Abstract
Background
Genome-wide association (GWA) studies have led to a paradigm shift in how researchers study the genetics underlying disease. Many GWA studies are now publicly available and can be used to examine whether or not previously proposed candidate genes are supported by GWA data. This approach is particularly important for the field of alcoholism because the contribution of many candidate genes remains controversial.
Methods
Using the Human Genome Epidemiology (HuGE) Navigator, we selected candidate genes for alcoholism that have been frequently examined in scientific articles in the past decade. Specific candidate loci as well as all the reported SNPs in candidate genes were examined in the Study of Alcohol Addiction: Genetics and Addiction (SAGE), a GWA study comparing alcohol dependent and non-dependent subjects.
Results
Several commonly reported candidate loci, including rs1800497 in DRD2, rs698 in ADH1C, rs1799971 in OPRM1 and rs4680 in COMT, are not replicated in SAGE (p> .05). Among candidate loci available for analysis, only rs279858 in GABRA2 (p=0.0052, OR=1.16) demonstrated a modest association. Examination of all SNPs reported in SAGE in over 50 candidate genes revealed no SNPs with large frequency differences between cases and controls and the lowest p value of any SNP was .0006.
Discussion
We provide evidence that several extensively studied candidate loci do not have a strong contribution to risk of developing alcohol dependence in European and African Ancestry populations. Due to lack of coverage, we were unable to rule out the contribution of other variants and these genes and particular loci warrant further investigation. Our analysis demonstrates that publicly available GWA results can be used to better understand which if any of previously proposed candidate genes contribute to disease. Furthermore, we illustrate how examining the convergence of candidate gene and GWA studies can help elucidate the genetic architecture of alcoholism and more generally complex diseases.
Keywords: Alcohol dependence, Candidate genes, GWAS, Genetics
INTRODUCTION
Genome-wide association (GWA) studies have revolutionized the search for common genetic variants that influence individual risk for complex diseases. Before this revolution, the discovery of genetic associations was dominated by candidate gene studies that used targeted gene approaches. Examination of these previous gene association studies demonstrates that most reported associations are not consistently replicated (Hirschhorn et al., 2002) and the strength of genetic associations in initial studies commonly erodes in subsequent research (Ioannidis et al., 2001). Despite this suggested irreproducibility, many candidate gene association studies continue to be published annually (Yu et al., 2008).
GWA studies rapidly evaluate millions of single nucleotide polymorphisms (SNPs) throughout the genome and therefore have the potential for identifying key variants in complex diseases. Since the publication of the first GWA study in 2005 (Klein et al., 2005), over 1000 GWA studies have established genetic associations of more than 200 traits, many of which are complex diseases. SNP-trait associations from published GWA studies are readily available to investigators through “A Catalog of Genome-Wide Association Studies” by the National Human Genome Research Institute (www.genome.gov/gwastudies). More recently, several datasets from GWA studies have also become available to the scientific community through the database of Genotypes and Phenotypes (dbGaP) maintained by NCBI (Mailman et al., 2007). These online scientific databases provide opportunities for investigators to access GWA data.
Online databases can specifically be used to evaluate whether genes that were previously suggested in candidate gene studies are replicated in GWA studies. Research by Siontis et al. demonstrates that only a few of previously proposed candidate loci of common diseases reached genome-wide significance in GWA studies (Siontis et al., 2010). The loci that did replicate, however, had important genetic effects and included variants implicated in Alzheimer’s disease and statin-induced myopathy. Similarly, a recent analysis by Obeidat et al. examined genetic associations with lung function measures to evaluate the role of previously associated genes in a large GWA study and clarified the role of many controversial associations (Obeidat et al., 2011). This approach of comparing candidate gene and GWA studies is powerful because it highlights which findings have consistent scientific evidence and therefore merit being pursued in future studies. These findings prompted us to examine whether proposed candidate genes associated with alcohol dependence are supported by GWA data.
Genetic and environmental factors contribute to individual susceptibility to alcohol dependence. Twin studies estimate that heritable influences explain 47-64% of variance in risk for alcohol dependence (Heath et al., 1997; Knopik et al., 2004). Several past research efforts have focused on targeted gene approaches to shed light on the genes that underlie these heritable influences. This has led to the proposal of hundreds of candidate genes that contribute to the development of alcohol dependence (Yu et al., 2008). A few GWA studies have also explored genes potentially involved in alcohol dependence (Bierut et al., 2010; Edenberg et al., 2010; Farrer et al., 2009; Heath et al., 2011; Hodgkinson et al., 2010; Joslyn et al., 2010; Treutlein et al., 2009; Zlojutro et al., 2011). Despite extensive candidate gene studies and several GWA studies, little consensus exists over which if any genes contribute to the genetic basis of alcohol dependence.
