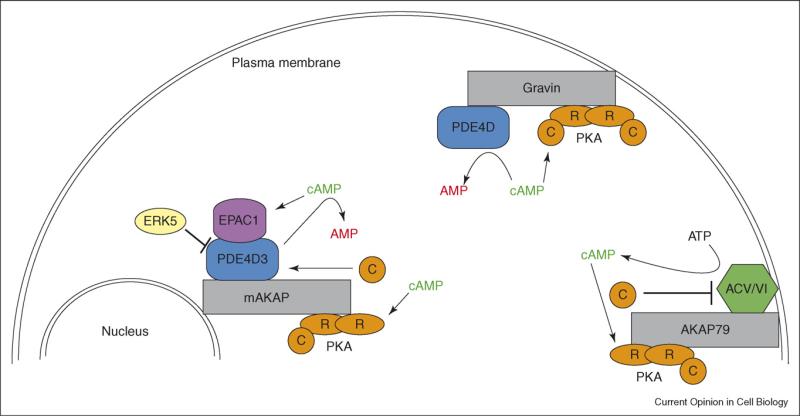
Figure 2.
Compartmentalized cAMP signaling. Schematic depicting the role of various AKAPs in regulating the synthesis and degradation of cAMP. These AKAP-mediated processes contribute to the generation of local cAMP gradients in the cell and ultimately the spatiotemporal dynamics of the cAMP effectors PKA and EPACs.
In terms of compartmentalized cAMP signaling and the integration of distinct regulatory inputs that scaffold proteins provide, the muscle-specific AKAP (mAKAP) deserves special note (Figure 2). In cardiomyocytes, mAKAPβ is targeted to the perinuclear membrane and assembles a negative feedback loop between PKA and PDE4D3 [27,28]. PKA phosphorylation of Ser13 and 54 in PDE4D3 strengthens the mAKAP/PDE interaction and enhances the PDE's catalytic efficiency [29,30]. These effects are countered by extracellular-signal-regulated kinases (ERKs). Phosphorylation of PDE4D3 on Ser579 by ERKs suppresses PDE activity [31,32]. This is potentially catalyzed by ERK5, which is also a component of the mAKAP complex [26••]. Furthermore, the mAKAP complex includes the cAMP-dependent guanine-nucleotide exchange factor Epac1 [26••]. Thus it appears that mAKAPβ functions as a cAMP signaling module that incorporates two cAMP effectors, PKA and Epac1, and integrates signals from both cAMP and ERK pathways to bi-directionally regulate cAMP signaling.
AKAPs can also shape upstream events in cAMP signaling. Both AKAP79 and Gravin are known to associate with the β2-adrenergic receptor and to contribute to its regulation by β-arrestins [33,34]. Recent studies on AKAP79, which is targeted to the plasma membrane, have shown that it interacts with both ACV and ACVI in HEK293 cells (Figure 2) [35•]. In an analogous scenario to that seen with AKAP–PDE complexes, the interaction between AKAP79 and the two ACs generates a negative feedback loop. PKA phosphorylation of ACV/VI suppresses cAMP production. Live-cell imaging experiments combining RNAi and the modified CNG-channel probe demonstrated that knockdown of AKAP79 led to sustained subplasmalemmal cAMP levels following β2-adrenergic stimulation. Control experiments in which AKAP79 levels were unaltered showed transient bursts of cAMP production at the plasma membrane. Further imaging studies using RNAi and AKAR2 showed a similar profile for PKA activity. When AKAP79 expression was knocked down, β2-adrenergic stimulation lead to prolonged PKA activity compared to control or rescue experiments. This work considered in combination with the AKAP– PDE studies illustrates the central role AKAPs play in the spatiotemporal resolution of cAMP signaling. They shape both downstream and upstream events in the cAMP pathway and provide a mechanism by which this ubiquitous second messenger can have localized effects.
