Abstract
Immunoepidemiological studies from endemic areas have revealed age-dependent resistance correlation with increased level of IgE and decreased level of IgG4 antibodies in responses to schistosomes’ soluble worm antigen. However, there have been limited studies on analyses of major antigens that provoke IgE and IgG4 immune response during chronic stage of schistosomiasis. In this study, for the first time, immunoproteomics approach has been applied to identify S. japonicum worm antigens in liquid fractions that are recognized by IgE and IgG4 antibody using plasma from chronically infected population. ProteomeLabPF 2D fractionated 1-D and 2-D fractions of SWA antigens were screened using pooled high IgE/IgG4 reactive plasma samples by dot-blot technique. In 1-D fractions, IgE isotype was detected by fewer antigenic fractions (43.2%). The most recognized isotype was IgG3 (79.5%) followed by IgG1 (75.0%) and IgG4 (61.4%). Liquid chromatography MS/MS protein sequencing of reactive 2-D fractions revealed 18 proteins that were identified, characterized and gene ontology categories determined. 2-D fractions containing proteins such as zinc finger, RanBP2-type, domain-containing protein were strongly recognized by IgE and moderately by IgG4 whereas fractions containing proteins such as ubiquitin-conjugating enzyme and cytosolic II 5'-nucleotidase strongly recognizing by IgG subclasses (IgG1, IgG3 and IgG4) but not IgE. By this study, a simple and reproducible proteomic method has been established to identify major immunoreactive S. japonicum antigens. It is anticipated that this will stimulate further research on the immunogenicity and protective potential of proteins identified as well as discovery of novel compounds that have therapeutic importance.
Keywords: Schistosoma japonicum, IgE, IgG4, Proteome, Mass Spectrometry, Genome
Introduction
The pathophysiology of schistosomiasis is mainly due to the immune response against tissue trapped eggs with consequent clinical manifestations being typical of the species infecting, intensity of worm burden as well as the immunity of the infected host. The variety of antigens released by dead worms or secreted by the worms or shed during the various developmental stages of the worm life cycle (cercariae, schistosomula, adult male and female, and eggs) provide strong sustained stimuli to the host’s humoral and T-lymphocyte-mediated immune responses [1]. In recent years, immune response regulation by the schistosomes during infections has been a topic of great concern. Particularly, the role of antibodies in resistance to reinfection. In schistosomiasis, the balance between IgE and IgG4 antibody isotypes is thought to play a role in resistance or susceptibility to infection. Immunoepidemiological studies from endemic areas have revealed age-dependent resistance correlation with specific antibody isotype responses to the schistosome antigens, particularly IgE responses to Schistosoma mansoni adult worm antigens (AWA). The IgE levels are low in children and high in adults, whereas for IgG4 the reverse has been reported [2–4]. Furthermore, since IgE and IgG4 can exhibit parallel specificity profile, it has been suggested that IgG4 subclass acts as a blocking antibody against killing of the parasites by inhibiting IgE antibody-dependent cellular cytotoxicity (ADCC) mediated by monocytes, platelets or eosinophiles. Similar effect has also been suggested for IgM and IgG2 antibodies [2, 5–8]. The IgG3 antibody level also correlated with susceptibility to and biomarkers in liver fibrosis [6]. The production of IgE is stimulated by interleukin-l3 (IL-13) and IL-4, and modulated by IL-12 and interferon-gamma (IFN-γ) while the production of IgG4 is also stimulated by IL-4 [4]. The IL-4-dependent production of IgE and IgG4 is blocked by IFN-γ, though the level required to block IL-4-dependent IgE production is much lower than that needed to block IgG4. In the sequential events of class switching, IgG4 is synthesized thereafter IgE, caused by sequential involvement of different lymphokines raising the possibility that development of protection against schistosomes would depend on population of lymphocytes producing cytokine [4, 9, 10].
In spite of many studies demonstrating importance of antibody-mediated protection against re-infection of schistosomes both in experimental and epidemiological models, many of the human schistosome vaccine research based on antibody-mediated protection have not progressed to the phase III clinical trials. This in part might be due to the limited understanding of protective anti-schistosome response against specific proteins [11]. Relatively, limited target antigens have been analyzed in the context of selective antibody isotype recognition for IgE or IgG4 especially in S. japonicum infection [2–4, 6]. Antigens that are IgE, IgG4 or both antibodies preferred can be very useful for studying mechanisms associated with antibody related resistance to schistosomiasis.
Many of the antigenic substances produced by the schistosomes at the various life cycle stages consist of proteins, glycoproteins and polysaccharides in nature [12]. So far, characterization of schistosome antigens has involved studying crude parasite extracts that had no detailed characteristics of reactive immunoglobulins. Some studies have also focused on proteins or glycoprotein components of schistosomes either directly or by cloning in bacteria systems [5, 13]. Although, elevated IgE level is important for development of resistance to reinfection in schistosomiasis, only a limited number of studies have been conducted to isolate and characterize IgE-specific antigens from S. mansoni [14] with a homologous antigen identified in S. haematobium [15] and S. japonicum [16]. Therefore, the antigenic source of variation in IgE antibody isotype-specific response to S. japonicum is limited.
The mass spectrometry (MS) based proteomics has facilitated identification of large numbers of proteins from complex biological systems. Proteomics has in recent years achieved improvements in platforms and the standard proteomics approaches rely on the second dimensional (2-D) separation of complex protein mixtures using second dimensional gel electrophoresis (2-DE) [17, 18]. In some cases, the 2-DE may be combined with difference in gel electrophoresis (DIGE) as a profiling platform and proteins are identified by ESI-MS/MS of trypsin-derived peptides. However, 2-DE has a number of shortcomings including limited loading capacity; inability to resolve proteins of extreme pIvalues; limitation in resolution of hydrophobic proteins and inability to resolve proteins of smaller molecular weights. Therefore, fractionating complex protein mixtures while maintaining intact proteins by liquid chromatography (LC) is most desirable for downstream analyses (top-down proteomics) [19].
The proteomeLabPF 2D instrument introduced by Beckman-Coulter (Beckman Coulter, Fullerton, CA, USA) features a rapid semi-automated 2-D HPLC system that uses two different methods to separate proteins; ion-exchange in the 1-D and non-porous reversed phase in the 2-D chromatography [20–22]. Unlike gel electrophoresis, it offers an added advantage that collected fractions are in liquid phase and can be utilized directly for any of various analytical procedures, such as enzymatic digests, mass spectrometer analysis, additional fractionation, western blot, or a combination of analytical tests. Additionally, it has been shown to be suitable for high-throughput large-scale analysis of intact proteins [23–26] and high loading capacity (up to 5 mg) than with gel electrophoresis, thus significantly increasing the sensitivity of protein identification. Liquid-based fractionation and separation systems offer great flexibility and can be suitable for large-scale proteomic profiling in a quantitative analysis [25, 26].
This study focused on isolating, identifying, and characterizing immunogenic S. japonicum proteins that are preferentially detected by IgE and IgG4 antibodies using serological proteomics approach. Identifying and characterizing antigenic proteins detected by the isotypes studied would contribute to understanding of schistosome-specific adaptive immunity. This also, highlights the importance of vaccine research focusing on induction of protective isotype-specific antibody response to specific peptides as a single protein from the parasite might possess undetermined antigenic determinants capable of stimulating various antibody productions.
Materials and Methods
Soluble Worm Antigen Preparation
Soluble worm antigen (SWA) extract was prepared from frozen Chinese strain S. japonicum adult worms following the procedures previously described [27] with slight modifications. Briefly, adult worms (600 mg) were homogenized in 3.25 ml cold Diethyl Ether (Wako Pure Chemical Industries, Ltd. Osaka, Japan). The homogenate was centrifuged at 2,000 g, 5 min to remove lipids together with the diethyl ether. Thereafter, the pellet was freeze-thawed several times in 3.5 ml of lysis buffer (6 M Urea, 2 M Thiourea, 10% Glycerol, 50 mM Tris-HCl, pH 7.8, 2% n-OG, 5 mM TCEP) mixed with 0.1 mM PMSF and 2 µg/ml Leupeptin. This was dialyzed in PBS (pH 7.5) containing 8 M Urea at 4°C with stirring. The homogenate was centrifuged at 20,000 g for 1 hr at 4°C and then filtered through 0.22 µm filter (Millex GP Filter Unit, Millipore Ireland Ltd. Tullagreen, Carrigtwohill Co Cork, Ireland). Protein concentration was determined by BCA Protein Assay Kit (Bio-Rad Laboratories Inc., Tokyo, Japan) and stored at –80°C until used.