The existence of many controversial candidate genes for alcoholism highlights the need for further research on whether or not these genes replicate in large datasets. Results from the Study of Alcohol Addiction: Genetics and Addiction (SAGE) have recently become available through dbGaP. SAGE compares DSM-IV alcohol dependent individuals and non-dependent, unrelated control subjects of European and African American descent. Using the SAGE data, we examined differences in SNP frequencies between cases and controls within previously reported candidate genes. These targeted candidate genes were selected using the Human Genome Epidemiology (HuGE) Navigator, a publicly searchable database established in 2001 of published genetic association and human genome epidemiological studies (Yu et al., 2008). The HuGE Navigator along with the SAGE results facilitated the systematic examination of candidate genes considered in many alcoholism studies over the last decade.
MATERIALS AND METHODS
Selection of Candidate Genes
The HuGE Navigator was developed using PubMed abstracts as the core data source and using data and text mining algorithms to develop a knowledge database (Yu et al., 2008). Each week since 2001, articles are systematically deposited in the database and represent a comprehensive list of recent articles. An automatic literature program screens PubMed for abstracts and then a genetic epidemiologist selects abstracts meeting inclusion criteria and indexes them. Phenopedia of the HuGE Navigator gives a disease-centered view of genetic association studies by providing information about genes studied in relation to a queried phenotype (Yu et al., 2010). Phenopedia was queried in July 2011 for Alcoholism and 584 genes were retrieved.
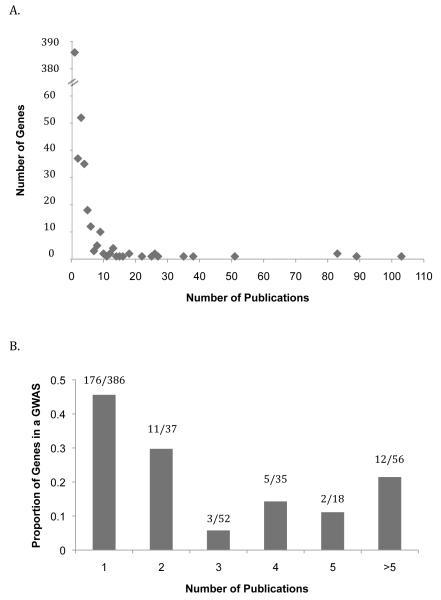
We focused our study on genes that have been frequently characterized by candidate gene studies. In primary analysis, over 90% of the genes associated with alcoholism in the HuGE database have 5 or fewer publications (528 out of 584 genes). The 56 candidate genes that have more than 5 publications vary substantially in the number of publications (6-103 publications). Figure 1A highlights that many genes have one or a few reported publications and there are some outliers that have been examined in many papers. This distribution may be explained in part by the fact that many of the genes with a low number of publications have been primarily identified in a GWA study and are not well characterized in targeted candidate gene studies. Figure 1B demonstrates that for almost 50% (176/386) of the genes with one publication that publication is itself a GWA study. Based on these preliminary observations, we narrowed our investigation to genes with more than 5 publications to focus our analysis on well-studied genes.
Fig. 1.
Characteristics of genes associated with Alcoholism in the Human Genome Epidemiology (HuGE) Navigator (A) Distribution of number of publications on genes; (B) Proportion of genes observed in at least one GWA study stratified based on number of publications. 4 genes were identified in 2 GWA study (these genes had 2, 2, 10 and 51 publications). All other genes were found in 1 or none GWA study. A total of 8 GWA studies on Alcoholism are listed in the HuGE database (Bierut et al., 2010; Edenberg et al., 2010; Farrer et al., 2009; Heath et al., 2011; Hodgkinson et al., 2010; Joslyn et al., 2010; Treutlein et al., 2009; Zlojutro et al., 2011).
Since the X chromosome is not included in the publicly available SAGE results, the two candidate genes on the X chromosome MAOA and HTR2C, which have 26 and 8 publications respectively, were excluded from the analysis. The 54 autosomal genes that had more than 5 publications were pursued using the SAGE dataset. For the remainder of this paper, we will only refer to the 54 autosomal candidate genes.
SAGE Data
SAGE is a case-control study that analyzed genetic data on over 3,800 phenotyped subjects funded as part of the Gene Environment Association Studies (GENEVA) initiative supported by the National Human Genome Research Institute (Bierut et al., 2010). Alcohol-dependent cases and controls were selected from three large datasets: the Collaborative Study on the Genetic of Alcoholism (COGA), the Family Study of Cocaine dependence (FSCD) and the Collaborative Genetic Study of Nicotine Dependence (COGEND). Cases are required to have a lifetime history of DSM-IV alcohol dependence. Controls are required to have been exposed to alcohol because alcohol use is necessary to develop dependence, but not to have met lifetime diagnosis criteria for alcohol dependence or dependence for illicit drugs. A common assessment was performed for all cases and controls in the three studies that was based on the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) (Bucholz et al., 1994). The common methodology of interview administration, question format and queried domains enabled phenotypic standardization across the three studies (Bierut et al., 2010). Characteristics of the cases and controls in the SAGE dataset are listed in Table 1 and additional information is available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1. The SAGE dataset is publicly searchable through the Genome Brower under the Analysis tab on this website.
Table 1.
