Abstract
Glioblastoma is the most common and aggressive primary brain tumor. MicroRNAs (miRNAs) are a set of noncoding RNA of about 20∼22 nt in length and they play regulatory roles such as regulating the expression of proteins. Altered miRNA expression is related to cancers, including glioblastoma. In this report, we used deep sequencing to explore the miRNA profiles of glioblastoma and normal brain tissues. We found 875 and 811 known miRNA and miRNA* in glioblastoma and normal brain tissue, respectively, representing the largest characterization of the miRNAs in GBM so far. 33 of them were upregulated in glioblastoma, including miR-21, which is well known as an oncomir, while 40 of them were downregulated. Using miR-10b, miR-124, miR-433, and miR-92b as examples, we verified the data by quantitative RT-PCR, suggesting that deep sequencing was able to capture the expression profiles of miRNAs. In addition, we found 18 novel miRNA and 16 new miRNA* in glioblastoma and normal brain tissues. This report provides a useful resource for future studies of the roles of miRNAs in the pathogenesis and early detection of glioblastoma.
Introduction
Glioma is the most common primary tumor of the central nervous system, comprising over 50% of primary brain tumors. The annual incidence of malignant gliomas is approximately 5 cases per 100,000 people (Wen and Kesari, 2008). Glioblastoma (GBM) is the most common and deadliest glioma. Due to its rapid growth and highly infiltrative nature, the median survival is approximately 14 months, even with aggressive surgery, radiation, and chemotherapy (Davis and McCarthy, 2001). An incomplete knowledge of the molecular biology, genetics, causes, and cellular origin of glioblastoma limits the development of improved therapeutics. The deep understanding of the molecular basis of glioblastoma and the search for molecular markers for early detection are fundamental to develop targeted treatments and to improve the outcome for glioblastoma.
miRNAs are a novel group of short RNAs, about 20∼22 nucleotide in length, which regulate gene expression through binding to the 3′UTR of target mRNAs, thereby either targeting the transcripts for degradation or blocking translation of the encoded proteins (Bartel, 2004). In some special condition, miRNA can also stimulate translation (Vasudevan et al., 2007). A pre-miRNA stem-loop can produce two mature miRNAs, one from the plus strand and one from the minus strand (miRNA*). In most cases, the miRNA* sequence is degraded, leaving the mature sequence to be incorporated into the RISC complex and to target mRNAs in a sequence-specific manner through the RNA interference pathway (He and Hannon, 2004). Sometimes, miRNA* activity can also contribute to the vertebrate miRNA targeting network (Yang et al., 2011). It has been predicted that about one-third of all mammalian genes were targets of miRNAs (Lewis et al., 2005). miRNAs can also be used for cancer classification (Lu et al., 2005) and detection (Mitchell et al., 2008), or as therapeutic targets (Kota et al., 2009).
Previously, miRNA arrays have been used to identify differentially expressed known miRNAs in GBM. For example, Ciafre et al. (2005) studied by microarray the global expression levels of 245 microRNAs in GBM and found that miR-221 was strongly upregulated in glioblastoma and miR-128, miR-181a, miR-181b, and miR-181c were downregulated in glioblastoma. Loftus et al. (2012) used miRNA arrays to analyze the miRNA expression profiles in matched populations of migrating cells versus migration-restricted cells in seven well-established glioma cell lines, and identified miR-23b as a key regulator of cell migration and invasion for GBM. The Cancer Genome Atlas (TCGA) also conducted miRNA array analysis of GBM (Network, 2008). Srinivasan et al. (2011) used the TCGA dataset and identified a ten-microRNA expression signature that predicts survival in glioblastoma.
With the advance of the next generation sequencing, Skalsky and Cullen (2011) used miRNA-seq to profile the miRNA expression profiles in GBM and nontumor brain tissues, and they identified 484 miRNAs. In this study, we used deep sequencing to profile the expression of miNRA in three glioblastoma and three normal brain tissues. We sequenced a total of 16.6 millions of sequences and identified 885 known miRNA and miRNA* found in at least two libraries. In addition, we also found isomiRs, novel miRNAs, and miRNA*. This represents, to our knowledge, the largest catalogue of miRNAs for brain and glioma tissues at this moment.