Measurement of Anti-worm Antibody Levels
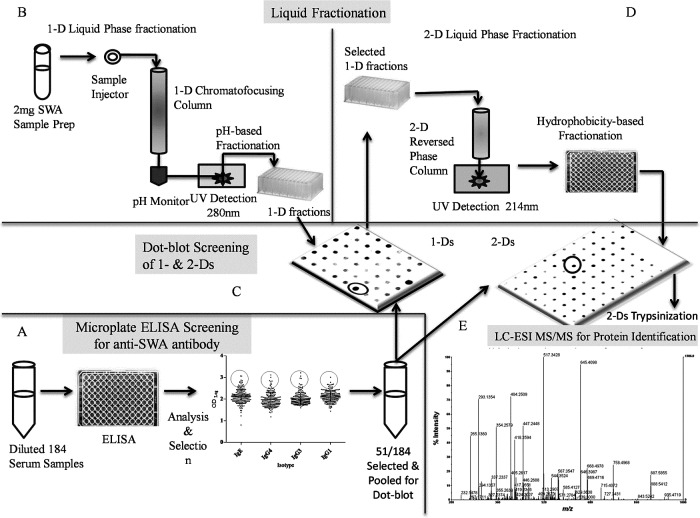
An ELISA was carried out using SWA to screen plasma samples obtained from individuals with liver fibrosis (n = 31 grade 0; n = 62 grade 1; n = 91 grade 2 and 3 individuals) due to schistosomiasis japonica as previously described [28, 29]. The project proposal including the reuse of the stored samples was processed to the Institutional Review Board at NEKKEN and was approved (No. 12081793). Five plasma samples originally confirmed by microscopy and ultrasound were included as positive controls. Three plasma samples were also included as negative controls which were obtained from healthy Japanese individuals without schistosomiasis history. Briefly, plates (Nunc-Immuno Plate, Nunc, Denmark) were coated with 5 µg/ml of SWA. After washing unbound antigens two times (2×) with PBS containing 0.05% Tween 20 (PBST, pH 7.4), the plates were blocked with 5% non-fat skimmed milk in PBST for 60 min at room temperature (RT) followed by 2× washing. Plasma samples were diluted 1:20 for detection of IgE and IgG4 and 1:800 for detection of IgG1 and IgG3 with 1% blocking solution at followed by incubation at 37°C for 60 min and then 3× washing. The procedure continued with 60 min incubation (37°C) with horseradish peroxidase-conjugated mouse anti-human IgG1, IgG3 (Southern Biotechnology Associates Inc., Birmingham, AL, USA), IgG4 (MP Biomedicals. LLc, France) or biotin-conjugated goat anti-human IgE (Invitrogen Corporation, Camarillo, CA, USA) in 1% blocking solution at 1:1000, 1:1000 1:400 or 1:400 respectively. For detection of IgE, the plates were further treated with 1:400 horseradish peroxidase-conjugated streptavidin (DakoCytomation, Copenhagen, Denmark). Finally, plates were developed with stabilized chromogen (SB01, Invitrogen) in the dark followed by addition of stop solution (1N H2SO4, WAKO). The OD was measured at 450 nm (iMark Microplate Absorbance Reader, Bio-Rad laboratories, Inc. Japan). The mean ODs obtained were Log-transformed after subtracting the mean ODs of the negatives and samples within the upper quartile (95 percentile) (51/184) were pooled for dot-blot reactivity against 1-D and 2-D fractionated SWA (Fig. 1A).
Fig. 1.
Workflow of the experiment. In order to obtain anti-worm antibody, plasma from chronically S. japonicum infected individuals were screened in ELISA system (A) and highly reactive samples pooled for dot-blot screening. Soluble worm antigen (SWA) was fractionated by Chromatofocusing (B) using ProteomeLab PF 2D (Beckman Coulter, Fullerton, CA, USA) followed by dot-blot screening (C). Reactive fractions were further fractionated by reversed phase chromatography which was again screened by dot-blot and reactive fractions trypsinized for ESI-MS/MS protein identification (E).
Buffer Exchanging and Chromatofocusing
The PD-10 desalting column containing 8.3 ml of Sephadex G-25 medium (85 to 260 µm particle size), (GE Healthcare Bio-Sciences K. K. Tokyo, Japan) was applied in buffer exchange of SWA before chromatofocusing following the manufacturer’s recommendation. Briefly, the PD-10 column was equilibrated with proprietary buffer, “ProteoSep Start” buffer (Eprogen, Darien, IL, USA) by allowing it to enter the packed bed completely. The flow-through was discarded. This was repeated with a total of 25 ml “ProteoSep Start” buffer. The 2 mg of SWA was resuspended in 1.25 ml of “ProteoSep Start” buffer, loaded onto the equilibrated PD-10 column and allowed to enter the column completely. The flow-through was again discarded. Elution was performed with 3.5 ml “ProteoSep Start” buffer added onto the column and the eluent collected into a new 15 ml tube under gravity and applied in 1-D Chromatofusing.
Chromatofocusing was performed using the ProteomeLab PF 2D protein separation system (Fig. 1B) with 32 Karat user interface software. The pH gradient was formed using two proprietary buffers: “ProteoSep Start” buffer ([essentially contained urea, Tris-HCl and n-OG at pH of 8.5) and “ProteoSep Elution” buffer (Eprogen, Darien, IL, USA) (essentially Urea, Polybuffer 74-HCl, n-OG and iminodiacetic acid, pH 4.0). The High Performance Chromatofocusing Column (A51685 ProteoSep HPCF Column, 250 mm × 2 mm, Eprogen. Darien, IL, USA) was treated according to the manufacturer’s instructions. Briefly, the column was washed with 10 volumes of autoclaved MilliQ water at a flow rate of 0.2 ml/min for 45 min and then equilibrated with 30 volumes of “ProteoSep Start” buffer for 130 min at 0.2 ml/min, ambient temperature. The buffer exchanged SWA sample was introduced with a manual injector into the column. Proteins bound to the strong anion exchanger in the HPCF column were eluted with a continuous decreasing pH from 8.5 to 4.0. Twenty minutes after sample injection, the valve automatically switched from “ProteoSep Start” buffer to “ProteoSep Elution” buffer at a flow rate of 0.2 ml/min over 95 min. The pH began to decrease after about 45 min. Fractions were automatically collected every 0.3 pH units into a 96-well deep-plate (Part No. 26700J, Beckman Coulter, Fullerton, CA, USA). At 170 min, the HPCF column was washed for 45 min with 10 column volumes of a third buffer of high ionic strength solution (1 M NaCl) and re-stored by 10 column volumes of distilled water for 45 min. The absorbance of the column effluent was monitored at 280 nm with an online pH flow cell. The percentage concentration eluted over the different pH conditions was estimated using the peak area of the fraction monitored at 280 nm, a wavelength at which the peak area is directly proportional to the quantity of the proteins [30]. The 1-D fractions obtained were screened by dot-blot assay and selected reactive fractions directly applied to the 2-D reversed phase unit.
Second Dimension Reversed Phase Chromatography
The 2-D separation (Fig. 1D) was performed using Reversed Phase High Performance Column (391106 PF 2D HPRP Column, Beckman Coulter, Fullerton, CA, USA) and two solvents, 0.1% TFA in water (Solvent A) and 0.08% TFA in ACN (Solvent B). At the end of each run, equilibration of the column was achieved with initial mobile phase (Solvent A) for 10 min followed by Solvent B for 5 min prior to each injection. All 2-D chromatography was conducted at a column temperature of 50°C and buffer flow rate of 0.75 ml/min with the absorption of the effluent monitored at 214 nm. From the selected 1-D fractions, 200 µl was automatically injected into the PF 2D HPRP column and ran for 6 min. The column was eluted at a flow rate of 0.75 ml/min with a 0–100% linear gradient of solvent A and solvent B for 35 min. Thereafter, Solvent B was continued for 5 min, followed by re-equilibration with 100% Solvent A for 10 min. The fractions were collected at a flow rate of 0.18 min into 96-well microplate (Product code 3363, Corning International K. K. Tokyo, Japan) placed in an automated fraction collector (Gilson FC 204 Fraction Collector, M & S Instruments Inc. Osaka, Japan). The 2-D fractions were stored at –80°C while some screened by dot-blot and some.