Sex, age, race, ethnicity and alcohol dependence characteristics of cases and controls in the Study of Alcohol Addiction: Genetics and Addiction (SAGE) dataset (Bierut et al., 2010).
| Characteristic | Cases n=1,897 | Controls n=1,932 | Total n=3,829 |
|---|---|---|---|
| Sex, n (%) | |||
| Males | 1.155 (60.9) | 606 (31.4)* | 1,761 (46.0) |
| Females | 742 (39.1) | 1,326 (68.6) | 2,068 (54.0) |
| Age, years | |||
| Mean ± SD | 39.0 ± 9.3 | 39.3 ± 9.1 | 19.2 ± 9.2 |
| Range | 18.0-77.0 | 18.0-65.0 | 18.0-77.0 |
| Self-reported race, n (%) | |||
| European-American | 1,235 (65.1) | 1,433 (74.2)* | 2,668 (69.5) |
| African-American | 662 (34.9) | 499 (25.8) | 1,161 (30.3) |
| Self-reported ethnicity, n (%) | |||
| Hispanic | 76 (4.0) | 56 (2.8) | 132 (3.4) |
| Alcohol dependence | |||
| Dia nosis n % | 1 897 100.0 | 0 0.0 * | 1 897 49.5 |
p<0.0001 for difference between cases and controls
The power Calculator for Association with Two Stage design (CATS) was used to determine what effect sizes the SAGE dataset is able to detect (Skol et al., 2006). Using a sample size of 1900 cases and 1900 controls and an alpha level of .05, we calculated different allele frequencies and risk ratios.
Examination of SNPs in Candidate Genes
The HuGE database was used to survey articles on the ten candidate genes that had the most publications (listed in Table 2). The most well established loci based on expert opinion of the literature for each of the top ten candidate genes was searched in the genome browser to test whether candidate loci that had been highly reported in candidate gene studies replicated in the SAGE dataset. Since allele A9 for SLC6A3 is a VNTR, we examined the two SNPs rs27072 and rs27048 as proxies because they have been found to be associated with similar withdrawal symptoms and are roughly in the same region of the gene as the VNTR (Le Strat et al., 2008). As the originators of the SAGE dataset, we were also able to compare the odds ratios and p values within the original three datasets (COGA, FSCD and COGEND) to verify whether there was any heterogeneity across the three contributing studies.
Table 2.
In the ten most frequently studied genes associated with alcoholism according to the Human Genome Epidemiology (HuGE) Navigator, the most well studied variants were examined in Study of Alcohol Addiction: Genetics and Addiction (SAGE).
| Candidate Genes |
Publications on Alcoholism association |
Commonly reported SNP |
Common Name of SNP | P value in SAGE |
Odds Ratio in SAGE (CI) |
|---|---|---|---|---|---|
| ALDH2 | 103 | rs671 | ALDH2*2 (Harada, 1982) |
- | - |
| ADH1B | 89 | rs1229984 | ADH1B*2/ADH2*2 (Thomasson, 1992) |
- | - |
| DRD2 | 83 | rs1800497 | TaqIA (Blum, 1990) |
0.09 | 1.1053 (.9845-1.2408) |
| SLC6A4 | 83 | rs4795541 | 5-HTTLPR/S allele (Sander, 1997) |
- | - |
| ADH1C | 51 | rs698 | ADH1C*2 (Thomasson, 1992) |
0.1452 | 1.0819 (.9732-1.2028) |
| OPRM1 | 38 | rs1799971 | Ala118Gly (Bond, 1998) |
0.1372 | .8823 (.7481-1.0407) |
| CYP2E1 | 35 | rs3813867 | CYPE1*c2 (Hayashi, 1991) |
- | - |
| GABRA2 | 27 | rs279858* | (Edenberg, 2004) | 0.0052 | 1.1572 (1.0445-1.2821) |
| COMT | 26 | rs4680 | Val158Met (Tiihonen, 1999) |
0.6328 | 1.0244 (.9278-1.1311) |
| SLC6A3 | 25 | ** | A9 (VNTR) (Dobashi, 1997) |
One of over 20 SNPs significantly associated with alcohol dependence (Edenberg et al., 2004). This SNP was examined because it was the only one in an exon.
Examined 2 SNPs, rs27072 and rs27048, as proxies (Le Strat et al., 2008)
The 54 candidate genes with more than 5 publications were identified and chromosomal regions containing the gene plus 10 kb both 5′ and 3′ of the gene were examined. These expanded regions were searched using the SAGE genome browser and SNPs within these regions with p<.05 were recorded.
For each candidate gene, all SNPs with p<.05 were queried together in SNP Annotation and Proxy Search (SNAP) to assess linkage disequilibrium (Johnson et al., 2008). These searches were performed using the 1000 genomes pilot 1 SNP dataset, an r2 ≥ .8, and a distance limit of 500. This analysis was performed with both the CEU and YPI population panels separately because of allele frequency differences between European American and African American subsets. All SNPs that had an r2 greater than 0.8 and at least one other variant in a group were considered a cluster.
