Dear Editor:
Breast cancer is a multifactorial disease. It is the first cause of mortality per cancer for the woman in the world (Jemal et al., 2011). The implication of specific genes, such as BRCA1 and BRCA2 tumor suppressor genes, has been shown in mammary carcinogenesis. In sporadic breast cancers, specific modifications of BRCA1 and BRCA2 mRNA expression have been reported too (Bernard-Gallon et al., 1999; Bieche et al., 1999). Epigenetic modifications such as promoter hypermethylation of the oncosuppressor genes BRCA1 and BRCA2 can play a role in the oncogenesis of cancer (Esteller et al., 2000). Indeed, hypermethylation of the CpG islands in the promoters of these genes involve their inactivation and therefore a higher risk of developing a tumor (Baylin and Ohm, 2006; Moelans et al., 2011; Rahmatpanah et al., 2009).
We have compared the DNA methylation rates of the BRCA1 and BRCA2 gene promoters using DNA isolated from blood samples from COSA (Cancer d'Ovaire et du Sein en Auvergne) patients suffering from breast or ovarian cancer and from a population of healthy women. To this aim, the QAMA method (Quantitative Analysis of Methylated Alleles) was used (Zeschnigk et al., 2004) and adapted to BRCA1 and BRCA2 genes (Bosviel et al., 2011). This method is based on bisulfite conversion of the nonmethylated cytosines and an analysis by qPCR using Taqman minor groove binder probes specific for methylated or nonmethylated target sites after conversion. Percentage of methylation is then obtained by calculating the difference between CT values of the methylated DNA targeting probe and the nonmethylated DNA targeting probe (ΔCT) and reporting the obtained values on a standard curve.
In a previous work, we reported that BRCA1 methylation is significantly decreased in ovarian cancer by comparison with the control group. The comparison between the two different populations did not show any significant difference regarding BRCA2 methylation but exhibited a trend in the decrease of BRCA2 promoter methylation in peripheral blood DNA of sporadic ovarian cancer (Bosviel et al., 2011).
Then we demonstrated a trend toward BRCA1 promoter hypermethylation in PBCs of sporadic breast cancer patients by comparison with controls (Bosviel et al., 2012). BRCA1 promoter methylation in PBCs corresponded to 47.1% with CI 95% [46.1; 48.1] in breast cancer patients and to 45.9% with CI 95% [45.0; 46.8] in controls. Association between methylation level and clinicopathological features were evaluated using statistical tests. BRCA1 promoter methylation in PBCs increased significantly in breast cancer patients by comparison with controls, with the age over 70 years old (p=0.022), in post menopausal status (p=0.013), with a BMI<20 (p=0.0095), or with a WHR≤76 .8 (p=0.0027). We also found an association of increased BRCA1 promoter methylation in PBCs with ACA/ACA genotype for the SNP Thr594Thr in ESR (estrogen receptor), known to be associated with breast cancer risk (p=0.092), due to the reduced presence of this genotype in this breast cancer case-control study.
Within this study, the objective was to compare the methylation of the CpG islands present in the BRCA2 promoter in the same population of women suffering from breast cancer compared to the control population. In total, 873 breast samples belonging to COSA and 980 control samples were converted and the methylation rates measured. The complete database of this study is available as supplementary data (supplementary data are available online at www.liebertonline.com/omi).
BRCA2 promoter methylation mean in PBCs is 16.9% (CI95% [16.3; 17.4]) in breast cancer patients and 16.2% (CI95% [15.7; 16.8]) in controls. The statistical analysis of the mean methylation rates obtained for the BRCA2 promoter did not reveal a significant difference (p=0.1) between the two populations (Fig. 1).
FIG. 1.
Average percentage of methylation of the promoter of BRCA2 in DNA from control patients and COSA (p=0.10).
Significant differences in methylation rates between patients and healthy women were, however, obtained for different subclasses (Table 1). It reached 17.3% in breast cancer patients older than 70 years by comparison with control patients (14.7%) with p=0.016. In the subclass with an early menopause (before 48 years), the level was respectively 16.6% in breast cancer patients versus 15.4% in control patients (p=0.028).
Table 1.