Integration of signaling through multiprotein AKAP complexes
In addition to compartmentalizing cAMP signaling, AKAPs also assemble multi-protein complexes with other signaling enzymes. Inspection of the complexes listed in Table 1 illustrates two common trends. First, AKAPs often incorporate both kinases and phosphatases into a single complex, indicating that some AKAPs anchor both the positive and negative regulators of a common phosphorylation site [36]. For example, AKAP79 targets PKA and protein-phosphatase2B (PP2B) activity towards Ser845 in the AMPA-type glutamate receptor1 (GluR1) subunit to bi-directionally regulate AMPA receptor currents [37,38]. Second, multiple signaling pathways can be integrated via a single AKAP complex.
Table 1.
Selected AKAP signaling complexes.
| AKAP (alternative name) | Signaling constituents | Subcellular site |
|---|---|---|
| mAKAP (AKAP6) Gravin (AKAP 250, AKAP12) |
PKA, EPAC1, protein phosphatase2A, ERK5, PDE4D3 PKA, PKC, PDE4D, β2-adrenergic receptor |
Perinuclear membrane Actin cytoskeleton |
| AKAP79 (AKAP150, AKAP5) | PKA, PKC, PP2B, ACV/VI, β-adrenergic receptor, AMPA receptors, NMDA receptors, KCNQ2 channels, L-type voltage-gated Ca2+ channels | Plasma membrane |
| AKAP–Lbc (AKAP13) WAVE1 DAKAP1 (AKAP1) |
PKA, PKC, PKD, 14-3-3, Rho PKA, Abl, Rac, Arp2/3, WRP PKA, protein phosphatase 1, BAD PKA, protein phosphatase 1, PDE4A |
Actin cytoskeleton Actin cytoskeleton Mitochondria Mitochondria |
| AKAP220 (AKAP11) | PKA, GSK3β, protein phosphatase 1 | Vesicles |
Abbreviations not defined in the text: AMPA, α-amino-5-hydroxy-3-methyl-4-isoxazole propionic acid; BAD, BCL2-antagonist of cell death; GSK3β, glycogen-synthase kinase3β; KCNQ2, Q2 voltage-gated potassium channel; NMDA, N-methyl-D-aspartate; WRP, WAVE-associated Rac-GAP protein.
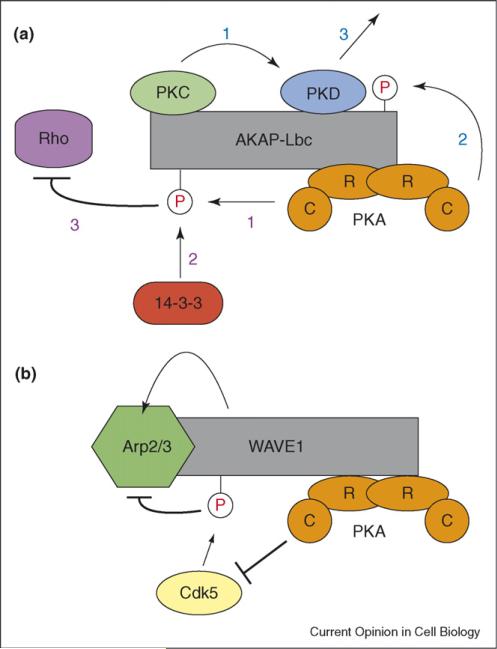
A prime example of the role AKAPs play in signal integration comes from AKAP–Lbc, which, in addition to being a guanine-exchange factor (GEF) for the small GTPase Rho, targets a multi-protein complex including PKA, protein kinase C (PKC) and protein kinase D (PKD) to the actin cytoskeleton (Figure 3a) [39,40]. AKAP–Lbc serves as an activation platform for PKD by colocalizing it near its upstream activating kinase, PKC [39]. This positions PKC so it can effectively phosphorylate PKD. PKA then promotes the release of active PKD by phosphorylating AKAP–Lbc at Ser2737 in the PKD binding domain. Anchored PKA also plays a regulatory role in Rho signaling. Phosphorylation of AKAP–Lbc at Ser1565 generates a 14-3-3 binding site [41,42•]. Binding of 14-3-3 suppresses the Rho-GEF activity of AKAP–Lbc and downregulates Rho activity. Inhibition of Rho potentially functions to suppress PKD activity, since one downstream effect of Rho activity is the stimulation of phospholipase C, an upstream activator of PKC. As a result, AKAP-Lbc not only serves as a platform for PKD activation but also mediates the integration of cAMP signaling with both the Rho and PKD pathways and potentially regulates the ability of Rho signaling to impinge on PKC activation.