RESULTS
The SAGE dataset contains half of the most commonly reported variants associated with the ten most well studied candidate genes (Table 2). Of the 5 candidate variants reported in SAGE, the only variant with a p< .05 is rs279858 in GABRA2 (p=.0052, OR=1.16). The commonly reported variants, rs1800497 in DRD2, rs698 in ADH1C, rs1799971 in OPRM1 and rs4680 in COMT, have p> .05. The minor allele for rs17999971 in OPRM1 trends towards being protective (OR=.88) while the minor alleles of rs1800497 in DRD2, rs698 in ADH1C and rs4680 in COMT trend toward being associated with alcohol dependence (OR=1.11,1.08,1.02 respectively). The effects of these associations are in the expected direction based on previous candidate gene studies (Blum et al., 1990; Bond et al., 1998; Hendershot et al., 2011; Ponce et al., 2008; Thomasson et al., 1991; Tiihonen et al., 1999; Tolstrup et al., 2008; Zhang et al., 2006). In addition, these effects in SAGE were similar to the findings in the individual three studies that contributed to SAGE: COGA, FSCD and COGEND. Across the three contributing studies, the odds ratios ranged from 1.07-1.13 for rs1800497 in DRD2, 1.06-1.11 for rs698 in ADH1C, 0.82-0.95 for rs1799971 in OPRM1, 1.09-1.17 for rs279858 in GABRA2 and 1.02-1.09 for rs4680 in COMT (data not shown).
Several commonly reported variants associated with alcoholism are not on the Illumina chip that was used to generate the SAGE dataset. These SNPs include rs671 in ALDH2, rs1229984 in ADH1B, rs4795541 in SLC6A4 and rs3813867 in CYP2E1. Since the A9 allele in SLC6A3 is a VNTR and therefore also not reported in SAGE, we examined two proxy SNPs (Le Strat et al., 2008). Neither of these two SNPs show a significant difference between the cases and controls (p=.8646 for rs27072 and p=.3842 for rs27048).
In every gene with more than 5 publications, few SNPs had impressive differences between cases and controls. Of the 2175 SNPs reported in the 54 genes with more than 5 publications, approximately 5% have a p<.05 (116/2175) and approximately 1% have a p<.01 (16/2175) (Table 3). The lowest p value of any variant was 0.0006 for rs925946, which is a SNP upstream of BDNF.
Table 3.
All genes with more than 5 publications in Human Genome Epidemiology (HuGE) Navigator were examined in Study of Alcohol Addiction: Genetics and Addiction (SAGE). Genes were expanded by 10 kb on both sides before they were queried in the SAGE database. MAOA and HTR2C were excluded because they were on the X chromosome.
| Candidate Genes |
Publications on Alcoholism |
SNPs recorded in dbSNP |
Total SNPs in SAGE |
SNPs in SAGE | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| p<.05 | p<.01 | p<.005 | p<.001 | ||||
| ALDH2 | 103 | 407 | 24 | - | - | - | - |
| ADH1B | 89 | 337 | 21 | - | - | - | - |
| DRD2 | 83 | 826 | 41 | 7 | 1 | - | - |
| SLC6A4 | 83 | 637 | 21 | 2 | - | - | - |
| ADH1C | 51 | 522 | 29 | 1 | - | - | - |
| OPRM1 | 38 | 3568 | 122 | 4 | - | - | - |
| CYP2E1 | 35 | 210 | 49 | - | - | - | - |
| GABRA2 | 27 | 1692 | 29 | 16 | 5 | 1 | - |
| COMT | 26 | 752 | 55 | - | - | - | - |
| SLC6A3 | 25 | 1322 | 38 | - | - | - | - |
| HTR2A | 22 | 1036 | 61 | 8 | - | - | - |
| HTR1B | 18 | 60 | 12 | 2 | 1 | - | - |
| DRD4 | 18 | 184 | 9 | - | - | - | - |
| BDNF | 16 | 624 | 29 | 10 | 2 | 2 | 1 |
| NPY | 15 | 241 | 19 | 2 | - | - | - |
| DRD3 | 14 | 693 | 31 | 3 | - | - | - |
| APOE | 13 | 106 | 12 | 1 | - | - | - |
| MTHFR | 13 | 324 | 49 | - | - | - | - |
| GABRA6 | 13 | 215 | 18 | - | - | - | - |
| TPH1 | 13 | 277 | 14 | - | - | - | - |
| GRIN2B | 12 | 5233 | 245 | 11 | 4 | 3 | - |
| CNR1 | 12 | 2778 | 27 | 5 | 2 | - | - |
| TPH2 | 11 | 1415 | 54 | 1 | - | - | - |
| ADH4 | 10 | 830 | 62 | 1 | - | - | - |
| CHRM2 | 10 | 1992 | 62 | 3 | - | - | - |
| CRHR1 | 9 | 1183 | 26 | 1 | - | - | - |
| ANKK1 | 9 | 218 | 26 | 4 | - | - | - |
| ALDH1A1 | 9 | 739 | 122 | 3 | - | - | - |
| DRD1 | 9 | 82 | 19 | - | - | - | - |
| GABRG2 | 9 | 991 | 30 | - | - | - | - |
| GABRB2 | 9 | 2699 | 80 | 1 | - | - | - |
| HTR1A | 9 | 45 | 9 | 4 | - | - | - |
| GSTM1 | 9 | 123 | 3 | - | - | - | - |
| OPRD1 | 9 | 585 | 20 | 1 | - | - | - |
| OPRK1 | 9 | 226 | 37 | - | - | - | - |
| GABRB3 | 8 | 23 | 104 | 9 | 1 | - | - |
| GABRA1 | 8 | 583 | 20 | - | - | - | - |
| DBH | 8 | 562 | 47 | 1 | - | - | - |
| ADH1A | 8 | 293 | 19 | 2 | - | - | - |
| ADH5 | 7 | 584 | 38 | - | - | - | - |
| GAD1 | 7 | 692 | 31 | 1 | - | - | - |
