Abstract
Introduction
Free-circulating DNA is present in minute amounts in plasma of healthy individuals, whereas increased levels are found in a number of malignant pathologies including non-small cell lung cancer (NSCLC). The objective of this research was the evaluation of the plasma DNA quantification capacity to distinguish between healthy subjects and non-small cell lung cancer (NSCLC) patients.
Materials and methods
Plasma samples were collected prospectively from 16 healthy volunteers and 30 untreated NSCLC patients (I-IIIA). Subsequently, free-circulating DNA extraction and quantitative real-time PCR analysis were performed.
Results
The values of plasma DNA concentration ranged from 0.9 up to 7.0 ng/ml in healthy individuals and from 1.5 up to 50 ng/ml in NSCLC patients before treatment. Cancer group showed several-fold higher mean free-circulating DNA concentration than that present in healthy subjects (mean 12.00 vs. 2.65 ng/ml; P < 0.001). A greater variability of plasma DNA concentrations was observed in NSCLC patients than in controls (SD 14.50 vs. 2.02, respectively). The area under the ROC curve was 0.87 (95% CI, 0.744 to 0.954, P < 0.001).
Conclusion
Non-small cell lung cancer is associated with elevated levels of cell-free DNA in plasma with respect to healthy controls. Real-time PCR method proved its utility in effective free-circulating DNA detection and quantification.
Keywords: DNA quantification, free-circulating DNA, lung cancer, plasma, real-time PCR
Introduction
The discovery of extracellular DNA circulating in plasma, serum, and other body fluids influences cancer genomics, as it allows non-invasive access to genetic material originating form diseased cells and tissues. Blood is easily and cheaply accessible material and its proper sampling doesn't require additional personnel training, therefore it is ideal for extensive research projects on the clinical value of free-circulating DNA in lung cancer [1].
Free-circulating DNA is present in minute amounts in the plasma of healthy individuals. Increased levels of circulating DNA have been found in a number of pathologies, such as cancer, stroke, trauma, myocardial infarction, autoimmune disorders, chronic inflammation, and pregnancy-associated complications [2]. The precise mechanism by which DNA is released into the bloodstream remains unknown. Recent reports demonstrated that a significant proportion of circulating DNA is of hematopoietic origin in healthy as well as in diseased individuals [3]. However, the detection of common genetic (e.g., mutations, microsatellite alterations) and epigenetic (e.g. promoter methylation) modifications in tumors and paired plasma/serum samples evidenced that in cancer patients part of the free-circulating DNA is of tumor origin [4].
It is believed that free-circulating DNA, by its apparent linkage with altered cell proliferation, apoptosis, necrosis, angiogenesis and other cancer-related phenomena, might provide us with valuable biomarkers for lung cancer diagnostics and therapy monitoring. Recent findings on correlation of plasma DNA levels in lung cancer patients with disease stage, cancer histopathology, disease progression rate and response to therapy evidence the importance of quantitative studies on free-circulating DNA [5]. Here, we present results of real-time qPCR plasma DNA measurement in non-small cell lung cancer patients and healthy controls as the promising introduction to further investigations.
Materials and methods
Collection of Plasma Samples
Blood samples were collected from 16 healthy volunteers and 30 patients with resectable (I-IIIA) non-small cell lung cancer (NSCLC) before any treatment. Blood was collected to 9-ml tubes containing EDTA as anticoagulant agent and processed within 1 h. Plasma was then separated from the cellular fraction by two centrifugations at 1000 × g for 10 min in 4°C, and banked at -80°C.
Extraction and Real-Time PCR Quantification of Plasma DNA
DNA was extracted from 0.5 ml plasma aliquots with QIAmp DNA Blood Midi kit (Qiagen, Germany) according to the manufacturer's instructions. DNA concentration was measured by quantitative real-time PCR using human β-actin reference gene (99 bp) as described previously [6]. DNA samples were quantified from genomic DNA standard curves. All samples were processed in duplicate, and the mean value was used for quantification.
Statistical Evaluation
Kruskal-Wallis analysis of variance by ranks was used to test statistical significance of differences between groups. The Receiver Operating Characteristics (ROC) curve analysis was performed to test the diagnostic potential of real-time PCR assay in discrimination between cancer disease patients and healthy controls.