Materials and Methods
Tissue sample collection and RNA isolation
Tissue samples were obtained after informed consent from adult patients diagnosed with glioblastoma de novo or other nontumor illness, freshly resected during surgery and immediately frozen in liquid nitrogen and then stored at −80°C for subsequent total RNA extraction. For the initial miRNA sequencing analysis, two samples (one GBM and one non-GBM brain tissue) were obtained from the Swedish Medical Center, Seattle, WA, and four samples (two GBMs and two non-GBM brain tissues) were collected from the Second Affiliated Hospital of Soochow University. For RT-PCR confirmation, additional samples were collected from the Second Affiliated Hospital of Soochow University. All patients gave informed consent prior to collection of specimens according to institutional guidelines. RNA was isolated from tissues using mirVana™ miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer's instructions. The RNA integrity was evaluated by Agilent 2100 BioAnalyzer.
Small RNA library construction and sequencing
For small RNA sequencing library construction, approximately 10 μg of total RNA was used and all the procedures were done according to Illumina miRNA sample preparation protocol. Briefly, RNA was purified by polyacrylamide gel electrophoresis (PAGE) to enrich molecules in the range of 16–30 nt, and proprietary adapters were ligated to the 5′ and 3′ terminal of the RNA. The ligated RNAs were reverse transcribed to cDNAs. The cDNA was amplified for 10 cycles by PCR to produce sequencing libraries. Small RNA libraries were sequenced using the high throughput sequencing technology developed by Illumina.
Sequence annotation and novel miRNA prediction
After filtering the adaptor and low quality sequences, we counted the occurrences of each unique sequence read and used only the unique sequences for further analysis. We aligned each sequence to the human miRNA precursor (Sanger miRBase 17.0) using BLAST (version 2.2.11). For every read, the longest perfect alignment with more than 18 bp was determined. According to the location of miRNA/miRNA* in miRNA precursor, we identified the sequence and added the count of sequence which matched the same miRNA/miRNA* as raw abundances. The sequence that matched precursor but did not belong to any miRNA/miRNA* in the database was identified as new miRNA*. The remaining sequences were mapped to human reference genome (NCBI build 36.1)(Guo et al., 2009), RepBase and Rfam (Gardner et al., 2010). The sequences that could be mapped to the human genome but did not have annotation information were used to detect candidate novel miRNA genes. In brief, 100 nucleotides of genomic sequence flanking each side of these sequences were extracted and the RNA secondary structures were predicted using RNAfold. Candidate novel miRNA was identified using the custom miRNA pipeline developed by Morin et al. (2008), who later converted this custom miRNA pipeline into a program called Novel miRNA detection 1.0 (http://www.bcgsc.ca/platform/bioinfo/software/novel-mirna-detection). We used the program with the default settings. In this program, two machine learning approaches were used: the first one, termed MiPred, relies on an RF algorithm (Jiang et al., 2007) that uses both structural and thermodynamic parameters, and the second one applies a SVM (support vector machine) classifier that uses parameters not used by the RF method. The intersection of the positive predictions from both methods was considered as reliable novel miRNA predictions.
Differential expression detection
The miRNA/miRNA* which had more than 3 tags in two samples of the six glioblastoma or normal tissues was used in the following analysis. Raw abundances in the libraries were normalized according to the formula: normalized abundance=raw abundance/total miRNA matched *1,000,000 (TPM: transcripts per million). Student t-test was used to measure the significance of differential expression. Sequences were deemed significantly differentially expressed if (1) the p value given by this method was <0.05, (2) the total count was greater than 50 TPM in at least one group of samples, and (3) there was at least a twofold change in sequence counts between the two groups.
Quantitative RT- PCR of miRNA
Differentially expressed miRNAs of hsa-miR-10b, miR-124, miR-433, and hsa-miR-92b were selected for validation. Expression of these mature miRNAs were assayed using stem-loop RT followed by PCR analysis as previously described(Chen et al., 2005). PCR was performed in triplicate for each sample. The relative amount of miRNA was normalized against U6 snRNA, and the fold change for each miRNA was calculated by the 2−ΔΔCt method.
