Abstract
BK polyomavirus infection is the important cause of virus-related nephropathy following kidney transplantation. BK virus reactivates in 30%–80% of kidney transplant recipients resulting in BK virus-related nephropathy in 1%–10% of cases. Currently, the molecular processes associated with asymptomatic infections in transplant patients infected with BK virus remain unclear. In this study we evaluate intrarenal molecular processes during different stages of BKV infection. The gene expression profiles of 90 target genes known to be associated with immune response were evaluated in kidney graft biopsy material using TaqMan low density array. Three patient groups were examined: control patients with no evidence of BK virus reactivation (n = 11), infected asymptomatic patients (n = 9), and patients with BK virus nephropathy (n = 10). Analysis of biopsies from asymptomatic viruria patients resulted in the identification of 5 differentially expressed genes (CD3E, CD68, CCR2, ICAM-1, and SKI) (P < 0.05), and functional analysis showed a significantly heightened presence of costimulatory signals (e.g., CD40/CD40L; P < 0.05). Gene ontology analysis revealed several biological networks associated with BKV immune control in comparison to the control group. This study demonstrated that asymptomatic BK viruria is associated with a different intrarenal regulation of several genes implicating in antiviral immune response.
1. Introduction
Innovations to immunosuppressive regimens have improved patient and kidney transplant survival rates; however, drug-induced immune suppression has also resulted in significant increases in complications associated with infections. BK polyomavirus (BKV) infections have emerged as an important cause of virus-related nephropathy following kidney transplantation in the era of modern immunosuppressive therapies [1]. BKV has been shown to reactivate in 30%–80% of kidney transplant recipients but only in 1%–10% of cases resulted in the development of BKV nephropathy (BKVN) associated with subsequent kidney graft deterioration and failure [2–5].
Recently, the polymerase chain reaction (PCR) has been used for routine monitoring of BKV replication in peripheral blood [5]. However, PCR screening has demonstrated that a majority of kidney transplant recipients are BKV positive in urine but not blood and never develop BKV nephropathy with graft function deterioration. Furthermore, it was shown that patients with asymptomatic viruria presented with significant BKV viral loads in kidney graft biopsy specimens [6], suggesting successful control of the BKV infection by the host immune system.
Generally, innate immunity and nonspecific effector mechanisms represent a first line of defense against infection, followed by the development of more specific, acquired immune responses. Active viral replication in tissues has been considered to be a trigger point for inflammation, and cellular immunity has been suggested to play a critical role in viral control. Recently, it was shown that patients with self-limited BKV reactivation without therapeutic intervention developed BKV-specific T cells and cleared BKV rapidly compared to patients suffering from BKV associated-nephropathy [7].
However, the process of successful BKV control on molecular level has not been fully described.
In this study, we investigated the intrarenal specific transcripts during a different outcome of BKV infection in kidney graft.
2. Materials and Methods
2.1. Patients
Based on the BKV monitoring study [8] and histopathological archive of the Institute for Clinical and Experimental Medicine, Prague, different patient groups were established as follows. (i) The asymptomatic viruria group (AV) (n = 9) presented with BK viral urine loads higher than 107/mL [9] on the day of protocol biopsy (3 months after transplant). These patients never developed BKV-associated nephropathy and were negative for BKV replication screening in both urine and blood 12 months after transplantation. Protocol kidney graft biopsies revealed normal findings free of rejection and patients had stable graft function. (ii) Control group (NCG) (n = 11) consisted of patients who were neither BKV positive in urine nor in serum at any time-point after kidney transplantation. None of the patients exhibited allograft rejection; protocol biopsy was normal and patients had stable f graft function. (iii) The BK virus nephropathy (BKVN) group consisted of patients (n = 10) with BKV-associated nephropathy. Eight patients presented with BKV-associated nephropathy prior to the start of the study and archived biopsy specimens for molecular biology were used for analysis. In 2 cases, BKV-associated nephropathy developed during the course of the study. Histological confirmation of BKV-associated nephropathy was defined as detection of viral cytopathic changes with intranuclear inclusion bodies, associated renal tubular epithelial cell injury, including tubular epithelial cell necrosis, and denudation of basement membranes, as well as positive immunohistochemical staining for the SV40 T large antigen. Patients' clinical and demographic variables are to be found in Table 1.
Table 1.
Demographics and transplant related variables.