Figure 3.
AKAP-mediated signal integration. (a) Schematic of the role AKAP–Lbc plays in integrating cAMP signaling with both PKD and Rho pathways. Activation of PKD (blue numbers): (1) AKAP-Lbc colocalizes PKD near PKC, its upstream activator; (2) PKC phosphorylates PKD; (3) Dissociation of active PKD from the AKAP platform is promoted by PKA phosphorylation of AKAP-Lbc. Rho inhibition (purple numbers): (1) PKA phosphorylation of AKAP-Lbc generates a 14-3-3 binding site; (2) binding of 14-3-3 downregulates the Rho-GEF activity of AKAP-Lbc and (3) inhibits Rho. (b) Schematic of the role WAVE1 plays in coordinating the bi-directional regulation of the Arp2/3 complex by Cdk5 and PKA. Cdk5 phosphorylation of WAVE1 leads to inhibition of Arp2/3. This is opposed by the activity of PKA.
More recently, the AKAP WAVE1 [43] (WASP-family verprolin homologous protein1) has been shown to integrate cAMP and cyclin-dependent kinase 5 (Cdk5) signaling to control actin dynamics and dendritic spine morphology (Figure 3b) [44••]. A key function of WAVE1 is to regulate actin-dependent morphological processes. [45] It does this by activating the actin-related protein (Arp2/3) complex. Using a combination of in vitro biochemistry and fluorescence microscopy, Kim et al. show that phosphorylation of WAVE1 by Cdk5 inhibits Arp2/3 activation and actin polymerization [44••]. Neurons cultured from WAVE1 knockout mice or in which WAVE1 had been knocked down with RNAi showed a significant decrease in mature dendritic spines. This phenotype was reversed by rescue with a dephosphorylation-mimic of WAVE1. In addition, the authors demonstrate that cAMP signaling reduces Cdk5 phosphorylation of WAVE1 and leads to an increase in dendritic spine density in a WAVE1-dependent manner. These findings indicate that WAVE1 integrates signals from cAMP and Cdk5 to bi-directionally regulate the Arp2/3 complex and actin polymerization. They also provide a molecular mechanism for the learning and memory deficiencies seen in WAVE1 knockout mice [46].
The way forward
At present, proteomic and biochemical techniques have been exhaustively used to identify the majority of AKAPs and to characterize the components of their signaling complexes. What is needed now is a better understanding of the role AKAPs play in cellular physiology. This in part entails the application of molecular and live-cell imaging techniques to the study of AKAP function. Recent efforts employing these techniques are beginning to glean details of the dynamic nature of AKAP complexes and how these state-dependent features contribute to cellular processes. Continued investigation of the role of AKAPs in PKA-mediated processes is also important. One potentially helpful advancement would be the development of methods for the identification of AKAP-dependent PKA substrates. This would provide a starting point for elucidating the downstream output of AKAP complexes. Furthermore, PKA is involved in a wide array of cellular processes, including metabolism, learning and memory, and exocytosis [47,48]. Several of these processes contribute to various disease states. PKA is implicated in pancreatic β-cell function and may provide a novel means for combating diabetes [49]. PKA is also important in the molecular mechanisms of learning and memory, making it an attractive target for therapies seeking to inhibit cognitive decline resulting from neurodegenerative diseases [50,51]. Identifying the role that AKAPs play in these processes could provide new therapeutic targets to treat diseases.