Results
miRNA sequencing of glioblastoma and normal brain tissues
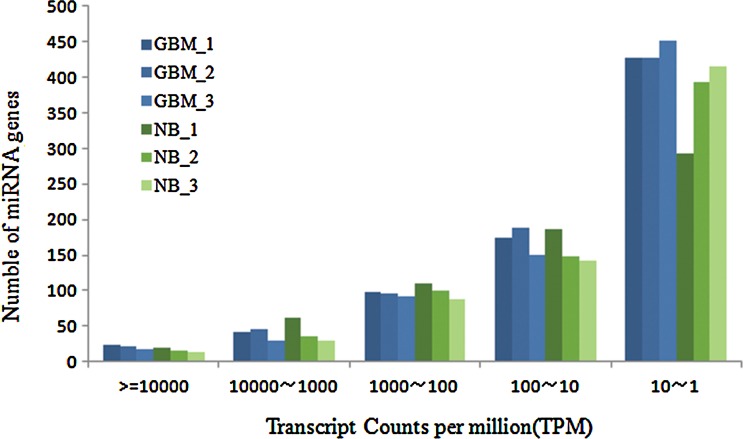
Small RNAs were isolated from three glioblastoma and three normal brain tissue samples and prepared for Illumina sequencing. After removing low quality and adaptor sequences, 1.2 to 3.8 millions sequencing reads of each sample were mapped to the human genome, totaling 16.6 million reads (Table 1). Most of them were mapping to miRNA precursor while some were mapping to mRNAs, repeat regions, tRNAs and unknown area. We found 885 known human miRNA and miRNA* in miRBase17.0, and they were expressed in at least 2 out of the 6 libraries. Among them, 875 were expressed in glioblastoma and 811 were expressed in normal brain tissue (Supplementary Table S1; Supplementary Data are available online at www.liebertonline.com/omi). The abundance value of each known miRNA was normalized using “transcripts per million (TPM)” in each small RNA library (see Methods). The abundance of miRNAs varied from several to hundreds of thousands TPM. Taking GBM tissue 1 as an example, 56.08% miRNAs were lowly expressed (<10 TPM), 40.92% miRNAs were expressed modestly (10–10,000 TPM), and only 3% miRNAs were expressed abundantly (>10,000 TPM) (Fig. 1, Supplementary Table S2).
Table 1.
Distributions of Sequence Counts for the Different RNA Classes
| GBM_1 | GBM_2 | GBM_3 | NB_1 | NB_2 | NB_3 | |
|---|---|---|---|---|---|---|
| miRNA | 2023066 | 2374826 | 3266646 | 1045787 | 2323031 | 3635038 |
| mRNA | 13726 | 39204 | 102034 | 12027 | 12934 | 16294 |
| Repeat | 5956 | 13955 | 10835 | 5770 | 6255 | 6086 |
| Unknown | 210246 | 463737 | 227200 | 143895 | 146902 | 170804 |
| scaRNA | 4950 | 6373 | 9658 | 5893 | 8755 | 11927 |
| snRNA | 669 | 3337 | 1881 | 776 | 899 | 1836 |
| tRNA | 10635 | 59317 | 29602 | 8049 | 11163 | 11642 |
| srpRNA | 161 | 526 | 211 | 27 | 65 | 95 |
| rRNA | 10610 | 28543 | 64404 | 3803 | 6163 | 11632 |
| scRNA | 10468 | 10969 | 10645 | 1479 | 7243 | 11488 |
| total | 2290487 | 3000787 | 3723116 | 1227506 | 2523410 | 3876842 |
FIG. 1.
The abundance of miRNA genes.
Differentially expressed miRNAs
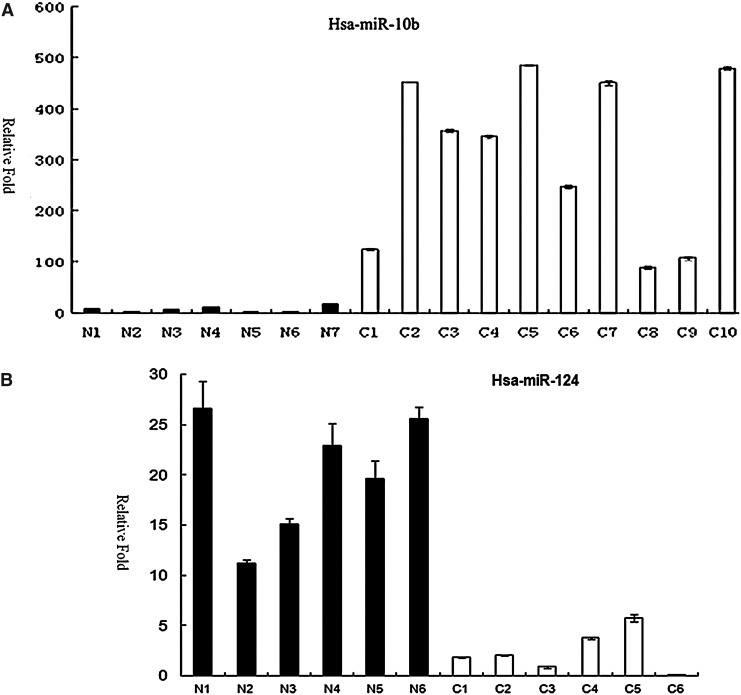
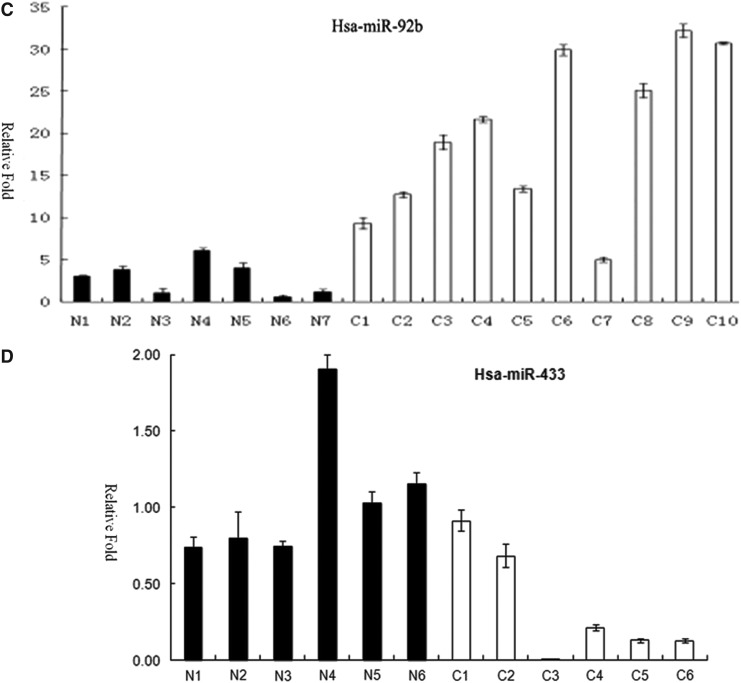