Results
Assay Performance and Plasma DNA Concentration
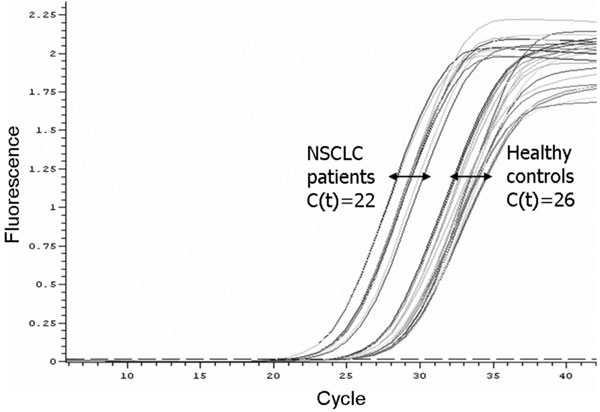
The concentration of DNA in plasma samples was measured by quantitative real-time PCR method using β-actin gene as the amplifying target. Linearity of product amplification assessed as the slope and correlation coefficient (r2) of the standard curve were -3.20 and 0.99, respectively. Generally, NSCLC patients demonstrated higher plasma DNA concentrations than controls (Figure 1). The values of plasma DNA concentration ranged from 0.9 up to 7.0 ng/ml in healthy individuals, and from 1.5 up to 64.4 ng/ml in NSCLC patients before any treatment. The Kruskal-Wallis analysis showed significant differences in mean plasma DNA levels between two groups studied (P < 0.001; Table 1). A higher standard deviation was observed in patients than in controls.
Figure 1.
Amplification plots of the real-time PCR for β-actin in plasma DNA from non-small cell lung cancer patients and healthy controls. The relative amount of plasma DNA is higher in patient samples compared with controls.
Table 1.
Descriptive statistics for the data of free-circulating DNA concentration (ng DNA per 1 ml plasma) analysis in plasma samples from healthy subjects and non-small cell lung cancer patients by real-time PCR.
| n | Mean | Median | Minimum | Maximum | SD | |
|---|---|---|---|---|---|---|
| Healthy controls | 16 | 2.65 | 1.86 | 0.91 | 7.00 | 2.02 |
| NSCLC patients | 30 | 12.00 | 6.91 | 1.49 | 64.40 | 14.50 |
Test Sensitivity and Specificity
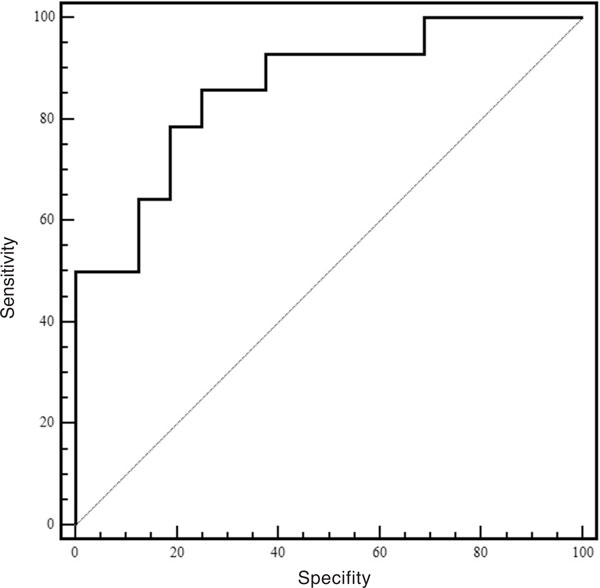
The ROC curve was built to assess the discriminative power of plasma DNA quantification. According to our data, the area under the ROC curve is 0.86 (95% CI, 0.67 to 0.96, P < 0.0001; Figure 2.). It was found that lower cut-off values increased the sensitivity of the assay but at the cost of specificity and vice versa (Table 2). If the value of 2.8 ng/ml was selected as a cut-off point, it corresponded to the highest average of sensitivity (86%) and specificity (75%). However, selecting the value of 7.0 ng/ml, the smallest cut-off point does not overlap with other cut-off values, decreased the sensitivity to 50%, but increased the specificity to 100%.
Figure 2.
ROC curve for the identification of non-small cell lung cancer using plasma DNA values.
Table 2.