| CG (n = 11) | BKVN (n = 10) | AV (n = 9) | P* value | |
|---|---|---|---|---|
| At baseline | ||||
| Recipient age (years) | 50 ± 10 | 44 ± 21 | 47 ± 10 | ns |
| Donor age (years) | 49.2 ± 13 | 47 ± 18 | 46 ± 15 | ns |
| Living donor (n, %) | 1/9% | 3/30% | 1/22% | ns |
| Gender (male, %) | 6/54% | 3/30% | 6/66% | ns |
| HLA mismatches | 3 ± 1 | 4 ± 1 | 2 ± 1 | ns |
| Peak level of PRA | 5 ± 7 | 44 ± 40 | 1 ± 1 | ns |
| CMV serostatus | ||||
| D−/R− | 1/9% | 1/10% | 1/11% | ns |
| D−/R+ | 1/9% | 1/10% | 1/11% | ns |
| D+/R− | 3/27% | 1/10% | 1/11% | ns |
| D+/R+ | 6/54% | 7/70% | 6/66% | ns |
| Dialysis before TX (months) | 19 ± 10 | 27 ± 15 | 32 ± 26 | ns |
|
| ||||
| At 3M biopsy | ||||
| Time after TX | 94.8 ± 5.8 | 343 ± 268 | 85.6 ± 13.5 | ns |
| BMI (kg/m2) | 23.5 ± 4.8 | 27.3 ± 4.3 | 27.2 ± 4.7 | ns |
| Immunosuppression | ||||
| Induction | 36% | 50% | 44% | ns |
| Tacrolimus | 73% | 80% | 89% | ns |
| Cyclosporin A | 27% | 0% | 0 | ns |
| mTOR | 18% | 40% | 11% | ns |
| Mycophenolate mofetil | 55% | 70% | 89% | ns |
| Prednisone | 73% | 90% | 89% | ns |
| CIT (deceased donor) | 20 ± 3 | 13 ± 9 | 19 ± 3 | ns |
| Serum creatinine (μmol/L) | 112 ± 60.3 | 210 ± 67.2 | 126 ± 35.3 | ** |
| Serum creatinine at 36-month followup | 122.6 ± 37.3 | NA | 144.1 ± 69.5 | ns |
Data shown as mean ± standard deviation if not indicated otherwise. ESRD: end-stage renal disease; CIT: cold ischemia time; PRA: panel reactive antibody; BMI: body mass index; TX: transplantation; **(P < 0.01) BKVN versus negative control group; ns: not significant; *P values for categorical data χ 2 or Fisherś test and for continuous variables Mann-Whitney U test (Student's t-test where appropriate).
Three months after transplantation, protocol kidney graft biopsies were performed in all patients from group I and II and a part of the biopsy specimen fixed for molecular biology analysis at a later date. The study protocol was approved by the Ethics Committee of the Institute for Clinical and Experimental Medicine in Prague and a written informed consent was obtained from all patients.
2.2. RNA Isolation and TaqMan Low Density Array (TLDA)
Small portions (~2 mm) of the cortical or juxtamedullary zone from biopsy specimens were immediately stored in RNA later (Ambion Corporation, Austin, TX). Renal tissues were homogenized; total RNA were extracted using StrataPrep Total RNA Microprep Kit (Stratagene, La Jolla, CA, USA) and reverse transcribed into complementary DNA (cDNA) as described elsewhere [10].
The gene expression profile of 90 candidate gene targets known to play roles in the elicitation of immune responses (e.g., genes involved in cytokine expression, costimulatory molecules, growth factors, chemokines, immune regulation, apoptosis markers, and ischemia markers) was determined using real-time RT-PCR (2−ΔΔCt) with GAPDH as internal control and cDNA from a control kidney serving as the calibrator in 30 renal biopsy specimen analyses. All evaluated genes are described in Table S1 in Supplementary Material available online at doi:10.1155/2012/972102. Each immune TLDA profile contains lyophilized gene expression reagents (primers and probes (FAM labeled)) in a preconfigured 384 well format. Two samples in duplicate were analyzed per card. Each loading port was filled with a 100 μL cDNA, nuclease free water, and 2X TaqMan universal PCR master mix. Following centrifugation, cards were sealed with a TLDA sealer (Applied Biosystems, Foster City, CA) to prevent cross-contamination. RT-PCR amplification was performed using an ABI Prism 7900 H.T. Sequence Detection system (Applied Biosystems). TLDA cards were analyzed as relative quantification (RQ) and RQ manager 1.2. software for automated data analysis was used (Applied Biosystems).
2.3. Statistical and Functional Analyses
Continuous variables were presented as the mean ± SD (standard deviation). Statistical analysis of categorical characteristics was performed using the χ 2 test, of continuous parametric and nonparametric variables using student's t-test and Mann-Whitney U tests. The Bonferroni correction was used when appropriate. Supervised hierarchical clustering was performed using the MeV (V.3) software in order to visualize results. Gene expression data were compared using the Mann-Whitney U test followed by the Bonferroni correction.
For functional analysis, large-scale data management was used to identify specific transcript patterns. In the set analysis, genes were grouped into sets determined by their annotation and then compared between defined groups. For the functional analysis, 2 types of set analyses were used. Since a limited number of genes were assessed, the fully coupled flux analysis that reflects the analysis of the pathway fragment was used as the first set analysis [11]. Fully coupled flux represents a gene network that corresponds to a pathway in which non-zero flux for one reaction implies a nonzero flux for a second reaction, and vice versa. This flux represents the strongest qualitative connectivity that can be identified in a network. The genes coupled by their enzymatic fluxes were shown to have similar expression patterns, share transcriptional regulators, and frequently reside in the same operon. The second gene set type analysis included genes sharing a common gene ontology [12]. For the use of functional analysis, gene expression data were log transformed and compared between groups with one-way analysis of variance (ANOVA) followed by the Bonferroni correction to appropriately account for multiple comparisons. Set-level analysis was carried out using XGENE.ORG [13, 14].
3. Results
3.1. Gene Expression Profiles
We first determined whether the 90 candidate target transcripts identified differed in their levels of expression between the groups. Using these criteria, cohorts of patients with asymptomatic viruria and the negative control group were established; we also examined archived biopsies from patients with histologically proven BKV-associated nephropathy.