We found that 73 miRNAs were differentially expressed between glioblastoma and normal brain tissues (Supplementary Table S3). 33 of them were upregulated in glioblastoma, including miR-21 that is well known as an oncomir (Chan et al., 2005), while 40 of them were downregulated. Among these 73 miRNAs, 16 of them were also detected in Saklsky's study as differentially expressed (Skalsky and Cullen, 2011). Additional 11 of them were found in other previous studies (Ciafre et al., 2005; Ferretti et al., 2009; Godlewski et al., 2008; Silber et al., 2008; Srinivasan et al., 2011). 46 miRNAs were reported for the first time as differentially expressed between GBM and normal brain tissues, including has-miR-1, hsa-miR-10b, hsa-miR-92b, has-miR-124, and has-miR-433 (Table S3). The 10 most upregulated and 10 most downregulated miRNAs were shown in Table 2. We took hsa-miR-10b, hsa-miR-92b, has-miR-124, and has-miR-433 as examples to validate the result in another set glioblastoma and normal brain tissues. The QRT-PCR results were consistent with the sequencing data (Fig. 2).
Table 2.
Top 10 Upexpressed and Top 10 Downexpressed miRNAs Between Glioblastoma and Normal Brain Libraries
| |
Count |
|
|
|||||
|---|---|---|---|---|---|---|---|---|
| microRNA | GBM_1 | GBM_2 | GBM_3 | NB_1 | NB_2 | NB_3 | P value | GBM/NB |
| hsa-miR-10b | 166 | 336 | 550 | 0 | 2 | 0 | 0.0010 | 433.29 |
| hsa-miR-96 | 10 | 42 | 41 | 1 | 0 | 0 | 0.0021 | 97.79 |
| hsa-miR-10b* | 7 | 50 | 12 | 0 | 0 | 0 | 0.0094 | 68.24 |
| hsa-miR-182 | 16 | 10 | 46 | 2 | 0 | 0 | 0.0053 | 37.73 |
| hsa-miR-135a* | 60 | 24 | 28 | 1 | 1 | 1 | 0.0004 | 36.26 |
| hsa-miR-21* | 36 | 244 | 51 | 12 | 8 | 1 | 0.0384 | 15.34 |
| hsa-miR-21 | 10424 | 57525 | 7533 | 2517 | 2089 | 956 | 0.0313 | 13.57 |
| hsa-miR-542-3p | 47 | 322 | 195 | 27 | 13 | 6 | 0.0287 | 12.20 |
| hsa-miR-148a | 78 | 221 | 104 | 28 | 4 | 3 | 0.0201 | 11.63 |
| hsa-miR-92b | 3305 | 9797 | 1834 | 1165 | 470 | 209 | 0.0405 | 8.10 |
| hsa-miR-433 | 300 | 19 | 298 | 1616 | 1210 | 1482 | 0.0013 | 0.14 |
| hsa-miR-7-1* | 15 | 11 | 9 | 81 | 71 | 42 | 0.0026 | 0.18 |
| hsa-miR-129* | 39 | 10 | 32 | 499 | 348 | 377 | 0.0031 | 0.07 |
| hsa-miR-628-5p | 34 | 62 | 17 | 434 | 256 | 287 | 0.0048 | 0.12 |