Test performance evaluation of plasma DNA quantification by real-time qPCR.
| Cut-off | Sensitivity | 95% CI | Specificity | 95% CI | +LR | -LR |
|---|---|---|---|---|---|---|
| ≥ 0.91 | 100 | 76.7-100.0 | 0 | 0.0-20.8 | 1.00 | |
| ≥ 1.98 | 92.9 | 66.1-98.8 | 62.5 | 35.5-84.7 | 2.48 | 0.11 |
| ≥ 2.78* | 85.7 | 57.2-97.8 | 75.0 | 47.6-92.6 | 3.43 | 0.19 |
| ≥ 3.36 | 78.6 | 49.2-95.1 | 81.3 | 54.3-95.7 | 4.19 | 0.26 |
| ≥ 5.39 | 64.3 | 35.2-87.1 | 87.5 | 61.6-98.1 | 5.14 | 0.41 |
| ≥ 7 | 50 | 23.1-76.9 | 100 | 79.2-100.0 | 0.50 |
*The cut-off value corresponding with the highest average of sensitivity and specificity. +LR is a positive and -LR is a negative likelihood ratio.
Discussion
The presence of free DNA in blood of patients with lung cancer has been intensively studied in recent years. Previously, we demonstrated the effective measurement of free-circulating DNA concentration in plasma of NSCLC patients by use of distinct fluorescence-based techniques [7]. In present study, we evaluated plasma DNA levels in untreated NSCLC patients with respect to healthy controls by use of real-time PCR amplification of β-actin housekeeping gene.
Among several available methods of DNA quantification, real-time PCR is regarded as the golden standard currently available for free-circulating DNA measurement. It allows simultaneous amplification and detection of specific DNA sequences by monitoring the fluorescence of dyes or probes introduced into the reaction in proportion to the amount of product formed. Consequently, real-time PCR is characterized by high accuracy, reproducibility, and time-effectiveness [8].
In our study, plasma specimen was chosen as a source of free-circulating DNA because ex vivo release of DNA from hematopoietic cells during the clotting process had been demonstrated [9]. Furthermore, EDTA was suggested to be a better anticoagulant than heparin or citrate for prospective blood processing prior to plasma DNA analysis [10].
To date, several research groups provided very interesting results of real-time PCR quantification studies on plasma DNA in lung cancer. Sozzi et al [11] applied quantitative real-time PCR of the human telomerase reverse transcriptase (hTERT) gene to measure cancer-derived DNA and to discriminate lung cancer patients (n = 100) from healthy smoking and nonsmoking controls (n = 100). Mean concentration of plasma DNA in patients was several-fold higher than level detected in controls (75.0 vs. 4.6 ng/ml). Very satisfactory sensitivity (90%), specificity (86%), positive predictive value (90%) and negative predictive value (90%) was obtained with cut-off DNA concentration value of 9.0 ng/ml. However, when the value of 25 ng/ml was used, the only cut-off point that did not overlap with other cut-off points, the sensitivity dropped down to 46%. Similarly, Paci et al [12] found the concentration of circulating hTERT DNA about four times higher in a group of 151 patients with lung cancer with respect to the 79 healthy controls (12.8 vs. 2.9 ng/ml). The area under the ROC curve was 0.79 (95% CI, 0.71-0.83). For the cut-off value of 2 ng/ml, the 86% sensitivity and 47% specificity was shown. Interestingly, in the study of Ludovini et al [3] plasma DNA levels of 76 lung cancer patients (mean 60 ng/ml, SD 99.8) were found significantly higher than levels observed in 66 controls (mean 6.0 ng/ml, SD 8.8; P < 0.0001). The highest accuracy of the real-time PCR assay was obtained at the cut-off point of 3.25 ng/ml, corresponding to a sensitivity and specificity of 80% and 61%, respectively.
The results of our quantitative real-time PCR assay are concordant with the presented above data. In fact, the mean plasma DNA concentration in untreated NSCLC patients is several-fold higher than value measured in healthy controls (12.00 vs. 2.65 ng/ml, respectively). Furthermore, the overlapping of the concentration ranges for patients and controls as well as greater SD values in cancer were frequently observed in all studies quantifying the plasma DNA. Selection of the optimal cut-off point in ROC analysis should therefore acknowledge such variability.
It has been suggested that ROC analysis takes at least 100 observations for meaningful conclusions [14]. However, a preliminary ROC analysis performed in our study might indicate the diagnostic potential for free-circulating DNA in lung cancer. The proposed cut-off point of 2.78 ng/ml distinguished between health and lung cancer with the highest 86% sensitivity and 75% specificity (Table 2) outmatching the score of Herrera et al [6], that didn't reach statistical significance for plasma DNA concentration in 25 non-small cell lung cancer patients and 11 healthy controls (14.6 vs. 10.6 ng/ml, respectively; P = 0.175). The highest detected DNA concentration in normal plasma (14.0 ng/ml) used as a cut-off identified 48% of lung cancer patients.