Using hierarchical clustering, different gene transcript profiles were identified among the study groups. Hierarchical clustering demonstrated that gene transcript profiles expressed in kidney graft tissues in patients not testing BKV positive were similar to profiles observed in patients presenting with asymptomatic BKV viruria who did not present with BKV positive blood samples or BKV-associated nephropathy. However, gene transcript profiles identified in patients with BKV-associated nephropathy formed a significantly different cluster (Figure 1).
Figure 1.
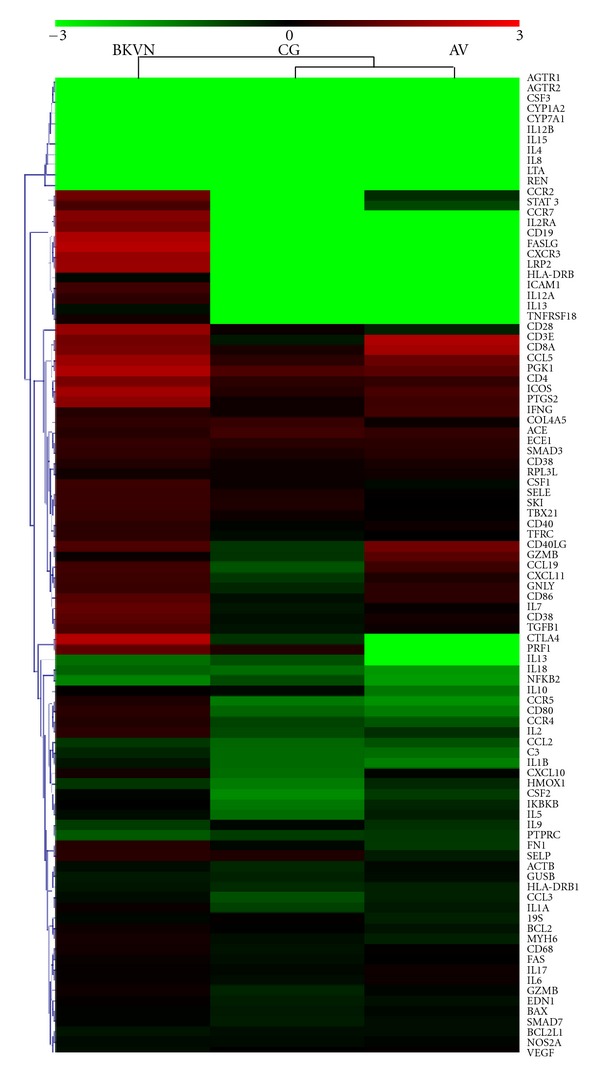
Hierarchical clustering of 3 studied groups. This heat map represents all 90 immune targets assayed (plus endogenous control). Medians of logarithmed relative quantification (RQ) values for each group were used to create the heat map. Red denotes genes with relative increased expression while green denotes genes with relative decreased expression.
3.2. Characterization of Intrarenal Gene Transcript Profiles in Patients with Asymptomatic BK Viruria
In order to identify the nature of intrarenal immune mechanisms associated with the control of BKV infections, intrarenal graft transcripts from patients with asymptomatic viruria or patients in the negative control group were analyzed. Five differentially expressed genes were identified, including upregulation of the T cell (CD3E) and macrophage (CD68) markers, in addition to genes encoding the receptor for the monocyte chemoattractant protein-1 (CCR2; a chemokine which specifically mediates monocyte chemotaxis) and the adhesion molecule, ICAM-1. The SKI protooncogene, involved in downregulation of TGF-β1 gene transcripts, was significantly downregulated in the asymptomatic BKV infection group (P < 0.05) (Table 2).
Table 2.
Genes with different regulation in asymptomatic viruria group.
| Gene |
Control group |
Asymptomatic viruria group |
P value* |
|---|---|---|---|
| CCR2 | 0 (0–6.38) |
0.3 (0–31.7) |
0.02 |
| CD3E | 0.53 (0.03–37.3) |
117 (0.15–1120) |
0.04 |
| CD68 | 0.61 (0.05–1.38) |
1.15 (0.72–4.96) |
0.04 |
| SKI | 2.32 (0.29–13.2) |
0.47 (0.07–13) |
0.04 |
| ICAM1 | 0.03 (0–0.08) |
0.34 (0.02–2.12) |
0.05 |
Data are shown as median (minimum–maximum) of relative quantity (RQ) of gene expression calculated with regard to the reference gene (GAPDH) and calibrator.
*P values calculated by Mann-Whitney U test followed by the Bonferroni adjustment.
Functional analysis revealed that patients with asymptomatic viruria exhibited significantly higher expression levels of the costimulatory signals CD40/CD40L (P < 0.05) compared to the negative control group. In addition, groups of genes sharing a common ontology were analyzed, revealing that several biological networks were involved in BKV immune control. These networks were involved primarily with B cell proliferation (BCL2, CD40, CD40L, IL10), T cell proliferation (CD28, CD3E, IL12A, IL4, PTPRC), transmembrane receptor tyrosine kinase pathways (CD4, CD8A, FN1), proteolysis (ACE, ECE1, GZMB, LTA, REN), protein kinase binding (CD3E, CD4, PTPRC), antiapoptotic processes (BCL2, BCL2L, CCL2, CD40L, FAS, IL10, IL1A), and leukocyte adhesion (CD40L, ICAM1) (Table 3).