| hsa-miR-935 | 27 | 9 | 27 | 550 | 254 | 175 | 0.0052 | 0.06 |
| hsa-miR-218 | 7 | 5 | 5 | 60 | 28 | 23 | 0.0053 | 0.15 |
| hsa-miR-31 | 32 | 55 | 32 | 656 | 226 | 218 | 0.0064 | 0.11 |
| hsa-miR-876-3p | 0 | 0 | 1 | 38 | 12 | 5 | 0.0083 | 0.02 |
| hsa-miR-1258 | 2 | 0 | 0 | 12 | 20 | 25 | 0.0093 | 0.05 |
| hsa-miR-132 | 23 | 29 | 26 | 137 | 57 | 89 | 0.0094 | 0.28 |
GBM/NB, glioblastoma compared to normal brain tissue.
FIG. 2.
Real-time PCR analysis of two upregulated and two downregulated miRNAs in a panel of normal brain tissues (the N series and indicated by black columns) and glioblastoma tissues (the C series and indicated by white columns). A. Upregulation of hsa-miR-10b in GBM compared with the normal brain tissues. B. Downregulation of hsa-miR-124 in GBM compared with the normal brain tissues. C. Upregulation of hsa-miR-92b in GBM compared with the normal brain tissues. D. Downregulation of hsa-miR-433 in GBM compared with the normal brain tissues. RNA input was normalized by human U6 snRNA. Y-axis, relative expression normalized to human U6 snRNA.
IsomiRs
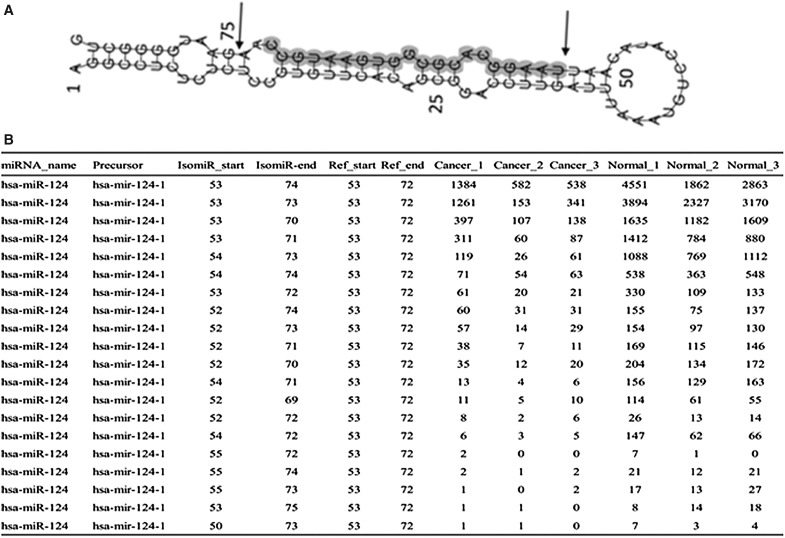
miRNAs are usually annotated as a single defined sequence. However, due to the imprecise and alternative cleavage of Dicer and Drosha during pre-miRNA hairpin processing, miRNAs frequently exhibit several length and/or sequence variants of the same miRNA. These variants, different from their reference mature sequence, are named isomiRs (Cloonan et al., 2012). Our data revealed that most of the mature miRNAs in the glioblastoma tissue and normal brain tissue samples retain isomiR sequence, such as hsa-miR-124 (Fig. 3). Among the 300 highest expressed miRNA, there are 190 miRNAs which the most abundance form of the isomiR is different from their mature reference miRNA, while only 110 miRNAs which the most abundant form of isomiR is the same as the mature reference miRNA (Supplementary Table S4). Notably, there are 23 miRNAs which the most abundant form of the isomiR have variations in their 5′ ends (Supplementary Table S5). These findings are of great importance and should be added to the annotations in the curated miRNA database.
FIG. 3.