Dot-blot Screening
The 44 1-D fractions as well as 80 2-D fractions (derived from each 1-D fraction) obtained were screened for reactivity against circulating anti-schistosome IgE, IgG4, Ig3 and IgG1 antibody isotypes in dot-blot assays in search of novel reactive proteins. The dot-blot was conducted using Bio-Dot SF Micro filtration apparatus (Bio Rad Laboratories, Inc., CA, USA) as previously described with modifications [27]. Briefly, 30 µl 1-D fraction was loaded onto polyvinylidene fluoride (PVDF) membrane (Amersham Hybond-P PVDF Membrane, GE Healthcare Bio-Sciences K. K. Tokyo, Japan) imbedded in transfer buffer (192 mM Glycine, 25 mM Tris, pH 7.4) [31] and fixed into the Bio-Dot SF Micro filtration apparatus. Following blocking, 30 µl of diluted pooled plasma (51 samples) (IgG1, 1:4000; IgG3, 1:4000; IgG4, 1:100; or IgE, 1:800) in TBS washing buffer (20 mM Tris, 137 mM NaC1, 0.01% Tween 20, pH 7.6) was applied to each respective well blotted with the fractions. Bound antibodies were incubated with respective conjugated enzymes using horseradish peroxidase-conjugated mouse anti-human IgG1, IgG3 (Southern Biotechnology Associates Inc.), IgG4 (MP Biomedicals, LLc) or biotin-conjugated goat anti-human IgE (Invitrogen) in dilutions of 1:16,000, 1:16,000 1:1,000 or 1:4,000 respectively. The IgE antibody bound membrane was further treated with 1:6,000 horseradish peroxidase-conjugated streptavidin (DakoCytomation). Blocking (5% skimmed milk/TBS washing buffer) and conjugate reaction of the membranes were conducted in a separate container. The reactivity was revealed by ImmunoStar Reagents (WAKO Pure Chemicals Industries, Ltd. Osaka, Japan) for chemiluminescence detection following the manufacturer’s protocol. Digital images were obtained by the Las-4000EPUV Mini with an interface Las-4000 Image Reader (Fujifilm Corporation. Tokyo, Japan). The acquired antigenic spots were further transformed into pixels units for quantification of the recognition intensity using ATTO Lane and Spot Analyzer 6.0 software (ATTO Corporation. Tokyo, Japan). In screening of 2-D fractions by dot-blot assay, similar steps were followed. Briefly, 50 µl of each fraction in transfer buffer after drying to remove most of the ACN and TFA to prevent interference of membrane blotting was loaded onto the PVDF membrane. Test plasma were diluted for IgG1, 1:25,000; IgG3, 1:15,000; IgG4, 1:400; and IgE, 1:1,000 and applied to each respect 80 wells blotted with the 2-D fractions. Bound antibody was again incubated with conjugated enzymes using horseradish peroxidase-conjugated mouse anti-human IgG1, IgG3, IgG4 and biotin-conjugated goat anti-human IgE accordingly, in dilutions of 1:30,000, 1:30,000 1:3,000 or 1:6,000 respectively. Two crude SWA and BSA (Sigma A4503, Sigma-Aldrich Co. MO, USA) spots were included in the entire test for positive and background control respectively. The IgG1 and IgG3 were included to aid in selection of IgG4 and IgE preferred fractions.
For the analysis of all the antigenic spots intensity, the background intensity was subtracted and then the value obtained divided by that of the positive control to generate a relative reactivity index of ‘1’ for positive control, ‘0’ for negative control. Using these criteria each antigenic spot was designated ‘not-reactive’, ‘weakly reactive’, ‘moderately reactive’ or ‘strongly reactive’ with respect to isotype recognition using the pixel analysis and manual verification. These were calculated for each isotype separately so that the spot intensity was standardized within the membrane. The intensity scoring rather than absolute values could be compared among the fractions. This approached was applied since it was not expected for any fraction to have equal intensity by the four isotypes for determining reactivity intensity across the antibody isotypes.
In-solution Tryptic Digestion
Fractions from basic, neutral and acidic regions were treated with trypsin prior to protein sequencing. Briefly, liquid fractions (50 to 150 µl) were precipitated at –80°C overnight in about 10-bed volume of pre-chilled acetone (WAKO) followed by centrifugation at 20,000 g for 30 min at 4°C. After removing most of the supernatant, the samples were speed-vacuumed to eliminate the remaining ACN and TFA together with the acetone. Then 15 µL of denaturation solution (8 M urea; 500 mM Tris-HCl, pH 8.5; 2.5 mM EDTA) was added and incubated for 10 min at 100°C followed by cooling at RT. Addition of 5 µl of reduction solution (40 mM DTT), (WAKO) in 25 mM NH4HCO3 (WAKO), incubated 1 hr at 56°C with shaking followed. Alkylation reaction performed with 5 µL of 250 mM iodoacetamide (Tokyo Chemical Industry Co, Ltd, Tokyo, Japan) in 25 mM NH4HCO3 and incubated in the dark for 45 min at 25°C with shaking. A volume of 180 µL 50 mM NH4HCO3 was added to dilute out the urea and to terminate all the reactions prior to trypsin proteolytic digestion. Trypsin proteolysis was conducted using 50 µl (10 µg/ml) sequence grade modified trypsin (Promega Corporation, Madison, WI, USA) in 50 mM NH4HCO3 overnight at 37°C followed by addition of 5 µl 5% LC-MS grade Formic Acid (WAKO) in LC-MS grade ultra pure water (WAKO) to terminate the reaction. The trypsinized peptides were again speed-vacuumed and resuspended in 25 µl of 0.3% formic acid before filtered in Spin-X centrifuge tubes (0.22 µm Nylon, Costar Corning Inc., Corning, NY, USA) which was span at 2,300 g for 30 sec. The filtrate was transferred into MS vials (0.3 ml, TPX Snap Vial, GL Sciences Inc, Tokyo, Japan) for loading onto the ESI-MS/MS system.
Mass Spectrometric Analysis and Database Search
The MS and tandem-MS (MS/MS) spectra of tryptic peptides (Fig. 1E) were obtained using the NanoFrontier nLC and NanoFrontier eLD Liquid Chromatography Mass Spectrometer (Hitachi High-technologies, Tokyo, Japan). The nanoLiquid Chromatography/ ElectroSpray Ionization/ Linear Ion Trap/ Time of Flight (nLC-ESI/LIT/TOF) and collision induced dissociation (CID) modes were used for MS detection and peptide fragmentation as described [32]. In the NanoFrontier nLC, the trypsinized peptides (1–10 µL) suspended in 0.3% formic acid were trapped on monolith trap column [C18-50-150 column, (0.05 mm I.D. × 150 mm L). Hitachi High-technologies] and separated on a packed nano-capillary column [NTCC-360/75-3-123, (0.075 mm I.D. × 100 mm L, particle diameter 3 µm), Nikkyo Technos Co., Ltd, Tokyo, Japan] at a flow rate of 200 nL/min. The peptides in the column were eluted using a stepwise ACN gradient (mobile phase A: 2% ACN, 0.1% formic acid; mobile phase B: 98% ACN, 0.1% formic acid, The A:B concentration gradient was 0.0 min: (A:B = 100:0%) → 60 min (0:100%). In the NanoFrontier eLD spectrometer, the eluted peptides were ionized with a capillary voltage of 1700 V and detected in a detector potential TOF range of 2050–2150 V.
Acquired MS and MS/MS spectra were converted into Mascot generic format (mgf) using a Data Processing software 2008 (Hitachi High-technologies) and subsequently searched in a locally established database for S. japonicum sequences downloaded from Chinese National Human Genome Centre at Shanghai (http://www.chgc.sh.cn/japonicum/Resources.html) (containing 12,657 predicted proteins) [33] using the MS/MS Ion Search provided by MASCOT Sequence Query sever version 2.3 (http://www.matrixscience.com). With the MASCOT search, the following search parameters were used, enzyme: trypsin, variable modifications: carbamidomethylation, carboxymethyl (C) and oxidation (M), mass values: monoisotopic, protein mass: unrestricted, peptide mass tolerance: ± 0.5 Da, fragment mass tolerance: ± 0.2 Da (CID data), maximum missed cleavages: 1 and Instrument type: ESI-TRAP. For MASCOT output, significant peptides were determined by the peptides score from the probability-based molecular weight search (MOWSE) which identifies proteins from the molecular weight of peptides created by the proteolytic digestion [34]. Peptide score > 27 indicate identity or extensive homology (p<0.05). Further stringency was added by eliminating any single peptide that could be assigned to more than one protein. To ensure a non-redundancy, the protein identifications were examined manually in the database for possible redundancies including multiple names and homologies.