Table 3.
Biological networks within graft tissue associated with asymptomatic BK viruria.
| GO term | Genes annotated | P value* | |
|---|---|---|---|
| Soluble fraction | GO:0005625 GO:0005625 |
ACTB, CCL3, CCR2, CD40LG, FAS, IL13, SELP | 0.0296 |
| B lymphocyte proliferation, B cell proliferation | GO:0042100 | BCL2, CD40, CD40LG, IL10 | 0.0364 |
| Transmembrane receptor tyrosine kinase pathway | GO:0007169 | CD4, CD8A, FN1 | 0.0394 |
| Proteolysis | GO:0006508 | ACE, ECE1, GZMB, LTA, REN | 0.0416 |
| Protein kinase binding | GO:0019901 | CD3E, CD4, PTPRC | 0.0426 |
| Platelet activation | GO:0030168 | CD40, CD40LG | 0.0428 |
| Positive regulation of T cell proliferation | GO:0042102 | CD28, CD3E, IL12A, IL4, PTPRC | 0.0444 |
| Antiapoptosis | GO:0006916 |
BCL2, BCL2L1, CCL2, CD40LG, FAS, IL10, IL1A
“IL1B,” “IL2," “TNF,” “TNFRSF18” |
0.0466 |
| Leukocyte cell-cell adhesion | GO:0007159 | CD40LG, ICAM1 | 0.0466 |
| T cell receptor complex | GO:0042101 | CD3E, CD4, CD8A | 0.0484 |
*P values by ANOVA followed by Bonferroni adjustment.
3.3. Identification of Intrarenal Gene Transcripts Associated with BKV-Associated Nephropathy
The BKVN group displayed differential regulation of 33/90 genes analyzed. The most differentially regulated genes were CCL2, CCL5, CCR2, CD4, CD68, FASL, GNLY, IL1B, IL2RA, IL8, PRF1, PTPRC, and TNF (all P < 0.01 compared to the negative control group). These genes are primarily involved in T cell signaling, chemotaxis, activation, and cytotoxicity (Table 4).
Table 4.
Genes with different regulation in polyoma BK nephropathy.
| Gene | Control group | BKVN group | P value* |
|---|---|---|---|
| C3 | 0.06 (0.02–0.6) | 0.375 (0.03–5.27) | 0.022 |
| CCL2 | 0.06 (0.01–0.12) | 0.22 (0.03–0.85) | 0.005 |
| CCL3 | 0.12 (0–0.65) | 0.71 (0.16–5.89) | 0.02 |
| CCL5 | 3.26 (0.41–45.4) | 66.9 (7.9–235) | 0.003 |
| CCR2 | 0 (0–6.38) | 20.7 (0–158) | 0.001 |
| CCR7 | 0 (0–659) | 35.3 (0–789) | 0.017 |
| CD19 | 0 (0–104) | 109 (0–2440) | 0.013 |
| CD28 | 1.36 (0–75.1) | 59.3 (0.74–448) | 0.012 |
| CD4 | 3.32 (0.22–15.1) | 27 (6.63–209) | 0.004 |
| CD68 | 0.61 (0.05–1.38) | 1.68 (0.97–4.79) | 0.001 |
| CD86 | 0.67 (0.01–16.9) | 10 (0.88–32.3) | 0.012 |
| CSF1 | 1.42 (0.02–4.89) | 5.17 (0.57–71.3) | 0.049 |
| CXCL10 | 0.05 (0–2.9) | 1.8 (0.06–3.76) | 0.015 |
| CXCR3 | 0 (0–34.3) | 61.2 (0–671) | 0.013 |
| EDN1 | 0.45 (0.14–1.32) | 1.18 (0.39–8.74) | 0.025 |
| FASL | 0 (0–67.2) | 145.72 (25.96–409.25) | 0.003 |
| GNLY | 0.36 (0.12–2.1) | 4.9 (1.45–52.2) | 0.001 |
| HLADRA | 0.39 (0.15–3.27) | 1.55 (0.503–7.6) | 0.021 |
| HLA-DRB1 | 0 (0–0.39) | 0.85 (0–85.6) | 0.036 |
| ICAM1 | 0.03 (0–0.8) | 0.235 (0.06–13.1) | 0.03 |
| IFNG | 2.65 (0–139) | 82.1 (0–519) | 0.023 |
| IL12B | 0 (0–3.83) | 3.49 (0–37.6) | 0.03 |
| IL1B | 0.17 (0–0.95) | 1.38 (0.3–3.67) | 0.002 |
| IL2RA | 0.14 (0–3.44) | 3.32 (0.47–19) | 0.009 |
| IL6 | 0.05 (0.01–3.49) | 0.72 (0.07–9.49) | 0.048 |
| IL8 | 0.55 (0–6.75) | 14.7 (0.11–53.4) | 0.004 |
| LTA | 0 (0–93.9) | 68.8 (0–903) | 0.036 |
| PRF1 | 8.7 (0–43.3) | 117 (1.4–4100) | 0.007 |
| PTPRC | 1.52 (0.55–24.6) | 47.6 (4.7–303) | 0.001 |
| TBX21 | 0 (0–15.2) | 7.14 (0–383) | 0.036 |
| TGFB | 0.73 (0.23–6.75) | 3.42 (0.73–13) | 0.025 |
| TNF | 0.61 (0.04–3.18) | 8.2 (0.95–30.7) | 0.002 |
| TNFRSF18 | 0 (0–1.77) | 1.7 (0–19.5) | 0.01 |
Data are shown as median (minimum–maximum) of relative quantity (RQ) of gene expression calculated with regard to the reference gene (GAPDH) and calibrator.