The isomiRs of hsa-miR-124. (A) The second structure of hsa-miR-124-1. The sequence between two arrays is the most abundant form of hsa-miR-124-1 isomiR and the highlighted sequence is the mature reference of hsa-miR-124. (B) The different isomiRs of has-miR-124. *the most abundant form of isomiR; #the mature reference of has-miR-124; IsomiR-start, the start position of isomiR in has-miR-124-1 precursor; IsomiR-end, the end position of isomiR in hsa-miR-124-1 precursor; Ref-start, the start position of mature reference hsa-miR-124 in hsa-miR-124-1 precursor; Ref-end, the end position of mature reference hsa-miR-124 in hsa-miR-124-1 precursor.
Novel miRNA and new miRNA*
One of the important advantages of using next-generation sequencing approach to miRNA study is that it allows identification of novel miRNAs. Using Novel_miRNA_detection (Morin et al., 2008), which was widely adopted for identification of miRNAs from deep-sequencing datasets, we found 18 novel miRNA genes (Table 3). The length of these miRNA varies from 20 to 24, which is consistent with known human miRNAs.
Table 3.
Genomic Location, Sequence, MFE, and Expression Level of 18 Novel miRNA
| |
|
|
|
|
Count |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| miRNA ID | Genomic Location | Sequence | MFE | Length | GBM_1 | GBM_2 | GBM_3 | NB_1 | NB_2 | NB_3 |
| hsa-novel-miR-01 | chr1:33570637:33570659:- | GGCTTCCTTGCTATCCATCCTCA | −24.95 | 23 | 3 | 1 | 1 | 1 | 1 | 2 |
| hsa-novel-miR-02 | chr1:66866775:66866794:+ | ATAGGACTCATATAGTGCCA | −39.97 | 20 | 47 | 16 | 13 | 14 | 9 | 6 |
| hsa-novel-miR-03 | chr2:207683011:207683032:- | ATTGTCCATTGTATCTGGGGAT | −56.58 | 22 | 49 | 125 | 28 | 14 | 26 | 11 |
| hsa-novel-miR-04 | chr3:146732600:146732622:- | ATAAGAATTCTGAGAAGTGTCAG | −86.52 | 23 | 1 | 9 | 5 | 1 | 3 | 6 |
| hsa-novel-miR-05 | chr3:190015310:190015330:+ | CATTGCATTTGTCTCGGTCTTA | −17.75 | 21 | 6 | 1 | 37 | 1 | 6 | 5 |
| hsa-novel-miR-06 | chr4:93842214:93842234:+ | CCTCGATGTTGGATCAGGACA | −46.05 | 21 | 2 | 16 | 2 | 3 | 3 | 8 |
| hsa-novel-miR-07 | chr4:156604722:156604745:- | TATGTCCTGATCCAACATCGAGGTC | −33.47 | 24 | 6 | 10 | 4 | 5 | 8 | 6 |
| hsa-novel-miR-08 | chr5:31686080:31686100:+ | ACTCAATAAATGTCTGTTGAA | −45.76 | 21 | 2 | 1 | 4 | 3 | 2 | 2 |
| hsa-novel-miR-09 | chr6:144076881:144076900:- | AAAAAACCGCAATTACTTTT | −44.55 | 20 | 1 | 2 | 1 | 1 | 2 | 1 |
| hsa-novel-miR-10 | chr7:142055000:142055022:+ | GACCTCGATGTTGGATCAGGACA | −50.21 | 23 | 2 | 3 | 8 | 2 | 1 | 8 |
| hsa-novel-miR-11 | chr8:3774059:3774080:- | AAGATGCTTTCTACTGCCCCCT | −41.18 | 22 | 1 | 1 | 2 | 6 | 3 | 18 |
| hsa-novel-miR-12 | chr9:33649040:33649062:+ | GACCTCGATGTTGGATCAGGACA | −48.25 | 23 | 2 | 3 | 8 | 2 | 1 | 8 |
| hsa-novel-miR-13 | chr16:858525:858545:- | GCTCTCACTGCAGTCCCCTGC | −79.44 | 21 | 3 | 2 | 1 | 1 | 2 | 4 |
| hsa-novel-miR-14 | chr16:14902938:14902959:+ | AGAAGGGGTGAAATTTAAACGT | −73.36 | 22 | 2 | 1 | 4 | 2 | 3 | 2 |
| hsa-novel-miR-15 | chr16:16301589:16301610:+ | AGAAGGGGTGAAATTTAAACGT | −73.36 | 22 | 2 | 1 | 4 | 2 | 3 | 2 |
| hsa-novel-miR-16 | chr16:18413263:18413284:- | ACGTTTAAATTTCACCCCTTCT | −64.97 | 22 | 2 | 1 | 4 | 2 | 3 | 2 |
| hsa-novel-miR-17 | chr17:872519:872540:- | TCCGAGCGACTCCGAGAGAGGC | −47.41 | 22 | 3 | 2 | 1 | 1 | 1 | 2 |
| hsa-novel-miR-18 | chr20:55367133:55367156:- | TATGTCCTGATCCAACATCGAGGTC | −28.33 | 24 | 6 | 10 | 4 | 5 | 8 | 6 |
MFE, minimum free energy.