Characterization of Identified Proteins
Identified S. japonicum proteins were characterized according to antigenic propensity, hydrophobicity, antigenic determinant, domains and gene ontology (GO) types with respect to their antibody isotype recognition. An estimate of hydrophobicity was determined from the grand average of hydropathicity index (GRAVY) [35] using ProtParam from ExPASy [36]. The GRAVY index is the average hydropathy score for all the amino acids in the protein. The positive GRAVY index indicates hydrophobic protein and negative, hydrophilic protein. The immune peptides or antigenic determinants and antigenic propensity were also estimated [37]. With reference to conserved domain sequences (CDS) that were similar to known genes and domains, the proteins were further characterized into GO terms and annotations (biological processes, molecular functions and cellular components) using UniProt-GOA server (ver. 106) (http://www.ebi.ac.uk/QuickGO/).
Statistical Analysis
The data generated were analyzed by Microsoft Excel and GraphPad Prism ver. 5. All the data were expressed as the mean ± SD. Differences between groups were analyzed for statistical significance by one-way analysis of variance and the t-test (Unpaired t test) using the GraphPad Prism. A p-value of <0.05 was considered as statistically significant.
Results
Fractions and Dot-blot Screening
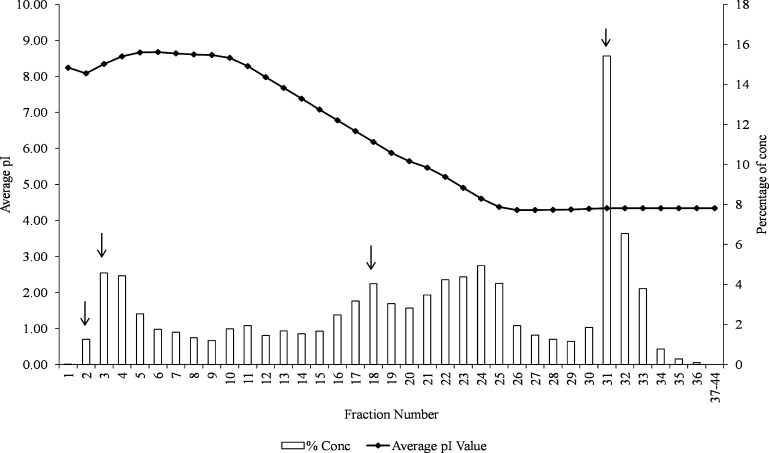
Protein fractionation by chromatofocusing is used to enrich proteins with similar isoelectric point (IEP or pI) and collected in one fraction [38]. Liquid fractionation of crude SWA into 1- and 2-D was achieved using the ProeteomLab PF 2D instrument. In the chromatofocusing, SWA proteins were separated and eluted into 44 fractions. Fig. 3 shows the fraction number against the elution profile (average pI) and percentage concentrations while Fig. 3, 2-D elution profiles of fractions 2, 3, 18 and 31. As expected, the basic proteins started to elute (fractions 1 to 17). The elution continued with the neutral proteins (18 to 20) followed by the weak and strong acidic proteins (21 to 44). Most of the proteins were eluted in fraction 31 (15.42%) with mainly acidic proteins. Fractions 37 to 44 had undetectable level of percentage concentration. The pIs of almost all the proteins identified in the higher pH gradient were consistent with the expected pI (Table 1). However, lower pH gradient showed a weak correlation to the expected pI range even though proteins with the expected pI were also found in these fractions (F31-H7). Such unexpected phenomenon is possible because the last few fractions can be enriched with proteins that carry post-translational modifications (PTM) such as phosphorylation which can cause a shift in their pI. Usually, this phenomenon is observed in 2-D electrophoresis and is useful for identifying proteins with different modification condition [38].
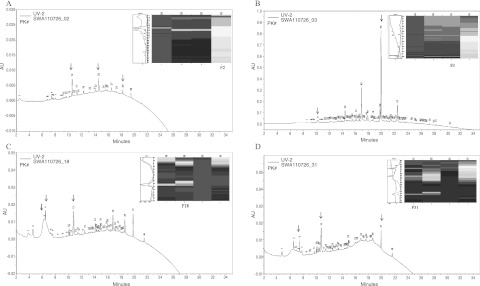
Fig. 3.
Second dimensional elution profile of 1-D fractions 2 (A), 3 (B), 18 (C) and 31 (D). 1-D fractions were run in the 2-D reversed phase chromatography using the ProteomeLab PF 2D. As shown is the elution profile with respect to time and absorbance units (AU). The elution gradient was achieved using two solvents, 0.1% TFA in water (A) and 0.08% TFA in ACN (B). The 2-D fractionation was run at column temperature of 50°C and a buffer flow rate of 0.75 ml/min with the absorption monitored at 214 nm. The column was eluted with a 0–100% linear gradient of solvent A and B for 35 min. Insert, 2-D UV difference maps obtained by ProteoVue of the fractions and the pH range; 31 (pH 4.29–4.34), 18 (pH 6.03–6.33), 3 (pH 8.22–8.45) and 2 (pH 8.45–8.47). Arrows indicate fractions from which proteins were sequenced. F2 P# = 14, 23, 29. F3 P# = 14, 23, 29. F18 P# = 3/4, 13. F31 P# = 6, 16, 47. P#, peak numbers.
Fig. 2.
Elution profile and percentage concentration of SWA into various fractions during chromatofocusing. Chromatofocusing was performed using the ProteomeLab PF 2D protein separation system. The pH gradient was formed using two proprietary buffers: “ProteoSep Start” buffer at a pH 8.5 and “ProteoSep Elution” buffer at a pH 4.0. As shown, the SWA was well fractionated with fraction 31 containing most of the void proteins. Proteins identified were obtained from fractions numbers indicated by arrows (2, 3, 18 and 3).
Table 1.
Fractions and identified proteins by ESI MS/MS. S. japonicum adult worms extract was liquid fractionated using ProteomeLab PF 2D system into 1-D by chromatofocusing and reversed phase chromatography followed which were screened by dot-blot. Immunoreactive 2-D fractions were subjected to ESI MS/MS and peptide identification using MASCOT server with a locally established S. japonicum protein sequences downloaded from S. japonicum database at http://www.chgc.sh.cn/japonicum/Resources.html. MP, MASCOT peptides score. *Total MASCOT Peptide Score.