*P values calculated by Mann-Whitney U test followed by the Bonferroni adjustment.
Functional analysis revealed that patients with BKVN had significantly higher expression levels of the flux for the FASL/FAS (P < 0.05) and CD28, CD80, CD86 (P < 0.05), signaling molecules associated with apoptosis, and costimulation. Moreover, a broad spectrum of molecular networks were shown to have an identical ontology, that is, genes associated with the positive regulation of NF-κB transcription and intracellular signal transduction (TNF, TGFB1), chemotaxis (CCL3, CCL5, IL18, IL1B, IL8, VEGF), activation of MAPK activity (IKBKB, TGFB1, TNF), and negative regulation of viral genome replication (CD80, IL8). A detailed description of ontology-related genes associated with BKV-associated nephropathy is listed in Table 5.
Table 5.
Biological networks in kidney graft tissues that are associated with BKVN.
| GO term | Genes annotated | P value* | |
|---|---|---|---|
| Negative regulation of transcription | GO:0048661 | EDN1, TNF | 0.003 |
| Positive regulation of NF kappa B transcription | GO:0016481 | TGFB1, TNF | 0.004 |
| Activation of MAPK activity | GO:0051092 | IKBKB, TGFB1, TNF | 0.005 |
| Positive regulation of phosphorylation | GO:0000187 | IL1B, TNF | 0.007 |
| Intracellular signal transduction | GO:0001934 | IL1B, TNF | 0.007 |
| Negative regulation of viral genome replication | GO:0007242 | CD80, IL8 | 0.008 |
| Signal transducer activity | GO:0045071 | CCL5, TNF | 0.010 |
| Organ morphogenesis | GO:0004871 | CCL2, CCL3, CCL5, HMOX1, IL12B, IL13, IL15, IL18, IL1A, IL1B, STAT3 | 0.011 |
| Positive regulation of transription, DNA dependent | GO:0009887 | CCL2, IL7, TGFB1, TNF | 0.011 |
| Transcription activator activity | GO:0045941 | CD80, CD86, TNF | 0.011 |
| Exocytosis | GO:0016563 | CD80, CD86, IKBKB, TGFB1 | 0.011 |
| Chemoattractant activity | GO:0006887 | CCL3, CCL5 | 0.013 |
| Angiogenesis | GO:0042056 | CCL3, CCL5 | 0.013 |
| Induction of positive chemotaxis | GO:0001525 | IL18, IL1B, IL8, VEGF | 0.013 |
| Regulation of cell adhesion | GO:0050930 | IL8, VEGF | 0.015 |
| Regulation of isotype switching | GO:0030183 | IL10, IL4 | 0.015 |
| Regulation of cell adhesion | GO:0045191 | IL10, IL4 | 0.015 |
| Response to oxidative stress | GO:0030155 | ICAM1, IL18, IL8 | 0.016 |
| Protein phosphorylation | GO:0006979 | CCL5, PTGS2 | 0.016 |
| Positive regulation of T helper 2 cell differentiation | GO:0006468 | CCL2, IKBKB, TGFB1 | 0.017 |
| Cellular component movement | GO:0045630 | CD86, IL6 | 0.018 |
| Positive regulation of B cell proliferation | GO:0006928 | ACTB, CCL3, CCL5, CXCR3, IFNG, IL13, IL8, PTGS2, STAT3 | 0.019 |
| Cellular calcium ion homeostasis | GO:0030890 | IL4, IL7, PTPRC | 0.019 |
| Chemokine activity | GO:0006874 | CCL19, CCL2, CCL3, CCL5 | 0.022 |
| Response to glucocorticoid stimulus | GO:0008009 | CCL19, CCL2, CCL3, CCL5, CXCL10, CXCL11, IL8 | 0.024 |
| Positive regulationof T cell proliferation | GO:0051384 | IL10, IL6, TNF | 0.024 |
| T cell differentiation | GO:0042102 | CD28, CD3E, IL12A, IL4, PTPRC | 0.024 |
| Defense response to virus | GO:0030217 | IL2, PTPRC | 0.025 |
| Induction of apoptosis by extracellular signals | GO:0051607 | BCL2, PTPRC | 0.026 |
| Negative regulation of transcription from RNA polymerase II promoter | GO:0008624 | CD38, FAS, FASLG | 0.027 |
| Positive regulation of protein kinase activity | GO:0000122 | SMAD3, STAT3, TNF | 0.030 |
| Response to virus | GO:0045860 | CD4, PTPRC | 0.033 |
| Negative regulation of cytokine secretion involved in immune response | GO:0009615 | CCL19, CCL5, IFNG, TNF | 0.033 |
| Negative regulation of interleukin-6 production | GO:0002740 | IL10, TNF | 0.033 |
| Receptor biosynthetic process | GO:0032715 | IL10, TNF | 0.033 |
| Positive regulation of cytokine production | GO:0032800 | IL10, TNF | 0.033 |
| Positive regulation of transcription from RNA polymerase II promoter | GO:0050715 | IL10, TNF | 0.033 |
| Positive regulation of isotype switching to IgG isotypes | GO:0045944 | IL4, IL6, SMAD3, TNF | 0.034 |
| Regulation of immune response | GO:0048304 | IFNG, IL4, TBX21 | 0.035 |
| Activation of caspase activity | GO:0050776 | IFNG, IL4, TBX21 | 0.035 |
| Receptor binding | GO:0006919 | BAX, SMAD3, TNF | 0.035 |
| Antigen processing and presentation | GO:0005102 | C3, REN | 0.038 |
| Activation a proapoptotic gene products | GO:0019882 | CD8A, IFNG | 0.038 |
| Cell cycle arrest | GO:0008633 | BCL2, FAS, FASLG | 0.039 |
| Protein kinase binding | GO:0007050 | IFNG, IL12A, IL12B, IL8, SMAD3, TGFB1 | 0.042 |