In summary, non-small cell lung cancer is associated with elevated levels of cell-free DNA in plasma with respect to healthy controls. Quantitative real-time PCR assay allows demonstrating this phenomenon.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
Supported by grant N401 174 31/3840, Ministry of Science and Higher Education, Republic of Poland.
References
- Chorostowska-Wynimko J, Szpechcinski A. The impact of genetic markers on the diagnosis of lung cancer: a current perspective. J Thorac Oncol. 2007;2(11):1044–51. doi: 10.1097/JTO.0b013e318158eed4. [DOI] [PubMed] [Google Scholar]
- Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids: A promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007;581(5):795–9. doi: 10.1016/j.febslet.2007.01.051. [DOI] [PubMed] [Google Scholar]
- Lui YY, Chik KW, Chiu RW, Ho CY, Lam CW, Lo YM. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sexmismatched bone marrow transplantation. Clin Chem. 2002;48(3):421–7. [PubMed] [Google Scholar]
- Gonzalez R, Silva JM, Sanchez A, Dominguez G, Garcia JM, Chen XQ, Stroun M, Provencio M, España P, Anker P, Bonilla F. Microsatellite alterations and TP53 mutations in plasma DNA of small-cell lung cancer patients: follow-up study and prognostic significance. Ann Oncol. 2000;11(9):1097–104. doi: 10.1023/A:1008305412635. [DOI] [PubMed] [Google Scholar]
- Pathak AK, Bhutani M, Kumar S, Mohan A, Guleria R. Circulating cell-free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin Chem. 2006;52(10):1833–42. doi: 10.1373/clinchem.2005.062893. [DOI] [PubMed] [Google Scholar]
- Herrera LJ, Raja S, Gooding WE, El-Hefnawy T, Kelly L, Luketich JD, Godfrey TE. Quantitative analysis of circulating plasma DNA as a tumor marker in thoracic malignancies. Clin Chem. 2005;51(1):113–8. doi: 10.1373/clinchem.2004.039263. [DOI] [PubMed] [Google Scholar]
- Szpechcinski A, Struniawska R, Zaleska J, Chabowski M, Orlowski T, Roszkowski K, Chorostowska-Wynimko J. Evaluation of fluorescence-based methods for total vs. amplificable DNA quantification in plasma of lung cancer patients. J Physiol Pharmacol. 2008;59(Suppl 6):675–81. [PubMed] [Google Scholar]
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2-3):95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Lee TH, Montalvo L, Chrebtow V, Busch MP. Quantitation of genomic DNA in plasma and serum samples: higher concentrations of genomic DNA found in serum than in plasma. Transfusion. 2001;41(2):276–82. doi: 10.1046/j.1537-2995.2001.41020276.x. [DOI] [PubMed] [Google Scholar]
- Lam NY, Rainer TH, Chiu RW, Lo YM. EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis. Clin Chem. 2004;50(1):256–7. doi: 10.1373/clinchem.2003.026013. [DOI] [PubMed] [Google Scholar]
- Sozzi G, Conte D, Leon M, Ciricione R, Roz L, Ratcliffe C, Roz E, Cirenei N, Bellomi M, Pelosi G, Pierotti MA, Pastorino U. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21(21):3902–8. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- Paci M, Maramotti S, Bellesia E, Formisano D, Albertazzi L, Ricchetti T, Ferrari G, Annessi V, Lasagni D, Carbonelli C, De Franco S, Brini M, Sgarbi G, Lodi R. Circulating plasma DNA as diagnostic biomarker in non-small cell lung cancer. Lung Cancer. 2008;64(1):92–7. doi: 10.1016/j.lungcan.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Ludovini V, Pistola L, Gregorc V, Floriani I, Rulli E, Piattoni S, Di Carlo L, Semeraro A, Darwish S, Tofanetti FR, Stocchi L, Mihaylova Z, Bellezza G, Del Sordo R, Daddi G, Crino L, Tonato M. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the Perugia multidisciplinary team for thoracic oncology. J Thorac Oncol. 2008;3(4):365–73. doi: 10.1097/JTO.0b013e318168c7d0. [DOI] [PubMed] [Google Scholar]
- Metz CE: Basic principles of ROC analysis. Semin Nucl Med. 1978;8(4):283–98. doi: 10.1016/S0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]