| HFE | 7 | 188 | 28 | 1 | - | - | - |
| GRIN1 | 6 | 513 | 18 | - | - | - | - |
| GAD2 | 6 | 784 | 49 | 1 | - | - | - |
| GABRB1 | 6 | 4354 | 111 | 2 | - | - | - |
| ADH7 | 6 | 288 | 32 | - | - | - | - |
| ADRA2A | 6 | 46 | 9 | - | - | - | - |
| CHRNA5 | 6 | 254 | 16 | - | - | - | - |
| POMC | 6 | 102 | 10 | - | - | - | - |
| SLC6A2 | 6 | 837 | 49 | 1 | - | - | - |
| CCKBR | 6 | 300 | 20 | 4 | - | - | - |
| CCKAR | 6 | 167 | 15 | - | - | - | - |
| TNF | 6 | 177 | 36 | 2 | - | - | - |
| CCK | 6 | 461 | 18 | 1 | - | - | - |
|
| |||||||
| Total (n=54) | 2175 | 116 | 16 | 6 | 1 | ||
In a few genes, a large proportion of the SNPs have modest frequency differences between cases and controls. In 10 out of the 54 genes examined, more than 10% of the SNPs have p<.05 and in 3 genes this portion exceeds 20%. Specifically, the proportion of SNPs in SAGE with p<.05 is 55% (16/29) in GABRA2, 24% (10/29) in BDNF and 44% (4/9) in HTR1A (Table 3). To test whether the large proportion of SNPs with small p values in these genes could be explained by linkage disequilibrium, we performed SNAP analyses.
Many variants clustered as defined by r2> 0.8 within the genes but the proportion of clusters containing SNPs with p<.05 remained quite similar with SNAP analyses in both CEU and YPI populations (data not shown). Of the variants with linkage disequilibrium data available in SNAP for the CEU population, 27 SNPs in GABRA2 broke down into 10 clusters of which 5 clusters had SNPs with p<.05 (50%), 24 SNPs in BDNF broke down into 9 clusters of which 4 clusters had SNPs with p<.05 (44%), and 6 SNPs in HTR1A broke down into 3 clusters of which 1 cluster had SNPs with p<.05 (33%). Generally fewer SNPs clustered in the YRI population than in the CEU population but the proportion of clusters containing SNPs with p<.05 was comparable between the two populations. In the YRI population, 25 SNPs in GABRA2 broke down into 16 clusters of which 12 clusters had SNPs with p<.05 (75%), 25 SNPs in BDNF broke down into 19 clusters of which 10 clusters had SNPs with p<.05 (53%), and 9 SNPs in HTR1A broke down into 5 clusters of which 2 cluster had SNPs with p<.05 (40%).
Power calculations demonstrate that the SAGE dataset has 90% power with an alpha level of .05 to detect a genetic variant with a minor allele frequency of .10 and an odds ratio of 1.25 or greater. The dataset also has 90% power with an alpha level of .05 to detect a variant with a minor allele frequency of .40 and an odds ratio of 1.15 or greater.
DISCUSSION
Over the last decade, hundreds of candidate genes have been proposed for alcoholism. We used local and global approaches to specifically investigate variants within the most widely studied of previously proposed candidate genes. Our primary finding is that most of these candidate genes are not strongly supported by GWA data. This observation reduces the likelihood that these previously proposed genes individually have a strong effect on the genetic risk of alcohol dependence. The results mirror prior work that most candidate loci in common diseases are not strongly replicated in GWA studies except for a few biologically important variants (Siontis et al., 2010; Obeidat et al., 2011).
Analysis of well-characterized loci that were previously proposed in candidate gene studies in a large GWA study on alcoholism, SAGE, reveals unimpressive differences between cases and controls at most loci. The frequently studied variants associated with alcoholism in DRD2, ADH1C, OPRM1 and COMT demonstrate insignificant frequency differences in SAGE (p>.05, Table 2). Although several studies implicate a biological role of these variants in alcoholism (Blum et al., 1990; Bond et al., 1998; Hendershot et al., 2011; Ponce et al., 2008; Thomasson et al., 1991; Tiihonen et al., 1999; Tolstrup et al., 2008; Zhang et al., 2006), our results reveal that these variants are not strongly associated with alcoholism in European and African ancestry populations. The only candidate that modestly replicated in SAGE, rs279858 in GABRA2, had a p-value of 0.0052 (OR=1.572). This finding was anticipated because a previous GWA study on the SAGE dataset demonstrated a similar association (Bierut et al., 2010). The replication of rs279858 in SAGE provides some support for future studies focused on the function of this variant and associated variants in GABRA2 (Edenberg et al., 2004).