| Cell adhesion | GO:0019901 | CD3E, CD4, PTPRC | 0.043 |
| JAK-STAT cascade | GO:0007155 | CCL2, CCL5, CD34, CD4, CXCR3, FN1, SELE, SELP | 0.044 |
| Plasma membrane | GO:0007259 | CCL2, CCR2, STAT3 | 0.046 |
| Neutrophil chemotaxis | GO:0005886 | ACE, AGTR1, AGTR2, CCR2, CCR4, CCR5, CCR7, CD19, CD28, CD34, CD38, CD3E, CD4, CD40, CD40LG, CD80, CD86, CD8A, CSF1, CXCR3, FAS, FASLG, ICAM1, ICOS, IL2RA, PTPRC, SELE, SELP, TFRC, TNF, TNFRSF18 | 0.046 |
| Positive regulation of interleukin-2 biosynthetic process | GO:0045086 | IFNG, IL1B, IL8 | 0.046 |
*P values by ANOVA followed by Bonferroni adjustment.
4. Discussion
Polyoma BK virus-associated nephropathy represents the one of the most challenging infectious complications associated with kidney transplantation [15].
A better understanding of the molecular processes associated with the immune control of BKV infections may facilitate improvement of clinical management strategies [16]. Evaluation of BKV in urine samples in this study was performed in a blinded fashion; therefore no changes in clinical management were carried out based on results obtained [8] since it would have influenced the transcript expression profiles present in biopsies collected 3 months after transplantation. To the best of our knowledge, this is the first molecular study in such patient cohort. This analysis demonstrated that effective BKV control 3 months after transplantation was associated with gene expression profiles with the potential of affecting cellular immune responses, including B and T cell signaling and anti-apoptotic gene networks whereas, in late BKV-associated nephropathy, the profound gene upregulation in networks covering T cell signaling, chemoattraction, activation, and cytotoxicity along with many other inflammatory networks were detected.
Specifically, 5 genes likely to be associated with the successful control of viral replication were identified. The heightened presence of T cells (CD3E), monocyte macrophages (CD68), and their chemoattractant chemokine CCR2 (as well as presence of adhesion molecule ICAM-1) were described. Previous studies have identified these molecules to be associated with viral infections [17, 18].
It is broadly known that T cell expansion and cytokine production are needed for the generation of effective antiviral immune responses [19]. CD8+ cytotoxic T cells secreting interferon-gamma (IFN-γ) or/and tumor necrosis factor alpha (TNF-α) are important components in mediating host immunity against viral infections and have been shown to play critical roles in BKV clearance [17]. In our study, however, the expression pattern of IFN-γ and TNF-α during asymptomatic viruria was just marginal (P < 0.1) that may reflect just a limited burden of immune injury. CD8+ T lymphocyte activation is tightly regulated, especially during primary responses elicited following positive and negative costimulation following BKV infections [20, 21]. In our study, molecules associated with costimulation were consistently upregulated in kidney tissues of asymptomatic viruria patients and also in biopsies from patients with confirmed BKV-associated nephropathy. Therefore, in order to further identify additional genes associated with protection from BKV infection, data from our study was further analyzed by carrying out functional analyses. Since a low number of genes were assessed compared to the number of genes that could be analyzed following a microarray analysis, we focused our research on describing smaller interactional and functional units using a gene ontology approach and network flux that corresponds to a part of the pathway.
Flux for CD40/CD40L costimulatory signal was significantly upregulated in biopsies from patients who successfully controlled BKV infection and presented with asymptomatic viruria. It is well known that activation of CD40 on antigen presenting cells following ligation of CD40L (expressed mainly on CD4+ T lymphocytes) contributes to proinflammatory responses necessary for eradication of infections caused by certain types of pathogens [22]. Studies focused on defining cellular immune responses with the potential of controlling BKV replication determined that the majority of the BKV-specific T cells expressed CD40L (CD154) [23].
Gene targets uncovered by gene ontology analysis identified genes typically associated with the elicitation of immune responses specific to viral infections where the interplay between B and T cell function and effective cellular proliferation represents a basic protective strategy.