| Fraction Well | Protein ID | Match to | Unique peptide | m/z | z | error (Da) | Peptide | MP Score | pI/Mw |
|---|---|---|---|---|---|---|---|---|---|
| F2-C3 | Sjc_0203170 | Chromatin licensing and DNA replication factor 1 | 1 | 530.81 | 2 | 0.0092 | DVIDLVKMK | 56 | 9.74/64520 |
| F2-C5 | Sjc_0213700 | Zinc finger, RanBP2-type,domain-containing | 1 | 450.29 | 2 | 0.0517 | INLSSLPR | 27 | 9.34/57532 |
| F2-C6 | Sjc_0213700 | Zinc finger, RanBP2-type,domain-containing | 1 | 450.29 | 2 | 0.0503 | INLSSLPR | 30 | 9.34/57532 |
| F3-E1 | Sjc_0034740 | Meiosis-specific, nuclear structural protein 1 | 1 | 689.85 | 2 | −0.0309 | RELEAINAYTAK | 40 | 6.79/45874 |
| Sjc_0203170 | Chromatin licensing and DNA replication factor 1 | 1 | 530.8 | 2 | −0.008 | DVIDLVKMK | 36 | 9.74/64520 | |
| Sjc_0058190 | NACHT and WD repeat domain-containing protein 1 | 1 | 590.33 | 2 | 0.0613 | MCEQLLKTR | 34 | 5.28/228171 | |
| Sjc_0037420 | Triple functional domain protein | 1 | 606.37 | 2 | 0.0134 | QFLAK | 30 | 6.13/173854 | |
| Sjc_0083690 | RNA Helicase | 1 | 606.37 | 2 | 0.0465 | MQLAK | 30 | 9.07/86337 | |
| Sjc_0040110 | Protein kinase | 1 | 699.86 | 2 | 0.0254 | ENFVLYDEIEK | 29 | 8.86/209937 | |
| Sjc_0302250 | BRO1 domain-containing protein BROX | 1 | 564.33 | 2 | 0.0046 | EKAGQAIAALR | 31 | 8.28/47921 | |
| Sjc_0111110 | Cell division control protein CDC7 | 2 | 464.79 | 2 | 0.1187 | LLEPCPEK | 32* | 9.5/36413 | |
| 716.92 | 2 | 0.0794 | TTDLNISENNRR | ||||||
| Sjc_0063180 | Centriolin | 3 | 572.88 | 2 | 0.1375 | GELEQIKAEK | 51* | 7.04/278991 | |
| 723.43 | 2 | 0.0615 | KISDQSELKLER | ||||||
| 730.45 | 2 | 0.2049 | RDYSLMRSCVR | ||||||
| Sjc_0046400 | Conductance calcium-activated potassium channel protein 2 | 2 | 481.32 | 2 | 0.1387 | FISLCNHK | 31* | 7.09/169207 | |
| 899.53 | 2 | 0.1672 | NLVTSVMGVLSDYMPR | ||||||
| Sjc_0009730 | CREB-binding protein | 2 | 679.52 | 2 | 0.153 | MILMR | 32* | 8.00/113822 | |
| 414.23 | 2 | 0.0905 | YTVCER | ||||||
| Sjc_0106490 | PDZ and LIM domain protein 3 | 3 | 564.31 | 2 | 0.0968 | VPMHPECLK | 30* | 9.08/35351 | |
| 506.97 | 2 | 0.2293 | VPMHPECLKCCK | ||||||
| 534.22 | 2 | 0.0493 | CCKCGIGLR | ||||||
| Sjc_0054640 | Prolyl-tRNA synthetase | 3 | 488.93 | 2 | 0.0718 | KGTQQGLRCCVR | 31* | 7.5/128901 | |
| 699.86 | 2 | 0.0117 | CKVEPHVRTGSK | ||||||
| 621.88 | 2 | 0.0013 | LALQNTVLSKR | ||||||
| F3-H6 | Sjc_0213700 | Zinc finger, RanBP2-type,domain-containing | 1 | 450.29 | 2 | 0.0415 | INLSSLPR | 30 | 9.34/57532 |
| F18-B1 | Sjc_0045150 | E3 ubiquitin-protein ligase HUWE1 | 1 | 466.78 | 2 | 0.0381 | RWTNLSR | 29 | 5.25/301151 |
| Sjp_0042440 | 5'-nucleotidase, cytosolic II | 1 | 384.8 | 2 | 0.1307 | VTSVHLL | 27 | 7.17/55038 | |
| F18-C3 | Sjc_0203170 | Chromatin licensing and DNA replication factor 1 | 1 | 530.83 | 2 | 0.0376 | DVIDLVKMK | 40 | 9.74/64520 |
| Sjc_0042100 | Ubiquitin-conjugating enzyme E2 N | 2 | 487.32 | 2 | 0.0935 | QNEAEALAK | 28* | 5.42/19728 | |
| 601.4 | 2 | 0.1123 | LGRICLDILK | ||||||
| F31-C3 | Sjc_0203170 | Chromatin licensing and DNA replication factor 1 | 1 | 530.8 | 2 | 0.0104 | DVIDLVKMK | 36 | 9.74/64520 |
| F31-E1 | Sjc_0001260 | Putative Serine/threonine-protein kinase C05D10.2 | 2 | 683.38 | 2 | 0.0558 | LCDFGLARSLK | 34* | 9.18/120799 |
| 590.36 | 2 | 0.1803 | CQNGNKINCK | ||||||
| F31-H7 | Sjc_0203170 | Chromatin licensing and DNA replication factor 1 | 1 | 530.81 | 2 | 0.0056 | DVIDLVKMK | 36 | 9.74/64520 |
| Sjc_0058190 | NACHT and WD repeat domain-containing protein 1 | 1 | 590.33 | 2 | 0.0763 | MCEQLLKTR | 31 | 5.28/228171 |
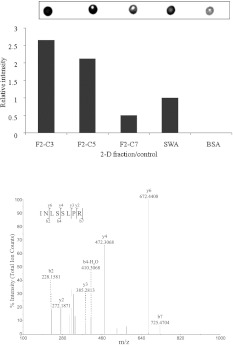
Four human circulating antibody isotypes (IgE, IgG1, IgG3 and IgG4) recognition by the 1-D fractions were evaluated using dot-blot ELISA. The reactivity intensity of each fraction was transformed into pixel unit and then expressed as relative reactivity intensity. The IgE was bound by 19 fractions (43.18%) with mean intensity of 0.22 ± 0.18 while IgG1 had 33 (75.00%) with mean intensity, 0.24 ± 0.23. For IgG3 and IgG4 there were 35 (79.55%) and 27 (61.36%) fractions detected with mean intensity of 0.28 ± 0.26 and 0.36 ± 0.30, respectively. The IgE isotype was detected by fewer fractions as compared to the remaining isotypes. The most detected isotype was IgG3 followed by IgG1 and IgG4 accordingly. It was observed that fractions 2 to 6 reacted with all the isotypes with varying intensity (Fig. 3). Likewise, the 2-D fractions (2, 3, 18 and 31) were further screened by dot-blot analysis to identify wells containing proteins that were IgE or IgG4 preferred (Fig. 3). The positive dot-blot fractions of such 2-D wells were selected from basic fractions (F2-C5, F2-C7, F2-C3; F3-E1, F3-H7), neutral fractions (F18-B1, F18-C3) and acidic fractions (F31-C3, F31-E1, F31-H7) which were processed for LC-MS/MS. Fig. 4A shows a representative 2-D dot-blot reactivity.
Fig. 4.
Representative 2-D fraction dot-blot reactivity and tandem mass spectrum. Represented by A, IgE reactivity intensity of F2-C3, F2-C5 and F2-C7 with crude SWA and BSA controls. B, a tryptic INLSSLPR peptide of Zinc finger, RanBP2-type, domain-containing protein, Sjc_0213700 (GenBank: CAX74641.1). The precursor ion was m/z 450.30(2+). Sjc_0213700 was sequenced from F2-C5 and F2-C7 with strong preference for IgE.
ESI-MS/MS Analysis
A total of 25 non-redundant proteins were identified by two-dimensional fractionation through ESI MS/MS analysis of 10 fractions (Table 1). A representative tandem mass spectrum is presented in Fig. 4B. Proteins identified with peptides obtained from the MS/MS mostly had a single confirmed peptide matches with a minimum Mascot score of 27. The 2-D wells (F2-C3, F2-C5 and F2-C7) originating from 1-D F2; 1-D F3 (F3-E1 and F3-H7); 1-D F18 (F18-B1 and F18-C3) and 1-D F31 (F31-C3, F31-E1 and F31-H7), yielded 3, 14, 4, 4 proteins respectively. Two proteins (chromatin licensing and DNA replication factor 1 and zinc finger, RanBP2-type, domain-containing) were sequenced from two sequential fractions (F2 and F3) and at the same time one of them (chromatin licensing and DNA replication factor 1), in other two non-sequential fractions (F18 and F31). Proteins with a wide range of molecular weight were identified from less than 20 kDa to more than 300 kDa (Table 1). The pI and the molecular weight profiles therefore indicate the benefit of protein fractionation which is an important aspect of protein profiling in identifying proteins with different biochemical properties. The majority of the S. japonicum proteins were recognized by IgE, IgG3 and IgG4 but not IgG1.
Characterization
It is worth noting that all the proteins identified have a GRAVY index within negative range (Table 2). Meaning the proteins are hydrophilic in consistent with 2-D reversed phase elution profile (Fig. 3). Meiosis-specific nuclear structural protein 1 (MNS1) was found to be the most hydrophilic (–1.306) and BRO1 domain-containing protein BROX, less hydrophilic (–0.177) protein. With respect to the number of possible epitope each protein have, ubiquitin-conjugating enzyme E2 N had 8 antigenic determinants, being the least yet with average antigenic propensity of 1.0386.
Table 2.