Acknowledgements
This work was supported by grant number GM42831 from the National Institutes of Health. The authors thank S. Taylor for providing the pdb coordinates of the PKA/DAKAP2 complex and M. Gold and D. Barford for assistance in the structural analysis of x-ray crystal data.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Carroll SB. Evolution at two levels: On genes and form. Plos Biol. 2005;3:1159–1166. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Ann Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 5.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 6.Tasken K, Aandahl EM. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 7.Carr DW, Stofkohahn RE, Fraser IDC, Bishop SM, Acott TS, Brennan RG, Scott JD. Interaction of the regulatory subunit (Rii) of Camp-dependent protein-kinase with Rii-anchoring proteins occurs through an amphipathic helix binding motif. J Biol Chem. 1991;266:14188–14192. [PubMed] [Google Scholar]
- 8.Newlon MG, Roy M, Morikis D, Hausken ZE, Coghlan V, Scott JD. Jennings PA: The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat Struct Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 9.Newlon MG, Roy M, Morikis D, Carr DW, Westphal R, Scott JD, Jennings PA. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. Embo J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Gold MG, Lygren B, Dokurno P, Hoshi N, McConnachie G, Tasken K, Carlson CR, Scott JD, Barford D. Molecular basis of AKAP specificity for PKA regulatory subunits. Mol Cell. 2006;24:383–395. doi: 10.1016/j.molcel.2006.09.006. [This article and [12••] were published simultaneously and describe the first x-ray crystal structures of the binding interface between PKA and an AKAP PKA-anchoring domain. The authors use this structural information to engineer a peptide inhibitor of PKA anchoring with improved PKA-subtype specificity.] [DOI] [PubMed] [Google Scholar]
- 11.Alto NM, Soderling SH, Hoshi N, Langeberg LK, Fayos R, Jennings PA, Scott JD. Bioinformatic design of A-kinase anchoring protein in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc Nat Acad Sci USA. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Kinderman FS, Kim C, von Daake S, Ma YL, Pham BQ, Spraggon G, Xuong NH, Jennings PA, Taylor SS. A dynamic mechanism for AKAP binding to RII isoforms of cAMP-dependent protein kinase. Mol Cell. 2006;24:397–408. doi: 10.1016/j.molcel.2006.09.015. [See [10••]. By modeling previous structure–function data onto the PKA/DAKAP2 crystal structure, the authors delineate possible molecular determinants that contribute to the PKA-subtype specificity of AKAPs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herberg FW, Maleszka A, Eide T, Vossebein L, Tasken K. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J Mol Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 14.Burns-Hamuro LL, Ma YL, Kammerer S, Reineke U, Self C, Cook C, Olson GL, Cantor CR, Braun A, Taylor SS. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc Nat Acad Sci USA. 2003;100:4072–4077. doi: 10.1073/pnas.2628038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlson CR, Lygren B, Berge T, Hoshi N, Wong W, Tasken K, Scott JD. Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J Biol Chem. 2006;281:21535–21545. doi: 10.1074/jbc.M603223200. [DOI] [PubMed] [Google Scholar]
- 16.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 17.Mongillo M, McSorley T, Evellin S, Sood A, Lissandron V, Terrin A, Huston E, Hannawacker A, Lohse MJ, Pozzan T, et al. Fluorescence resonance energy transfer-based analysis of cAMP dynamics in live neonatal rat cardiac myocytes reveals distinct functions of compartmentalized phosphodiesterases. Circ Research. 2004;95:67–75. doi: 10.1161/01.RES.0000134629.84732.11. [DOI] [PubMed] [Google Scholar]