The activation of cellular mechanisms in response to BKV infections represents an injury-repair immune response with fibrosis development as a consequence [9]. In the current study, patients with asymptomatic viruria (who never developed BKV-associated nephropathy) presented with normal graft function at the 36-month followup. This meant that successful immune control of viral infection was likely associated with limited or transient cytokine upregulation compared to BKV-associated nephropathy where the burden of injury initiated fibrosis development.
Currently, there is limited information regarding the molecular processes associated with BKV-associated nephropathy. The upregulation of large number of genes involved in cell cycle and proliferation was shown in vitro in BKV infected primary kidney epithelial cells that suggests stimulatory nature of BKV proteins [24]. In vivo, the analysis of genes upregulated in renal allografts affected by BKV-associated nephropathy identified proinflammatory genes (CD8 and related molecules associated with graft fibrosis) similar to the profile observed during cases of acute rejection; however, expression levels were larger in magnitude [25]. In our study, intraparenchymal upregulation of 33 genes was observed in BKV-associated nephropathy, confirming previous results [25], as well as demonstrating a similar expression profile to that observed during acute rejection [25–27]. Moreover, using the functional analysis approach, another 50 biological processes were described in kidneys affected by BKV-associated nephropathy. Using hierarchical clustering, gene expression in BKV-associated nephropathy formed clearly different group.
In this study, the quantitative PCR analysis was performed. Compared to microarray-based analyses, this technique was fast and quantitative, and the results were more reliable. More specific tools for the study of specific immune responses associated with BKV infections would include ELISPOT and multiparameter flow cytometry analyses [7, 17, 28, 29].
5. Conclusion
In conclusion, this study demonstrated that asymptomatic BKV viruria reflecting successful immune system control of viral infections was associated with specific gene transcripts and immune processes, specifically transcripts associated with B lymphocyte signaling and costimulation. Furthermore, the degree of associated immune responses was much higher in patients presenting with BKV-associated nephropathy.
Supplementary Material
The gene expression profile of 90 candidate gene targets known to play a role in the elicitation of immune responses (e.g., genes involved in cytokine expression, costimulatory molecules, growth factors, chemokines, immune regulation, apoptosis and ischemia markers) was determined using real-time RT-PCR.
Conflict of Interests
The author declare that there is no conflict of interests.
Ethical Approval
Multicenter Ethics Committee of the Thomayer Hospital and Institute for Clinical and Experimental Medicine, Prague, approved the study protocol.
Acknowledgments
The authors would like to thank F. Zelezny and M. Holec for their help with data analysis using the X-gene platform. In addition, the authors are indebted to the patients and nurses for their cooperation and help. This work received a Grant from the Internal Grant Agency from the Ministry of Health (NS/9714/2008 and institutional support 00023001) and MZO 00023001.
References
- 1.Nickeleit V, Hirsch HH, Binet IF, et al. Polyomavirus infection of renal allograft recipients: from latent infection to manifest disease. Journal of the American Society of Nephrology. 1999;10(5):1080–1089. doi: 10.1681/ASN.V1051080. [DOI] [PubMed] [Google Scholar]
- 2.Costa C, Bergallo M, Astegiano S, et al. Monitoring of BK virus replication in the first year following renal transplantation. Nephrology Dialysis Transplantation. 2008;23(10):3333–3336. doi: 10.1093/ndt/gfn289. [DOI] [PubMed] [Google Scholar]
- 3.Hariharan S. BK virus nephritis after renal transplantation. Kidney International. 2006;69(4):655–662. doi: 10.1038/sj.ki.5000040. [DOI] [PubMed] [Google Scholar]
- 4.Alméras C, Foulongne V, Garrigue V, et al. Does reduction in immunosuppression in viremic patients prevent BK virus nephropathy in de novo renal transplant recipients? A prospective study. Transplantation. 2008;85(8):1099–1104. doi: 10.1097/TP.0b013e31816a33d4. [DOI] [PubMed] [Google Scholar]
- 5.Hirsch HH. BK virus: opportunity makes a pathogen. Clinical Infectious Diseases. 2005;41(3):354–360. doi: 10.1086/431488. [DOI] [PubMed] [Google Scholar]
- 6.Costa C, Bergallo M, Sidoti F, et al. Polyomaviruses BK- and JC-DNA quantitation in kidney allograft biopsies. Journal of Clinical Virology. 2009;44(1):20–23. doi: 10.1016/j.jcv.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Schachtner T, Muller K, Stein M, et al. BK virus-specific immunity kinetics: a predictor of recovery from polyomavirus BK-associated nephropathy. American Journal of Transplantation. 2011;11(11):2443–2452. doi: 10.1111/j.1600-6143.2011.03693.x. [DOI] [PubMed] [Google Scholar]