Biochemical properties and immunoreactivity pattern of the proteins. Dot-blot assay was performed for each second dimension fraction containing the proteins identified. The reactivity was quantified into pixels unit and graded according to reactivity intensity with respect to that of the crude parasite antigen to obtain relative reactive intensity. Using the relative reactive intensity, each reactive spot was scored as ‘weak reactivity’ (−), ‘moderate reactivity’ (±) or ‘strong reactivity’ (+). In addition to reactive pattern, proteins were characterized using GRAVY index (ProtParam tool), number of antigenic determinant contained in the full amino acid length and the antigenic propensity where >1 indicates high antigenic character. In this Table antibody reactivity pattern is not repeated for proteins from the same fraction well.
| Fraction Well | Protein identified | GRAVY index | Amino acids | Antigenic determinants | Antigenic propensity | Antibody reactivity pattern | |||
|---|---|---|---|---|---|---|---|---|---|
| IgE | IgG1 | IgG3 | IgG4 | ||||||
| F2-C3 | Chromatin licensing/DNA replication factor 1 | −0.348 | 578 | 20 | 1.0414 | + | − | − | + |
| F2-C5 | Zincfinger, RanBP2-type,domain-containing | −0.986 | 513 | 21 | 1.0108 | + | − | − | ± |
| F2-C7 | Zinc finger, RanBP2-type,domain-containing | −0.986 | 513 | 21 | 1.0108 | ± | − | − | ± |
| F3-E1 | Meiosis-specific, nuclear structural protein 1 | −1.306 | 376 | 11 | 0.9936 | + | ± | + | + |
| Chromatin licensing/DNA replication factor 1 | −0.348 | 578 | 20 | 1.0414 | + | ± | + | + | |
| NACHT/WD repeat domain-containing protein 1 | −0.282 | 2005 | 81 | 1.0271 | |||||
| Triple functional domain protein | −0.414 | 1554 | 68 | 1.0336 | |||||
| RNA Helicase [EC:3.6.1.-] | −0.409 | 762 | 25 | 1.0308 | |||||
| Protein kinase [EC:2.7.1.-] | −0.22 | 1921 | 63 | 1.0397 | |||||
| BRO1 domain-containing protein BROX | −0.177 | 426 | 16 | 1.0459 | |||||
| Cell division control protein CDC7 | −0.333 | 330 | 13 | 1.0363 | |||||
| Centriolin | −0.847 | 2444 | 92 | 1.0139 | |||||
| Small conductance calcium-activated potassium channel protein 2, putative | −0.393 | 1536 | 58 | 1.0294 | |||||
| CREB-binding protein | −0.708 | 986 | 29 | 1.0269 | |||||
| PDZ and LIM domain protein 3 | −0.674 | 317 | 13 | 1.0199 | |||||
| Prolyl-tRNA synthetase [EC6.1.1.15] | −0.473 | 1135 | 41 | 1.0295 | |||||
| F3-H7 | Zinc finger, RanBP2-type, domain-containing | −0.986 | 513 | 21 | 1.0108 | − | − | ± | − |
| F18-B1 | E3 ubiquitin-protein ligase HUWE1 | −0.423 | 2720 | 105 | 1.0231 | − | + | + | + |
| 5'-nucleotidase, cytosolic II | −0.2 | 476 | 21 | 1.0368 | |||||
| F18-C3 | Chromatin licensing/DNA replication factor 1 | −0.348 | 578 | 20 | 1.0414 | − | + | + | + |
| Ubiquitin-conjugating enzyme E2 N | −0.228 | 173 | 8 | 1.0386 | |||||
| F31-C3 | Chromatin licensing/DNA replication factor 1 | −0.348 | 578 | 20 | 1.0414 | − | + | + | + |
| F31-E1 | Putative serine/threonine-protein kinase C05D10.2 | −0.774 | 1061 | 39 | 1.0118 | + | + | + | + |
| F31-H7 | Chromatin licensing/DNA replication factor 1 | −0.348 | 578 | 20 | 1.0414 | − | − | − | − |
| NACHT/WD repeat domain-containing protein 1 | −0.282 | 2005 | 81 | 1.0271 | |||||
The GO terms for the annotated proteins in terms of biological processes, molecular function and cellular components were identified for almost all the proteins (Table S1). In terms of biological processes, some are associated with DNA metabolic and physiological processes including nucleotide metabolic process (GO:0009117), cell division (GO:0051301), mitotic cell cycle (GO:0000278), RNA interference (GO:0016246) and protein physiological processes such as potassium ion transport (GO:0006813), ATP binding (GO:0005524) and protein modification process (GO:0006464). Others were associated with regulation, development or stress response including cell differentiation (GO:0030154), reproduction (GO:0000003) and response to oxidative stress (GO:0006979) and so on. In terms of cellular components, the proteins were found to be associated with cytoplasm (GO:0005737), intracellular (GO:0005622), nucleus (GO:0005634), plasma membrane (GO:0005886) and extracellular region (GO:0005576). The PDZ and LIM domain protein 3 expressed in the cytoplasm is associated with response to oxidative stress (GO:0006979). In addition, a membrane protein, Small conductance calcium-activated potassium channel (SK) protein 2 with Calmodulin binding domain (CaMBD) (GO:0015269), has been found to be a secreted protein [39–42].
Discussion
Following skin penetration by cercariae, S. japonicum adult worms migrate to the hepatic portal system, where they mature and survive for many years where the female occasionally migrating to the smallest venules to lay eggs [43]. The adult schistosomes are constantly exposed to the host immune system with antibodies produced against fractions of the worms. These antibodies are often used as potential diagnostic tools. Several immunoepidemiological studies have examined antibody isotype responses to schistosomal protein extracts in the form of isolated proteins, recombinant proteins or crude antigen [44, 45]. Many studies have also presented a global proteomics approach using 2-D gels to identify major S. japonicum excretory and secretory proteins as well as adult worm and egg extracts [46–50]. In this study, for the first time, proteomics approach was extended to identifying S. japonicum proteins in ProteomeLab FP 2D derived liquid fractions reactive to antibody isotypes in plasma samples from S. japonicum-infected population. ESI MS/MS was applied to sequence fractions containing immunreactive proteins. In all, 18 proteins were identified; characterized and GO categories determined to enhance understanding of the immunological significance of these proteins.
In IgE, IgG1, IgG3 and IgG4 antibody isotype recognition of 1-D fractions, it has been shown that some fractions from the adult S. japonicum proteome are preferentially recognized by certain isotypes. For instance, IgE isotype was detected by fewer antigenic fractions (43.2%). The most recognized isotype was IgG3 (79.5%) followed by IgG1 (75.0%) and IgG4 (61.4%) accordingly. The IgG3 response was directed against a larger repertoire of antigens in the fractions. This suggests that there were fewer dominant antigens stimulating IgE response. Earlier report [51, 52] showed where IgE reactivity to glycolipids extracted from schistosome eggs (SEA) or adult worms (SWA) was more than IgG4 and that proteins alone do not constitute the major binding targets of IgE, and that this isotype is substantially directed towards carbohydrate moieties portion of glycolipids on proteins present in SEA or AWA. Weiss et al. [53] showed that a carbohydrate epitope recognized by a monoclonal antibody that was raised against the cercarial glycocalyx was present on glycoproteins and glycolipids of various schistosomes’ life cycle stages. This might explain why IgG4 recognized more proteins than IgE in the 1-D dot-blot.
Furthermore, zinc finger, RanBP 2-type, domain-containing protein was strongly recognized by IgE but moderately by IgG3 and IgG4 and weakly by IgG1 indicating that it might play less role in IgG subclasses directed immune response. The antigens recognized strongly by IgE are of interest as such antigens could be associated with development of resistance to schistosomiasis [2–4, 6]. The E3 ubiquitin-protein ligase, ubiquitin-conjugating enzyme E2 N and 5'-nucleotidase, cytosolic II were strongly recognizing by IgG subclasses (IgG1, IgG3 and IgG4) but not IgE suggesting these enzymes might be of IgG subclasses preferred. The serine/threonine-protein kinase with a relatively higher antigenic propensity of 1.0118 was sequenced from a single 2-D well strongly reactive with all the four isotypes indicating strong preference for IgE, IgG1, IgG3 as well as IgG4 suggesting shared antigenic determinates or multiple epitopes. This highlights the importance of vaccine research focusing on induction of protective isotype-specific antibody response to specific peptides since a single protein from the parasite might possess undetermined antigenic determinants capable of stimulating various antibody productions. Therefore, further investigations employing peptide mapping techniques will be essential in determining specific antigenic determinants for the isotypes.