- 18.Rich TC, Fagan KA, Tse TE, Schaack J, Cooper DMF, Karpen JW. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc Nat Acad Sci USA. 2001;98:13049–13054. doi: 10.1073/pnas.221381398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Ma YL, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Nat Acad Sci USA. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts β-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [This article describes the development of a second generation FRET reporter for measuring PKA activity, in which the PKA phosphorylation site in the probe has been altered to allow monitoring of both phosphorylation and dephosphorylation of the reporter.] [DOI] [PubMed] [Google Scholar]
- 21.Perry SJ, Baillie GS, Kohout TA, McPhee I, Magiera MM, Ang KL, Miller WE, McLean AJ, Conti M, Houslay MD, et al. Targeting of cyclic AMP degradation to β2-adrenergic receptors by β-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 22.Rich TC, Tsf TE, Rohan JG, Schaack J, Karpen JW. In vivo assessment of local phosphodiesterase activity using tailored cyclic nucleotide-gated channels as cAMP sensors. J Gen Physiol. 2001;118:63–77. doi: 10.1085/jgp.118.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tasken KA, Collas P, Kemmners WA, Witczak O, Conti M, Tasken K. Phosphodiesterase 4D and protein kinase A type II constitute a signaling unit in the centrosomal area. J Biol Chem. 2001;276:21999–22002. doi: 10.1074/jbc.C000911200. [DOI] [PubMed] [Google Scholar]
- 24.Asirvatham AL, Galligan SG, Schillace RV, Davey MP, Vasta V, Beavo JA, Carr DW. A-kinase anchoring proteins interact with phosphodiesterases in T lymphocyte cell lines. J Immunol. 2004;173:4806–4814. doi: 10.4049/jimmunol.173.8.4806. [DOI] [PubMed] [Google Scholar]
- 25.Willoughby D, Wong W, Schaack J, Scott JD, Cooper DMF. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. Embo J. 2006;25:2051–2061. doi: 10.1038/sj.emboj.7601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Dodge-Kafka KL, Soughayer J, Pare GC, Michel JJC, Langeberg LK, Kapiloff MS, Scott JD. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 2005;437:574–578. doi: 10.1038/nature03966. [Using a combination of biochemistry and live-cell imaging, the authors show that in cardiomyocytes mAKAP functions as a cAMP signaling module that bi-directionally regulates local cAMP concentrations and mediates the spatiotemporal activity of PKA and EPAC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapiloff MS, Schillace RV, Westphal AM, Scott JD. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J Cell Sci. 1999;112:2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- 28.Dodge KL, Khouangsathiene S, Kapiloff MS, Mouton R, Hill EV, Houslay MD, Langeberg LK, Scott JD. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. Embo J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel JJC, Dodge KL, Wong W, Mayer NC, Langeberg LK, Scott JD. PKA-phosphorylation of PDE4D3 facilitates recruitment of the mAKAP signalling complex. Biochem J. 2004;381:587–592. doi: 10.1042/BJ20040846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sette C, Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase - Involvement of serine 54 in the enzyme activation. J Biol Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 31.MacKenzie SJ, Baillie GS, McPhee I, Bolger GB, Houslay MD. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases – the involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J Biol Chem. 2000;275:16609–16617. doi: 10.1074/jbc.275.22.16609. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann R, Baillie GS, MacKenzie SJ, Yarwood SJ, Houslay MD. The MAP kinase ERK2 inhibits the cyclic AMP-specific phosphodiesterase HSPDE4D3 by phosphorylating it at Ser579. Embo J. 1999;18:893–903. doi: 10.1093/emboj/18.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser IDC, Cong M, Kim J, Rollins EN, Daaka Y, Lefkowitz RJ, Scott JD. Assembly of an A kinase-anchoring protein-β2-adrenergic receptor complex facilitates receptor phosphorylation and signaling. Curr Biol. 2000;10:409–412. doi: 10.1016/s0960-9822(00)00419-x. [DOI] [PubMed] [Google Scholar]