- 8.Girmanova E, Brabcova I, Bandur S, Hribova P, Skibova J, Viklicky O. A prospective longitudinal study of BK virus infection in 120 Czech renal transplant recipients. Journal of Medical Virology. 2011;83(8):1395–1400. doi: 10.1002/jmv.22106. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005;79(10):1277–1286. doi: 10.1097/01.tp.0000156165.83160.09. [DOI] [PubMed] [Google Scholar]
- 10.Platzer C, Ode-Hakim S, Reinke P, Docke WD, Ewert R, Volk HD. Quantitative PCR analysis of cytokine transcription patterns in peripheral mononuclear cells after anti-CD3 rejection therapy using two novel multispecific competitor fragments. Transplantation. 1994;58(2):264–268. [PubMed] [Google Scholar]
- 11.Notebaart RA, Teusink B, Siezen RJ, Papp B. Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Computational Biology. 2008;4(1, article e26) doi: 10.1371/journal.pcbi.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holec M, Zelezny F, Klema J, Tolar J. Integrating multiple-platform expression data through gene set features. Proceedings of the 5th International Conference on Bioinformatics Research and Applications (ISBRA '09); 2009. [Google Scholar]
- 14.Holec M, Zelezny F, Klema J, Tolar J. Cross-genome knowledge-based expression data fusion. Proceedings of the International Conference on Bioinformatics, Computational Biology, Genomics and Chemoinformatics (BCBGC '09); 2009. [Google Scholar]
- 15.Rhee J, Al-Mana N, Freeman R. Immunosuppression in high-risk transplantation. Current Opinion in Organ Transplantation. 2009;14(6):636–642. doi: 10.1097/MOT.0b013e328332a405. [DOI] [PubMed] [Google Scholar]
- 16.Babel N, Volk HD, Reinke P. BK polyomavirus infection and nephropathy: the virus-immune system interplay. Nature Reviews Nephrology. 2011;7(7):399–406. doi: 10.1038/nrneph.2011.59. [DOI] [PubMed] [Google Scholar]
- 17.Comoli P, Hirsch HH, Ginevri F. Cellular immune responses to BK virus. Current Opinion in Organ Transplantation. 2008;13(6):569–574. doi: 10.1097/MOT.0b013e3283186b93. [DOI] [PubMed] [Google Scholar]
- 18.Christensen JE, Thomsen AR. Co-ordinating innate and adaptive immunity to viral infection: mobility is the key. Acta Pathologica, Microbiologica et Immunologica Scandinavica. 2009;117(5-6):338–355. doi: 10.1111/j.1600-0463.2009.02451.x. [DOI] [PubMed] [Google Scholar]
- 19.Mueller K, Schachtner T, Sattler A, et al. BK-VP3 as a new target of cellular immunity in BK virus infection. Transplantation. 2011;91(1):100–107. doi: 10.1097/tp.0b013e3181fe1335. [DOI] [PubMed] [Google Scholar]
- 20.van Kooten G, Banchereau J. CD40-CD40 ligand. Journal of Leukocyte Biology. 2000;67(1):2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 21.Pandiyan P, Hegel JKE, Krueger M, Quandt D, Brunner-Weinzierl MC. High IFN-γ production of individual CD8 T lymphocytes is controlled by CD152 (CTLA-4) Journal of Immunology. 2007;178(4):2132–2140. doi: 10.4049/jimmunol.178.4.2132. [DOI] [PubMed] [Google Scholar]
- 22.Munroe ME. Functional roles for T cell CD40 in infection and autoimmune disease: the role of CD40 in lymphocyte homeostasis. Seminars in Immunology. 2009;21(5):283–288. doi: 10.1016/j.smim.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W, Sharma M, Martinez J, et al. Functional characterization of BK virus-specific CD4+ T cells with cytotoxic potential in seropositive adults. Viral Immunology. 2007;20(3):379–388. doi: 10.1089/vim.2007.0030. [DOI] [PubMed] [Google Scholar]
- 24.Abend JR, Low JA, Imperiale MJ. Global effects of BKV infection on gene expression in human primary kidney epithelial cells. Virology. 2010;397(1):73–79. doi: 10.1016/j.virol.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mannon RB, Hoffmann SC, Kampen RL, et al. Molecular evaluation of BK polyomavirus nephropathy. American Journal of Transplantation. 2005;5(12):2883–2893. doi: 10.1111/j.1600-6143.2005.01096.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann SC, Hale DA, Kleiner DE, et al. Functionally significant renal allograft rejection is defined by transcriptional criteria. American Journal of Transplantation. 2005;5(3):573–581. doi: 10.1111/j.1600-6143.2005.00719.x. [DOI] [PubMed] [Google Scholar]
- 27.Mannon RB, Kirk AD. Beyond histology: novel tools to diagnose allograft dysfunction. Clinical journal of the American Society of Nephrology. 2006;1(3):358–366. doi: 10.2215/CJN.01681105. [DOI] [PubMed] [Google Scholar]
- 28.Schachtner T, Müller K, Stein M, et al. BK virus-specific immunity kinetics: a predictor of recovery from polyomavirus BK-associated nephropathy. American Journal of Transplantation. 2011;11(11):2443–2452. doi: 10.1111/j.1600-6143.2011.03693.x. [DOI] [PubMed] [Google Scholar]
- 29.Seemayer CA, Seemayer NH, Dürmüller U, et al. BK virus large T and VP-1 expression in infected human renal allografts. Nephrology Dialysis Transplantation. 2008;23(12):3752–3761. doi: 10.1093/ndt/gfn470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The gene expression profile of 90 candidate gene targets known to play a role in the elicitation of immune responses (e.g., genes involved in cytokine expression, costimulatory molecules, growth factors, chemokines, immune regulation, apoptosis and ischemia markers) was determined using real-time RT-PCR.