When examined globally, none of the well-studied candidate genes demonstrate impressive variant differences between cases and controls. More specifically, only one SNP reported in SAGE (rs925946 upstream of BDNF, p=0.0006) in the 54 candidate genes had a p value less than 0.0009, a corrected p value for the number of genes (.05/54= 0.0009). Additionally, the overall number of variants with p<.05 and p<.01 is close to that predicted by chance considering the total number of SNPs examined in all proposed candidate genes. Although the individual p values for variants in the examined candidate genes are modest, a few candidate genes have a large portion of SNPs with p<.05 (Table 3). The results support further research into whether GABRA2, which was the candidate gene with the largest proportion of SNPs with p< .05 (55%), contributes to risk of developing alcohol dependence. BDNF and HTR1A also had more than one fifth of SNPs with p<.05, indicating that these genes merit further investigation to elucidate their potential contribution to alcohol dependence.
Lack of replication in SAGE does not exclude the possibility that some previously proposed candidate genes and specific loci are biologically important. Several of the most well studied candidate loci for alcoholism were not available in SAGE, including rs671 in ALDH2, rs1229984 in ADH1B, rs4795541 in SLC6A4 and rs3813867 in CYP2E1. A recent study that specifically genotyped rs1229984 in SAGE reported that the minor allele has a significant protective effect on alcohol dependence (p=6.6× 10−10) (Bierut et al., 2011). Because rs1229984 is common in Asians but rare in European Americans, this variant in ADH1B was not genotyped in the original GWA study. This highlights that GWA studies may miss important variants because of lack of coverage of SNPs that are uncommon in European American populations. Additionally, GWA studies cannot assess all forms of inheritance that can be associated with candidate genes such as insertion/deletion mutations, copy number repeats and epigenetic changes. Although SAGE is a valuable tool, it cannot exclude the possibility that aspects of genes contribute to genetic risk of alcohol dependence.
Even though the well studied candidate variants in DRD2, ADH1C, OPRM1 and COMT were not significantly associated with alcohol dependence in SAGE, their odds ratios were in the expected direction based on previous candidate gene studies. More specifically, the odds ratio of 0.088 for rs1799971 in OPRM1 supports previous studies that the minor allele variant is protective against alcohol dependence (Bond et al., 1998; Zhang et al., 2006) while the odds ratios greater than 1 for rs1800497 in DRD2, rs698 in ADH1C, and rs4680 in COMT supports previous studies that the minor allele of these variants are more common in alcohol dependent individuals (Blum et al., 1990; Hendershot et al., 2011; Ponce et al., 2008; Thomasson et al., 1991; Tiihonen et al., 1999; Tolstrup et al., 2008). The fact that these odds ratios are in the expected direction but did not pass a threshold of .05 for significance may suggest that these variants have a small contribution to alcohol dependence and this study lacked the power to detect the association.
Our study design had several strengths. First, the literature search for candidate genes included all genetic associations irrespective of ethnicity and criteria for alcoholism. By including all genes with the most genetic association study publications, we comprehensively examined previously identified genes associated with alcoholism in a large GWA study on alcoholism. Second, the SAGE dataset has the power to detect associations of small magnitude. SAGE included more than 3,800 subjects and had 90% power to detect a genetic variant with an odds ratio of 1.25 for a risk locus with 10% minor allele frequency. Third, our findings in SAGE regarding the well-characterized loci were found to be very similar to the results in the three independent datasets that contributed to SAGE, which indicates that there is no heterogeneity across these datasets. Fourth, our approach used data that is available to the scientific community and can be easily replicated in future studies of other phenotypes.
Despite these strengths, the selection of candidate loci and genes based on number of publications retrieved by the HuGE Navigator Phenopedia has some limitations. One limitation is that no data suggests that the potential significance of a given gene is directly proportional to the number of publications. Despite this, we felt that the number of publications is an indicator of research efforts devoted to a given gene. By selecting genes with the most publications, we sought to capture well-studied genes that had been the focus of the field in the past. A second potential limitation is that we did not exclude publications based on the same datasets. Because we used a low threshold of greater than 5 publications in the initial analysis, however, we are confident that we did not exclude any genes that have been examined in many studies. Additionally, the most well-studied loci of the ten genes with the most publications were selected based on expert opinion and were felt to be unambiguously widely studied even if the exact order may not be reflective of the number of data sets published on the genes.