Demographics of Breast Cancer Patients and Controls and Their Association with BRCA2 Promoter Methylation in Peripheral Blood Cells
| |
Breast Cancer Patients |
Control |
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number 873 | Average 16.9 | SD 8.0 | 95%CI 16.3–17.4 | Comparison in Category | Number 980 | Average 16.2 | SD 8.1 | 95%CI 15.7–16.8 | Comparison in Category | Patients/Controls Comparisonp=0.10 | |
| Age | |||||||||||
| <50 years | 252 | 17.2 | 7.6 | 16.3–18.1 | 142 | 16.8 | 8.2 | 15.4–18.1 | p=0.39 | ||
| [50–60[ years | 241 | 16.6 | 7.8 | 15.6–17.6 | p=0.66 | 437 | 16.5 | 8.0 | 15.7–17.2 | p=0.23 | p=0.57 |
| [60–70[ years | 221 | 16.4 | 7.9 | 15.4–17.5 | 304 | 16.2 | 8.3 | 15.2–17.1 | p=0.45 | ||
| ≥70 years | 159 | 17.3 | 8.8 | 16.0–18.7 | 97 | 14.7 | 7.5 | 13.2–16.2 | p=0.016 | ||
| Menopausal status | |||||||||||
| Premenopausal | 265 | 17.1 | 7.8 | 16.2–18.0 | 246 | 16.3 | 8.0 | 15.3–17.3 | p=0.18 | ||
| Postmenopausal | 552 | 16.7 | 8.0 | 16.1–17.4 | p=0.52 | 734 | 16.2 | 8.1 | 15.6–16.8 | p=0.72 | p=0.24 |
| Menopausal Age | |||||||||||
| ≤48 years | 170 | 16.6 | 7.3 | 15.5–17.7 | 196 | 15.4 | 8.4 | 14.2–16.6 | p=0.028 | ||
| ]48–50] years | 117 | 16.9 | 7.6 | 15.5–18.3 | p=0.63 | 186 | 16.7 | 8.6 | 15.5–18.0 | p=0.076 | p=0.45 |
| ]50–53] years | 136 | 15.6 | 7.6 | 14.3–16.8 | 191 | 15.8 | 7.5 | 14.8–16.9 | p=0.72 | ||
| >53 years | 123 | 18.0 | 9.4 | 16.4–19.7 | 159 | 17.2 | 7.8 | 16.0–18.4 | p=0.62 | ||
| BMI | |||||||||||
| <20 | 97 | 16.9 | 8.1 | 15.3–18.5 | 134 | 15.4 | 8.1 | 14.0–16.7 | p=0.094 | ||
| [20–25[ | 394 | 17.6 | 8.1 | 16.8–18.4 | p=0.091 | 541 | 16.5 | 8.0 | 15.8–17.2 | p=0.29 | p=0.019 |
| [25–30[ | 216 | 15.9 | 7.5 | 14.9–16.9 | 220 | 16.4 | 8.4 | 15.3–17.5 | p=0.82 | ||
| ≥30 | 106 | 16.3 | 7.6 | 14.9–17.8 | 85 | 15.5 | 7.8 | 13.9–17.2 | p=0.48 | ||
| WHR | |||||||||||
| ≤76.8 | 105 | 17.2 | 8.4 | 15.6–18.8 | 320 | 17.1 | 8.6 | 16.2–18.1 | p=0.74 | ||
| ]76.8–81.7] | 163 | 16.7 | 7.4 | 15.6–17.9 | p=0.83 | 266 | 15.7 | 7.6 | 14.8–16.6 | p=0.052 | p=0.13 |
| ]81.7–87.5] | 196 | 17.1 | 7.6 | 16.0–18.2 | 238 | 15.4 | 7.8 | 14.4–16.4 | p=0.0046 | ||
| >87.5 | 272 | 16.6 | 8.0 | 15.7–17.6 | 144 | 16.9 | 8.4 | 15.6–18.3 | p=0.80 | ||
BMI, Body Mass Index; WHR, Waist-to-Hip Ratio; 95%CI, 95% Confidence Interval; SD, Standard deviation.
For 2 populations with a Gaussian distribution. Student τ test was used for equal variance population; otherwise, Z-score test (n≥30) was used. To compare more than 2 populations, variance analysis was performed. Finally, Kruskal-Wallis test was performed for non-Gaussian population comparisons.
Then, breast cancer patients showing a normal BMI (Body Mass Index) [20–25[ exhibited a BRCA2 promoter methylation in PBCs of 17.6% versus 16.5% for control patients (p=0.019).