There were also proteins found with strong immunogenic activity to IgE, IgG3 and IgG4 but not IgG1. Some of these proteins were the Small Conductance calcium-activated potassium channel protein 2, BRO1 domain-containing protein BROX both of which are membrane associated proteins and PDZ/LIM domain containing protein 3. Notwithstanding, it is possible that such proteins might have multiple isotype-specific or shared antigenic determinants within each protein. The PDZ/LIM domain containing proteins are known to be both interaction modules associated with proteins of diverse functions [20, 54].
Proteins associated with nucleus, or cytosolic component are not immediately and directly exposed to the immunity of the definitive host and hence might not evoke immune response. However, Harn, et al. [55], identified glycolytic enzymes, triosephosphate isomerase and glyceraldehyde-3-phosphate dehydrogenase to be expressed on the surface of larval schistosomes and it appears very unlikely that these fundamentally cytosolic proteins are exposed at the external surface of the parasites. However, evidence from other cell systems showed that both glycolytic enzymes and various heat shock proteins can bind to cytoskeletal structures such as actin, microfilaments and microtubules [56, 57] where they can evoke immune response from the host. In addition, small conductance calcium-activated potassium channel (SK) protein 2 with calmodulin binding domain (CaMBD), a membrane associate protein that can also be secreted has been reported to function in host-parasite interaction [39, 41, 42].
In this study, attempt was made to identify and characterize potential IgE and IgG4 immunoreactive proteins employing immunoproteomics approach and related identified proteins with their biochemical properties and gene ontology types. A number of proteins were sequenced from immunoreactive fractions. It is anticipated that, immunoreactive proteins identified herein will stimulate further studies to evaluate their immunogenicity through recombinant protein expression, immunomoderation properties in terms of protective potentials and novel compounds that have therapeutic importance.
Acknowledgements
DB is grateful for the financial support from Japan International Cooperation Agency (JICA). This research received further financial support from Grants Kakenhi (22406009, 23590489) and the Global Centre of Excellence (GCOE), Nagasaki University, Japan. Special thanks are addressed to Noguchi Memorial Institute for Medical Research, University of Ghana-Legon, Ghana and Department of Immunogenetics, Institute of Tropical Medicine, Nagasaki University, Japan, for their cooperation.
Abbreviations
- BCA
Bicinchoninic acid
- TCEP
Tris (2-Carboxyethyl) Phosphine Hydrochloride
- n-OG
n-Octylglucoside
References
- 1.Boros DL. Immunopathology of Schistosoma mansoni infection. Clin Microbiol Rev 1989; 2: 250–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunne DW, Butterworth AE, Fulford AJC, Kariuki HC, Langley JG, Ouma JH, Capron A, Pierce RJ, Sturrock RF. Immunity after treatment of human schistosomiasis: association between IgE antibodies to adult worm antigens and resistance to reinfection. Eur J Immunol 1992; 22: 1483–1494 [DOI] [PubMed] [Google Scholar]
- 3.Grogan JL, Kremsner PG, van Dam GJ, Deelder AM, Yazdanbakhsh M. Anti-schistosome IgG4 and IgE at 2 years after chemotherapy: infected versus uninfected individuals. J Infect Dis 1997; 176: 1344–1350 [DOI] [PubMed] [Google Scholar]
- 4.Hagan P, Blumenthal UJ, Dunn D, Simpson AJG, Wilkins HA. Human IgE, IgG4 and resistance to reinfection with Schistosoma haematobium. Nature 1991; 349: 243–245 [DOI] [PubMed] [Google Scholar]
- 5.Li Z, King CL, Ogundipe JO, Licate LS, Blanton RE. Preferential recognition by human IgE and IgG4 of a species-specific Schistosoma haematobium serine protease inhibitor. J Infect Dis 1995; 171: 416–422 [DOI] [PubMed] [Google Scholar]
- 6.Demeure CE, Rihet P, Abel L, Ouattara M, Bourgois A, Dessein AJ. Resistance to Schistosoma mansoni in humans: influence of the IgE/IgG4 balance and IgG2 in immunity to reinfection after chemotherapy. J Infect Dis 1993; 168: 1000–1008 [DOI] [PubMed] [Google Scholar]
- 7.Nara T, Iizumi K, Ohmae H, Sy OS, Tsubota S, Inaba Y, Tsubouchi A, Tanabe M, Kojima S, Aoki T. Antibody isotype responses to paramyosin, a vaccine candidate for schistosomiasis, and their correlations with resistance and fibrosis in patients infected with schistosoma japonicum in leyte, The Philippines. Am J Trop Med Hyg 2007; 76: 384–391 [PubMed] [Google Scholar]
- 8.Ndhlovu P, Cadman H, Vennervald BJ, Christensen NO, Chidimu M, Chandiwana SK. Age-related antibody profiles in Schistosoma haematobium infections in a rural community in Zimbabwe. Parasite Immunol 1996; 18: 181–191 [DOI] [PubMed] [Google Scholar]
- 9.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol 2004; 16: 257–275 [DOI] [PubMed] [Google Scholar]
- 10.Satoguina JS, Weyand E, Larbi J, Hoerauf A. T regulatory-1 cells induce IgG4 production by B cells: role of IL-10. J Immunol 2005; 174: 4718–4726 [DOI] [PubMed] [Google Scholar]
- 11.Hagan P, Sharaf O. Schistosomiasis vaccines. Expert Opin Biol Ther 2003; 3: 1271–1278 [DOI] [PubMed] [Google Scholar]
- 12.Cummings RD, Nyame AK. Glycobiology of schistosomiasis. FASEB J 1996; 10: 838–848 [DOI] [PubMed] [Google Scholar]
- 13.Webster M, Fulford AJC, Braun G, Ouma JH, Kariuki HC, Havercroft JC, Gachuhi K, Sturrock RF, Butterworth AE, Dunne DW. Human immunoglobulin E responses to a recombinant 22.6-kilodalton antigen from Schistosoma mansoni adult worms are associated with intensities of reinfection after treatment. Infect Immun 1996; 64: 4042–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitzsimmons CM, Stewart TJ, Hoffmann KF, Grogan JL, Yazdanbakhsh M, Dunne DW. Human IgE response to the Schistosoma haematobium 22.6 kDa antigen. Parasite Immunol 2004; 26: 371–376 [DOI] [PubMed] [Google Scholar]
- 15.Webster M, Fulford AJ, Braun G, Ouma JH, Kariuki HC, Havercroft JC, Gachuhi K, Sturrock RF, Butterworth AE, Dunne DW. Human immunoglobulin E responses to a recombinant 22.6-kilodalton antigen from Schistosoma mansoni adult worms are associated with low intensities of reinfection after treatment. Infect Immun 1996; 64: 4042–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santiago ML, Hafalla JC, Kurtis JD, Aligui GL, Wiest PM, Olveda RM, Olds GR, Dunne DW, Ramirez BL. Identification of the Schistosoma japonicum 22.6-kDa antigen as a major target of the human IgE response: similarity of IgE‐binding epitopes to allergen peptides. Int Arch Allergy Immunol 1998; 117: 94–104 [DOI] [PubMed] [Google Scholar]
- 17.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics 2005; 5: 826–827 [DOI] [PubMed] [Google Scholar]
- 18.Schlautman JD, Rozek W, Stetler R, Mosley RL, Gendelman HE, Ciborowski P. Multidimensional protein fractionation using ProteomeLab PF2DTM for profiling amyotrophic lateral sclerosis immunity: A preliminary report. Proteome Sci 2008; 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics 2006; 5: 26–34 [DOI] [PubMed] [Google Scholar]
- 20.Levreri I, Musante L, Petretto A, Bruschi M, Candiano G, Melioli G. Separation of human serum proteins using the Beckman-Coulter PF 2D system: analysis of ion exchange-based first dimension chromatography. Clin Chem Lab Med 2005; 43: 1327–1333 [DOI] [PubMed] [Google Scholar]
- 21.Barre O, Solioz M. Improved protocol for chromatofocusing on the ProteomeLab PF 2D. Proteomics 2006; 6: 5096–5098 [DOI] [PubMed] [Google Scholar]
- 22.Kang X, Frey DD. Chromatofocusing of peptides and proteins using linear pH gradients formed on strong ion-exchange adsorbents. Biotechnol Bioeng 2004; 87: 376–387 [DOI] [PubMed] [Google Scholar]