A pre-miRNA stem-loop can produce two mature miRNAs, one from the plus strand and one from the minus strand (miRNA*). In most cases, the miRNA* sequence is degraded and make it difficult to detect. Thus, there is less miRNA* than miRNA in the miRBase. Nonetheless, with the advantage of deep sequencing, we found 16 new miRNA* of the known miRNA precursors (Table 4).
Table 4.
16 New miRNA* Identified in Glioblastoma and Normal Brain Tissues
| |
|
|
Count |
|||||
|---|---|---|---|---|---|---|---|---|
| Known miRNA Precursor | Sequence | Length | GBM_1 | GBM_2 | GBM_3 | NB_1 | NB_2 | NB_3 |
| hsa-miR-539 | AUCAUACAAGGACAAUUUCUUUU | 23 | 429 | 54 | 928 | 894 | 552 | 1461 |
| hsa-miR-382 | AAUCAUUCACGGACAACACUUU | 22 | 131 | 52 | 633 | 336 | 231 | 315 |
| hsa-miR-1307 | UCGACCGGACCUCGACCGGCU | 21 | 187 | 431 | 75 | 39 | 23 | 36 |
| hsa-miR-212 | ACCUUGGCUCUAGACUGCUUAC | 22 | 26 | 44 | 120 | 110 | 92 | 249 |
| hsa-miR-204 | GCUGGGAAGGCAAAGGGACGUUC | 23 | 162 | 178 | 2 | 67 | 78 | 65 |
| hsa-miR-301a | GCUCUGACUUUAUUGCACUACU | 22 | 109 | 183 | 82 | 52 | 50 | 49 |
| hsa-miR-181b-1 | CUCACUGAACAAUGAAUGCA | 20 | 175 | 111 | 77 | 32 | 51 | 36 |
| hsa-miR-3676 | AGGAGAUCCUGGGUUCG | 17 | 44 | 176 | 38 | 24 | 25 | 13 |
| hsa-miR-758 | AUGGUUGACCAGAGAGCACACG | 22 | 10 | 0 | 31 | 66 | 30 | 33 |
| hsa-miR-98 | CUAUACAACUUACUACUUUCC | 21 | 20 | 34 | 27 | 16 | 14 | 23 |
| hsa-miR-873 | GGAGACUGAUGAGUUCCCGGGA | 22 | 8 | 0 | 5 | 58 | 29 | 25 |
| hsa-miR-135a-2 | AUGUAGGGAUGGAAGCCAUGA | 21 | 6 | 4 | 7 | 54 | 26 | 27 |
| hsa-miR-511-1 | AAUGUGUAGCAAAAGACAGA | 20 | 13 | 82 | 4 | 8 | 10 | 4 |
| hsa-miR-1271 | AGUGCCUGCUAUGUGCCAGGCA | 22 | 27 | 50 | 22 | 8 | 2 | 6 |
| hsa-miR-381 | AGCGAGGUUGCCCUUUGUAU | 20 | 5 | 1 | 6 | 37 | 19 | 20 |
| hsa-miR-487a | GUGGUUAUCCCUGCUGUGUUCG | 22 | 4 | 0 | 10 | 21 | 14 | 22 |
Discussion
There are 1733 miRNA and miRNA* in miRBase version 17 (Kozomara and Griffiths-Jones, 2011). In our study, we found 875 and 811 known human miRNA and miRNA* in glioblastoma and normal brain tissue, respectively. 423 miRNAs from Skalsky'study (Skalsky and Cullen, 2011) were found in our data (Supplementary Table S1). The abundance of miRNAs varies greatly among the miRNAs that we identified with let-7 family, miR-103a, −140-3p, −101, and −221, as examples of the high abundance miRNAs (Supplementary Table S1). miRNAs play important roles in glioblastoma and altered expression of miRNAs have been implicated in glioblastoma (Ciafre et al., 2005; Niyazi et al., 2011; Skalsky and Cullen, 2011). In our study, we showed that the expression of 73 miRNA genes changed significantly in glioblastoma compared to normal brain tissues. Many previously identified differentially expressed miRNAs were also identified, such as miR-21, miR-10b (Ciafre et al., 2005) and miR-31, which were shown to be downregulated in GBM and inhibited glioblastoma cell invasion (Hua et al., 2012). Moreover, 46 miRNAs were identified for the first time as differentially expressed.