Concerning the increase in the WHR (Waist-to-Hip Ratio) ]81.7%–87.5%], the BRCA2 promoter methylation in PBCs was 17.1% in COSA patients by comparison to control patients (15.4%) with p=0.0046. So, an android distribution of fat tissue revealed a higher methylation rate of the BRCA2 promoter region.
Conversely, relationship between clinicopathological characteristics of tumors in subclasses and BRCA2 promoter methylation in PBCs of breast cancer patients demonstrated no statistical significant difference for any of the analyzed clinical parameters (Table 2).
Table 2.
Relationship Between Clinicopathological Characteristics of Tumors and BRCA2 Promoter Methylation in PBCs of Breast Cancer Patients
| |
Breast Cancer Patients |
||||
|---|---|---|---|---|---|
| Number 873 | Average 16.9 | SD 8.0 | 95%CI 16.3–17.4 | Comparison in the Categorya | |
| SBR degrees | |||||
| 1 | 244 | 17.1 | 8.9 | 15.9–18.2 | p=1.00 |
| 2 | 290 | 16.7 | 8.0 | 15.8–17.6 | |
| 3 | 129 | 16.5 | 7.2 | 15.3–17.8 | |
| Histological type | |||||
| IDC | 599 | 16.7 | 7.9 | 16.1–17.4 | p=0.80 |
| ILC | 105 | 17.1 | 7.1 | 15.7–18.4 | |
| other | 168 | 17.0 | 8.7 | 15.7–18.3 | |
| Tumor size | |||||
| Infraclinic | 127 | 16.9 | 7.5 | 15.6–18.2 | p=0.92 |
| <2 cm | 157 | 16.2 | 7.7 | 15.0–17.4 | |
| 2–5 cm | 150 | 16.2 | 6.6 | 15.2–17.3 | |
| >5 cm | 26 | 16.7 | 8.2 | 13.4–20.0 | |
| Epidermal invasion | 26 | 16.8 | 9.3 | 13.2–20.4 | |
| Lymph node metastasis | |||||
| N0 | 306 | 16.5 | 7.1 | 15.7–17.3 | p=0.20 |
| N1–N2 | 111 | 15.8 | 7.9 | 14.3–17.3 | |
95%CI, 95% confidence interval; SD, standard deviation; SBR, Scarff-Bloom-Richardson; IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; N0, no invasive lymph node, and N1–N2, invasive lymph nodes. aStudent t test.
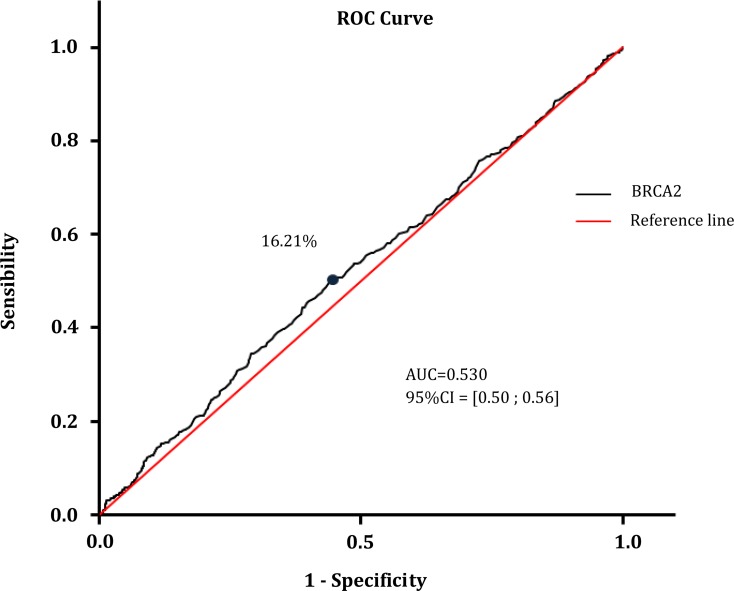
Sensibility and specificity of the blood-based assay to distinguish breast cancer cases from controls, using BRCA2 methylation, were calculated. Reliability of the methylation of a gene such as BRCA2 to serve as a sensitive and specific blood-based breast cancer test was assessed with significant different methylation patterns between cancerous and normal PBCs. To evaluate the applicability of BRCA2 methylation in circulating DNA from PBCs as a marker for breast cancer, receiver operating characteristic (ROC) curve analysis was used (Fig. 2). The best cut-off point (16.21% of methylation) and the corresponding sensitivity (52.3%), specificity (55.8%), area under curve (AUC=0.530) and confidence interval were calculated (Table 3). The positive predictive value (PPV) was 51.4% and negative predictive value (NPV) was 56.9% in case of a cut-off point at 16.21% for the methylation. The Chi2 test (12.68) between the risk and the disease was found significant (p=0.00037). The Relative Risk (RR=1.19; 95% CI [1.08; 1.31] ) and the Odd Ratio (OR=1.39; 95% CI [1.16; 1.67]) to develop the disease by comparison with the reference fixed at 1, were calculated and were found included in their respective asymptotic 95% CI.