Beyond limitations in our selection of candidate genes, the SAGE dataset has limitations that restrict the interpretation of our results. First, some of the most well studied variants were not covered in SAGE and therefore could not be assessed. Second, the X chromosome is not included in the publically available SAGE results so we were unable to investigate genes on the X chromosome. Specifically, two candidate genes on the X chromosome, MAOA and HTR2C that had 26 and 8 publications respectively, were not assessed. Third, SAGE is limited in its power to identify genotyped variants on the GWA chip that have small effect sizes. Despite the fact that the SAGE dataset was relatively large when it was originally published, identifying common variants with small effect sizes (<1.1) remains challenging and we are unable to rule out the possibility of real but modest effects of these genes. Forth, variants that are uncommon (1%-5%) or rare (<1%) in the study population may also not be detected in SAGE because of their individually small contribution to overall alcoholism. Fifth, the SAGE dataset primarily consists of European Americans (69.5%), African Americans (30.3%) and a few Hispanics (3.4%) (Table 1) and association findings may be different in other populations such as Asians. Some of the genes and variants examined in this analysis are more well studied and have a higher frequency in Asian Ancestry than in European and African Ancestry populations, such as the Asp40 allele of the candidate variant rs1799971 in OPRM1 (Arias et al., 2006), and therefore may have a more impressive effect in studies that focus on Asian ancestry populations. Sixth, our analysis did not examine the effects of combinations of genes or the effect of different environmental factors. Analysis of multiple genes and populations enriched for specific environmental risk factors will likely explain a greater degree of the genetic risk of alcoholism. Despite these limitations, this analysis demonstrates that GWA studies are a powerful technique for verifying the importance of genes and particular variants that have been previously identified in the candidate gene era.
In summary, we provide evidence that for alcohol dependence, several extensively studied candidate loci and genes are not replicated in a large GWA study, indicating that these variants do not individually have a large contribution to risk of developing alcohol dependence in European and African ancestry populations. Our analysis was unable to rule out the possibility that some variants and genes are important for risk of alcoholism due to lack of coverage. Recent work demonstrates that at least one highly reported variant rs1229984 in ADH1B that is not reported in SAGE is significantly associated with alcoholism (Bierut et al., 2011), suggesting the possible importance of further research on highly supported variants that cannot be assessed in SAGE. Our approach may also have missed variants that have a real but small individual contribution to overall inheritance of alcoholism.
This analysis demonstrates that targeted candidate gene studies and GWA studies each provide important information and studying the convergence of these two experimental designs has the potential to advance understanding of the etiology of alcohol dependence and more generally complex diseases. While GWA studies provide important information about the genetic contribution of common variants to complex diseases across populations, hypothesis driven candidate gene studies are also important to assess variants of lesser significance that may be missed because of the strict p value thresholds required for the large number of comparisons in GWA studies. Incorporating knowledge from both GWA and candidate gene studies will help clarify the role of genetics in complex disease and guide future research.
Our study also shows how the HuGE Navigator and dbGaP databases can be used as tools by researchers to easily access and analyze information on candidate genes and GWA data. Beyond alcoholism, the HuGE Navigator provides an easy way for investigators to search over 2,000 diseases and 10,000 genes for summary information and primary articles about genetic associations and human genome epidemiology (Yu et al., 2008). The dbGaP database provides access to results of over 100 studies examining phenotype and genotype associations, including 40 GWAS studies on different diseases. Since dbGaP currently contains a limited number of GWA studies, researchers examining phenotypes not available in dbGaP may benefit from directly contacting the authors of relevant GWA studies and meta analysis. Because of this easy accessibility, researchers who intend to perform future candidate gene studies should reference the HuGE navigator to assess background information and use dbGaP and existing GWA data to test whether their gene of interest is replicated in GWA data. Candidate gene studies need replication to meet scientific standards. Simple dbGaP analyses may help to focus future research on genes that are supported by GWA data and therefore more likely to be biologically important for human disease.
ACKNOWLEDGEMENTS
We thank Nancy Saccone for comments and criticisms and Louis Fox for technical assistance. This work was supported by a grant from the National Institute on Drug Abuse K02 DA021237 to Laura J Bierut. Emily Olfson was supported by T32 GM007200: National Research Service Award Medical Scientist Training Grant from the National Institute of General Medical Sciences.
This work was supported by K02 DA021237 from the National Institute on Drug Abuse (PI: L. Bierut), U01 HG004422: Study of Addiction: Genetics and the Environment (PI: L. Bierut) from the National Human Genome Research Institute, and U10 AA008401: The Collaborative Study on the Genetics of Alcoholism (PI: L. Bierut) from the National Institute on Alcohol Abuse and Alcoholism. Emily Olfson was supported by T32 GM007200: National Research Service Award Medical Scientist Training Grant from the National Institute of General Medical Sciences (PI: W. Yokoyama).