With the advantage of deep sequencing, the isomiRs could be detected by miRNA sequencing analysis (Morin et al., 2008; Zhou et al., 2012). In this study, we found that isomiRs exist in miRNAs. Furthermore, for about 63% of the 300 highest expressed miRNA, their isomiRs are expressed at higher levels than their reference mature miRNAs (Supplementary Table S4). The functional consequences of this observation remain to be studied. The seed sequence of an miRNA plays an important role in target recognition (John et al., 2004) and variations in isomiR sequence occurring at the 5′ terminal may result in altered binding repertoire of targets compared to their mature reference counterparts. Finally, we identified 18 completely novel miRNAs, suggesting there are additional miRNA genes in human genome remained to be uncovered.
Conclusion
Using deep sequencing, we constructed by far the largest dataset of miRNAs in glioblastoma and normal brain tissues. We also identified 46 differential expressed miRNAs for the first time between GBM and normal brain tissues. We further showed that the most abundant form of isomiR for 23 miRNAs each has variations in its 5′ end. Taking together, this dataset provides a useful resource for future studies of the roles of miRNAs in the pathogenesis and early detection of glioblastoma.
Supplementary Material
Acknowledgments
This work was supported by Grant 81072060 from the National Natural Science Foundation of China.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Chan JA. Krichevsky AM. Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chen C. Ridzon DA. Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA. Galardi S. Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Cloonan N. Wani S. Xu Q, et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2012;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FG. McCarthy BJ. Current epidemiological trends and surveillance issues in brain tumors. Expert Rev Anticancer Ther. 2001;1:395–401. doi: 10.1586/14737140.1.3.395. [DOI] [PubMed] [Google Scholar]
- Ferretti E. De Smaele E. Po A, et al. MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124:568–577. doi: 10.1002/ijc.23948. [DOI] [PubMed] [Google Scholar]
- Gardner PP. Daub J. Tate J, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2010;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J. Nowicki MO. Bronisz A, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Guo C-J. Pan Q. Li D-G. Sun H. Liu B-W. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766–778. doi: 10.1016/j.jhep.2008.11.025. [DOI] [PubMed] [Google Scholar]
- He L. Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hua D. Ding D. Han X, et al. Human miR-31 targets radixin and inhibits migration and invasion of glioma cells. Oncol Rep. 2012;27:700–706. doi: 10.3892/or.2011.1555. [DOI] [PubMed] [Google Scholar]
- Jiang P. Wu H. Wang W. Ma W. Sun X. Lu Z. MiPred: Classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–W344. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B. Enright AJ. Aravin A. Tuschl T. Sander C. Marks DS. Human MicroRNA Targets. PLos Biol. 2004;3:e264. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J. Chivukula RR. O'Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A. Griffiths-Jones S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP. Burge CB. Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Loftus JC. Ross JT. Paquette KM, et al. miRNA expression profiling in migrating glioblastoma cells: Regulation of cell migration and invasion by miR-23b via targeting of Pyk2. PLoS One. 2012;7:e39818. doi: 10.1371/journal.pone.0039818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Getz G. Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mitchell PS. Parkin RK. Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD. O'Connor MD. Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network, Cancer Genome Atlas Res. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyazi M. Zehentmayr F. Niemoller O, et al. MiRNA expression patterns predict survival in glioblastoma. Radiat Oncol. 2011;6:153. doi: 10.1186/1748-717X-6-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J. Lim DA. Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL. Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6:e24248. doi: 10.1371/journal.pone.0024248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S. Patric IR. Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PLoS One. 2011;6:e17438. doi: 10.1371/journal.pone.0017438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S. Tong Y. Steitz JA. Switching from repression to activation: MicroRNAs can up-regulate translation. Science. 2007:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Wen PY. Kesari S. Malignant gliomas in adults. New Eng J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- Yang J-S. Phillips MD. Betel D, et al. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Arcila ML. Li Z, et al. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40:5864–5875. doi: 10.1093/nar/gks247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.