FIG. 2.
ROC curve analysis using DNA samples extracted from PBCs for discriminating between breast cancer patients and normal subjects based on methylation pattern of the BRCA2 gene.
Table 3.
ROC Curve Analysis of DNA Samples Extracted from PBCs Based on Methylation Proportion of the BRCA2 Gene
| AUC* | 0.53 |
| Sensibility | 52.50% |
| Specificity | 55.80% |
| Positive predictive value | 51.40% |
| Negative predictive value | 56.90% |
| Chi2 | 12.68 |
| p Chi2 | 0.00037 |
| Relative Risk (RR) | 1.19 |
| Asymptotic 95% CI** | [1.08; 1.31] |
| Odd Ratio (OR) | 1.39 |
| Asymptotic 95% CI | [1.16; 1.67] |
AUC, area under the curve; **CI, confidence interval (lower-upper bound).
The presenting data are promising for development of a blood-based screening method for breast cancer that relies on pathologic methylation changes, corroborating other results with different genes (Radpour et al., 2011). Further studies comparing tissue specific and blood-based methylation markers might provide valuable information as prognostic and predictive markers for breast cancer, as well as for developing novel targeted therapeutic strategies.
Rémy Bosviel, Julie Durif, Jiaoli Guo,
Mourad Mebrek, Fabrice Kwiatkowski,
Yves-Jean Bignon, and
Dominique J. Bernard-Gallon
Supplementary Material
Acknowledgments
This study was supported by “La Ligue Nationale Française de Lutte Contre le Cancer” (Puy-de-Dôme, Allier and Cantal). Rémy Bosviel is a recipient of a grant from the Auvergne Regional Council/CPER 2008+FEDER n°32316 – 0930FDBG – 106NL.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Baylin SB. Ohm JE. Epigenetic gene silencing in cancer. A mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bernard-Gallon DJ. De Latour MP. Rio PG, et al. Subcellular localization of BRCA1 protein in sporadic breast carcinoma with or without allelic loss of BRCA1 gene. Int J Oncol. 1999;14:653–661. doi: 10.3892/ijo.14.4.653. [DOI] [PubMed] [Google Scholar]
- Bieche I. Nogues C. Lidereau R. Overexpression of BRCA2 gene in sporadic breast tumours. Oncogene. 1999;18:5232–5238. doi: 10.1038/sj.onc.1202903. [DOI] [PubMed] [Google Scholar]
- Bosviel R. Garcia S. Lavediaux G, et al. BRCA1 promoter methylation in peripheral blood DNA was identified in sporadic breast cancer and controls. Cancer Epidemiol. 2012;36:e177–182. doi: 10.1016/j.canep.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Bosviel R. Michard E. Lavediaux G. Kwiatkowski F. Bignon YJ. Bernard-Gallon DJ. Peripheral blood DNA methylation detected in the BRCA1 or BRCA2 promoter for sporadic ovarian cancer patients and controls. Clin Chim Acta. 2011;412:1472–1475. doi: 10.1016/j.cca.2011.04.027. [DOI] [PubMed] [Google Scholar]
- Esteller M. Silva JM. Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Jemal A. Bray F. Center MM. Ferlay J. Ward E. Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Moelans CB. Verschuur-Maes AH. Van Diest PJ. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol. 2011;225:222–231. doi: 10.1002/path.2930. [DOI] [PubMed] [Google Scholar]
- Radpour R. Barekati Z. Kohler C, et al. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PLoS One. 2011;6:e16080. doi: 10.1371/journal.pone.0016080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmatpanah FB. Carstens S. Hooshmand SI, et al. Large-scale analysis of DNA methylation in chronic lymphocytic leukemia. Epigenomics. 2009;1:39–61. doi: 10.2217/epi.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M. Bohringer S. Price EA. Onadim Z. Masshofer L. Lohmann DR. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): Analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.