- 23.Palm A, Hakansson L, Marko-Varga G. Microcolumn Chromatofocusing Using Slurry-Packed Mono P Anion-Exchanger Beads as Chromatographic Matrix. Chromatographia 2003; 58: 707–712 [Google Scholar]
- 24.Ruelle V, Falisse-Poirrier N, Elmoualij B, Zorzi D, Pierard O, Heinen E, De Pauw E, Zorzi W. An immuno-PF2D-MS/MS proteomic approach for bacterial antigenic characterization, to Bacillus and beyond. J Proteome Res 2007; 6: 2168–2175 [DOI] [PubMed] [Google Scholar]
- 25.Lee HJ, Kang MJ, Lee EY, Cho SY, Kim H, Paik YK. Application of a peptide-based PF 2D platform for quantitative proteomics in disease biomarker discovery. Proteomics 2008; 8: 3371–3381 [DOI] [PubMed] [Google Scholar]
- 26.Emily IC, Hewel J, Felding-Habermann B, Yates JR. Large Scale Protein Profiling by Combination of Protein Fractionation and Multidimensional Protein Identification Technology (MudPIT). Mol Cell Proteomics 2006; 5: 53–56 [DOI] [PubMed] [Google Scholar]
- 27.Abdel-Hafeez EH, Kikuchi M, Watanabe K, Ito T, Yu C, Chen H, Nara T, Arakawa T, Aoki Y, Hirayama K. Proteome approach for identification of schistosomiasis japonica vaccine candidate antigen. Parasitol Int 2009; 58: 36–44 [DOI] [PubMed] [Google Scholar]
- 28.Hirayama K, Chen H, Kikuchi M, Yin T, Gu X, Liu J, Zhang S, Yuan H. HLA-DR-DQ alleles and HLA-DP alleles are independently associated with susceptibility to different stages of post-schistosomal hepatic fibrosis in the Chinese population. Tissue Antigens 1999; 53: 269–274 [DOI] [PubMed] [Google Scholar]
- 29.Hirayama K, Chen H, Kikuchi M, Yin T, Itoh M, Gu X, Zhang S, Yuan H. Glycine-Valine dimorphism at the 86th amino acid of HLA-DRB1 influenced the prognosis of postschistosomal hepatic fibrosis. J Infect Dis 1998; 177: 1682–1686 [DOI] [PubMed] [Google Scholar]
- 30.Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics 2006; 5: 26–34 [DOI] [PubMed] [Google Scholar]
- 31.Reis BS, Bozzi A, Prado FLS, Pereira MCN, Ferreira FE, Godoy P, Moro L, Pedroso EP, Leite MF, Goes AM. Membrane and extracellular antigens of Paracoccidioides brasiliensis (Mexo): Identification of a 28-kDa protein suitable for immunodiagnosis of paracoccidioidomycosis. J Immunol Methods 2005; 307: 118–126 [DOI] [PubMed] [Google Scholar]
- 32.Okamoto K, Endo Y, Inoue S, Nabeshima T, Nga PT, Guillermo PH, Yu F, Loan do P, Trang BM, Natividad FF, Hasebe F, Morita K. Development of a rapid and comprehensive proteomics-based arboviruses detection system. J Virol Methods 2010; 167: 31–36 [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y, Zheng H, Chen Y, Zhang L, Wang K, Guo J, Huang Z, Zhang B, Huang W, Jin K, et al. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 2009; 460: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999; 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- 35.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982; 157: 105–132 [DOI] [PubMed] [Google Scholar]
- 36.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: John M. Walker, ed. The Proteomics Protocols Handbook. NJ: Humana Press; 2005. p. 571–607
- 37.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic deteminants on protein antigens. FEBS J 1990; 276: 172–174 [DOI] [PubMed] [Google Scholar]
- 38.Zhu K, Zhao J, Lubman DM, Miller FR, Barder TJ. Protein pI Shifts due to Posttranslational Modifications in the Separation and Characterization of Proteins. Anal Chem 2005; 77: 2745–2755 [DOI] [PubMed] [Google Scholar]
- 39.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-Conductance, Calcium-Activated Potassium Channels from Mammalian Brain. Science 1996; 20: 1709–1714 [DOI] [PubMed] [Google Scholar]
- 40.Liao Q, Yuan X, Xiao H, Liu C, Lv Z, Zhao Y, Wu Z. Identifying Schistosoma japonicum excretory/secretory proteins and their interactions with host immune system. PLoS One 2011; 6: e23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guillou F, Roger E, Mone Y, Rognon A, Grunau C, Théron A, Mitta G, Coustau C, Gourbal BE. Excretory/secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol Biochem Parasitol 2007; 155: 45–56 [DOI] [PubMed] [Google Scholar]
- 42.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 2004; 338: 1027–1036 [DOI] [PubMed] [Google Scholar]
- 43.Ward RO. Some surgical aspects of urinary bilharziasis. Proc R Soc Med 1945; 39: 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viana IR, Correa-Oliveira R, Carvalho OS, Massara CL, Colosimo E, Colley DG, Gazzinelli G. Comparison of antibody isotype responses to Schistosoma mansoni antigens by infected and putative resistant individuals living in an endemic area. Parasite Immunol 1995; 17: 297–304 [DOI] [PubMed] [Google Scholar]
- 45.Bosompem KM, Arishima T, Yamashita T, Ayi I, Anyan WK, Kojima S. Extraction of Schistosoma haematobium antigens from infected human urine and generation of potential diagnostic monoclonal antibodies to urinary antigens. Acta Trop 1996; 62: 91–103 [DOI] [PubMed] [Google Scholar]
- 46.Liu F, Cui SJ, Hu W, Feng Z, Wang ZQ, Han ZG. Excretory/Secretory proteome of the adult developmental stage of human blood fluke, Schistosoma japonicum. Mol Cell Proteomics 2009; 8: 1236–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knudsen GM, Medzihradszky KF, Lim K-C, Hansell E, McKerrow JH. Proteomic Analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics 2005; 4: 1862–1875 [DOI] [PubMed] [Google Scholar]
- 48.Zhong Z, Zhou HB, Li XY, Luo QL, Song XR, Wang W, Wen HQ, Yu L, Wei W, Shen J. Serological proteome-oriented screening and application of antigens for the diagnosis of Schistosomiasis japonica. Acta Trop 2010; 116: 1–8 [DOI] [PubMed] [Google Scholar]
- 49.Braschi S, Curwen RS, Ashton PD, Verjovski-Almeida S, Wilson A. The tegument surface membranes of the human blood parasite Schistosoma mansoni: A proteomic analysis after differential extraction. Proteomics 2006; 6: 1471–1482 [DOI] [PubMed] [Google Scholar]
- 50.Rihet P, Demeure CE, Dessein AJ, Bourgois A. Strong serum inhibition of specific IgE correlated to competing IgG4, revealed by a new methodology in subjects from a S. mansoni endemic area. Eur J Immunol 1992; 22: 2063–2070 [DOI] [PubMed] [Google Scholar]
- 51.Van Der Kleij D, Tielens AGM, Yazdanbakhsh M. Infection and immunity, recognition of schistosome glycolipids by Immunoglobulin E: Possible role in immunity. Infect Immun 1999; 67: 5946–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss JB, Magnani JL, Strand M. Identification of Schistosoma mansoni glycolipids that share immunogenic carbohydrate epitopes with glycoproteins. J Immunol 1986; 136: 4275–4282 [PubMed] [Google Scholar]
- 53.Te Velthuis AJW, Isogai T, Gerrits L, Bagowski CP. Insights into the molecular evolution of the PDZ/LIM family and identification of a novel conserved protein motif. PLoS One 2007; 7: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka T, Yamamoto Y, Muromoto R, Ikeda O, Sekine Y, Grusby MJ, Kaisho T, Matsuda T. PDLIM2 inhibits T helper 17 cell development and granulomatous inflammation through degradation of STAT3. Sci Signal 2011; 6: ra85. [DOI] [PubMed] [Google Scholar]
- 55.Harn DA, Gu W, Oligino LD, Mitsuyama M, Gebremichael A, Richter D. A protective monoclonal antibody specifically recognizes and alters the catalytic activity of schistosome triose-phosphate isomerase. J Immunol 1992; 148: 562–567 [PubMed] [Google Scholar]
- 56.Gitlits VM, Toh BH, Loveland KL, Sentry JW. The glycolytic enzyme enolase is present in sperm tail and displays nucleotide-dependent association with microtubules. Eur J Cell Biol 2000; 79: 104–111 [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Tolan DR, Pagliaro L. Metabolic compartmentation in living cells: structural association of aldolase. Exp Cell Res 1997; 237: 445–451 [DOI] [PubMed] [Google Scholar]