REFERENCES
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83(3):262–8. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI, Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107(11):5082–7. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, Briggs AH, Cohn JB. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA. 1990;263(15):2055–60. [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, Gong J, Schluger J, Strong JA, Leal SM, Tischfield JA, Kreek MJ, Yu L. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr., Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–58. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr., O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34(5):840–52. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Kranzler HR, Yu Y, Weiss RD, Brady KT, Anton R, Cubells JF, Gelernter J. Association of variants in MANEA with cocaine-related behaviors. Arch Gen Psychiatry. 2009;66(3):267–74. doi: 10.1001/archgenpsychiatry.2008.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27(6):1381–96. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW. A Quantitative-Trait Genome-Wide Association Study of Alcoholism Risk in the Community: Findings and Implications. Biol Psychiatry. 2011 doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot CS, Lindgren KP, Liang T, Hutchison KE. COMT and ALDH2 polymorphisms moderate associations of implicit drinking motives with alcohol use. Addict Biol. 2011 doi: 10.1111/j.1369-1600.2010.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Enoch MA, Srivastava V, Cummins-Oman JS, Ferrier C, Iarikova P, Sankararaman S, Yamini G, Yuan Q, Zhou Z, Albaugh B, White KV, Shen PH, Goldman D. Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proc Natl Acad Sci U S A. 2010;107(19):8695–700. doi: 10.1073/pnas.0908134107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslyn G, Ravindranathan A, Brush G, Schuckit M, White RL. Human variation in alcohol response is influenced by variation in neuronal signaling genes. Alcohol Clin Exp Res. 2010;34(5):800–12. doi: 10.1111/j.1530-0277.2010.01152.x. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopik VS, Heath AC, Madden PA, Bucholz KK, Slutske WS, Nelson EC, Statham D, Whitfield JB, Martin NG. Genetic effects on alcohol dependence risk: re-evaluating the importance of psychiatric and other heritable risk factors. Psychol Med. 2004;34(8):1519–30. doi: 10.1017/s0033291704002922. [DOI] [PubMed] [Google Scholar]
- Le Strat Y, Ramoz N, Pickering P, Burger V, Boni C, Aubin HJ, Ades J, Batel P, Gorwood P. The 3′ part of the dopamine transporter gene DAT1/SLC6A3 is associated with withdrawal seizures in patients with alcohol dependence. Alcohol Clin Exp Res. 2008;32(1):27–35. doi: 10.1111/j.1530-0277.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeidat M, Wain LV, Shrine N, Kalsheker N, Soler Artigas M, Repapi E, Burton PR, Johnson T, Ramasamy A, Zhao JH, Zhai G, Huffman JE, Vitart V, Albrecht E, Igl W, Hartikainen AL, Pouta A, Cadby G, Hui J, Palmer LJ, Hadley D, McArdle WL, Rudnicka AR, Barroso I, Loos RJ, Wareham NJ, Mangino M, Soranzo N, Spector TD, Glaser S, Homuth G, Volzke H, Deloukas P, Granell R, Henderson J, Grkovic I, Jankovic S, Zgaga L, Polasek O, Rudan I, Wright AF, Campbell H, Wild SH, Wilson JF, Heinrich J, Imboden M, Probst-Hensch NM, Gyllensten U, Johansson A, Zaboli G, Mustelin L, Rantanen T, Surakka I, Kaprio J, Jarvelin MR, Hayward C, Evans DM, Koch B, Musk AW, Elliott P, Strachan DP, Tobin MD, Sayers I, Hall IP. A comprehensive evaluation of potential lung function associated genes in the SpiroMeta general population sample. PLoS One. 2011;6(5):e19382. doi: 10.1371/journal.pone.0019382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce G, Hoenicka J, Jimenez-Arriero MA, Rodriguez-Jimenez R, Aragues M, Martin-Sune N, Huertas E, Palomo T. DRD2 and ANKK1 genotype in alcohol-dependent patients with psychopathic traits: association and interaction study. Br J Psychiatry. 2008;193(2):121–5. doi: 10.1192/bjp.bp.107.041582. [DOI] [PubMed] [Google Scholar]
- Siontis KC, Patsopoulos NA, Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18(7):832–7. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48(4):677–81. [PMC free article] [PubMed] [Google Scholar]
- Tiihonen J, Hallikainen T, Lachman H, Saito T, Volavka J, Kauhanen J, Salonen JT, Ryynanen OP, Koulu M, Karvonen MK, Pohjalainen T, Syvalahti E, Hietala J. Association between the functional variant of the catechol-O-methyltransferase (COMT) gene and type 1 alcoholism. Mol Psychiatry. 1999;4(3):286–9. doi: 10.1038/sj.mp.4000509. [DOI] [PubMed] [Google Scholar]
- Tolstrup JS, Nordestgaard BG, Rasmussen S, Tybjaerg-Hansen A, Gronbaek M. Alcoholism and alcohol drinking habits predicted from alcohol dehydrogenase genes. Pharmacogenomics J. 2008;8(3):220–7. doi: 10.1038/sj.tpj.6500471. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(7):773–84. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Clyne M, Khoury MJ, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26(1):145–6. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40(2):124–5. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, Zvartau E, Gelernter J. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15(6):807–19. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Jr., Rice JP, Schuckit MA, Foroud T, Edenberg HJ, Porjesz B, Almasy L. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156(1):44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]