- 34.Malbon CC, Tao JC, Wang HY. AKAPs (A-kinase anchoring proteins) and molecules that compose their G-protein-coupled receptor signalling complexes. Biochem J. 2004;379:1–9. doi: 10.1042/BJ20031648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Bauman AL, Soughayer J, Nguyen BT, Willoughby D, Carnegie GK, Wong W, Hoshi N, Langeberg LK, Cooper DMF, Dessauer CW, et al. Dynamic regulation of cAMP synthesis through anchored PKA-Adenylyl cyclase V/VI complexes. Mol Cell. 2006;23:925–931. doi: 10.1016/j.molcel.2006.07.025. [This work represents the first identification of an interaction between an AKAP and an adenyl cyclase and illustrates how the AKAP79 complex generates a negative feedback loop to downregulate cAMP production.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauman AL, Scott JD. Kinase- and phosphatase-anchoring proteins: harnessing the dynamic duo. Nat Cell Biol. 2002;4:E203–E206. doi: 10.1038/ncb0802-e203. [DOI] [PubMed] [Google Scholar]
- 37.Colledge M, Dean RA, Scott GK, Langeberg LK, Huganir RL, Scott JD. Targeting of PKA to glutamate receptors through a MAGUK-AKAP complex. Neuron. 2000;27:107–119. doi: 10.1016/s0896-6273(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 38.Tavalin SJ, Colledge M, Hell JW, Langeberg LK, Huganir RL, Scott JD. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J Neurosci. 2002;22:3044–3051. doi: 10.1523/JNEUROSCI.22-08-03044.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carnegie GK, Smith FD, McConnachie G, Langeberg LK, Scott JD. AKAP-Lbc nucleates a protein kinase D activation scaffold. Mol Cell. 2004;15:889–899. doi: 10.1016/j.molcel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Diviani D, Soderling J, Scott JD. AKAP-Lbc anchors protein kinase A and nucleates Gα12-selective Rho-mediated stress fiber formation. J Biol Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 41.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, et al. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 42•.Diviani D, Abuin L, Cotecchia S, Pansier L. Anchoring of both PKA and 14-3-3 inhibits the Rho-GEF activity of the AKAP–Lbc signaling complex. Embo J. 2004;23:2811–2820. doi: 10.1038/sj.emboj.7600287. [These investigations identify a PKA phosphorylation site on AKAP–Lbc that generates a 14-3-3 binding site, illustrating the capacity of AKAP–Lbc to integrate cAMP and Rho signaling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. Embo J. 2000;19:4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ryu SH, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442:814–817. doi: 10.1038/nature04976. [This study provides the first evidence for the role of PKA in the WAVE1 complex and demonstrates that WAVE1 integrates the activity of Cdk5 and PKA to regulate the Arp2/3 complex and dendritic spine maturation.] [DOI] [PubMed] [Google Scholar]
- 45.Stradal TEB, Rottner K, Disanza A, Confalonieri S, Innocenti M, Scita G. Regulation of actin dynamics by WASP and WAVE family proteins. Trends Cell Biol. 2004;14:303–311. doi: 10.1016/j.tcb.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Nat Acad Sci USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmen GY, Victor SM. Signalling mechanisms regulating lipolysis. Cell signal. 2006;18:401–408. doi: 10.1016/j.cellsig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen PV, Woo NH. Regulation of hippocampal synaptic plasticity by cyclic AMP-dependent protein kinases. Prog Neurobiol. 2003;71:401–437. doi: 10.1016/j.pneurobio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Furman B, Pyne N, Flatt P, O'Harte F. Targeting β-cell cyclic 3′5′ adenosine monophosphate for the development of novel drugs for treating type 2 diabetes mellitus. J Pharm Pharmacol. 2004;56:1477–1492. doi: 10.1211/0022357044805. [DOI] [PubMed] [Google Scholar]
- 50.Arnsten AFT, Ramos BP, Birnbaum SG, Taylor JR. Protein kinase A as a therapeutic target for memory disorders: rationale and challenges. Trends Molec Medicine. 2005;11:121–128. doi: 10.1016/j.molmed.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Churcher I. Tau therapeutic strategies for the treatment of Alzheimer's disease. Curr Topics Med Chem. 2006;6:579–595. doi: 10.2174/156802606776743057. [DOI] [PubMed] [Google Scholar]