Abstract
Cryptococcus gattii is an environmentally occurring pathogen that is responsible for causing cryptococcosis marked by pneumonia and meningoencephalitis in humans and animals. C. gattii can form long-term associations with trees and soil resulting in the production of infectious propagules (spores and desiccated yeast). The ever expanding reports of clinical and environmental isolation of C. gattii in temperate climates strongly imply C. gattii occurs world-wide. The key ability of yeast and spores to enter, survive, multiply, and exit host cells and to infect immunocompetent hosts distinguishes C. gattii as a primary pathogen and suggest evolution of C. gattii pathogenesis as a result of interaction with plants and other organisms in its environmental niche. Here we summarize the historical literature on C. gattii and recent literature supporting the world-wide occurrence of the primary pathogen C. gattii.
Keywords: Cryptococcus; Cryptococcosis, meningitis; fungal infection; molecular typing; pneumonia; fungal virulence; VG groups; pathogenesis; emerging outbreak, same-sex mating; opposite sex mating; HIV/AIDS
Introduction
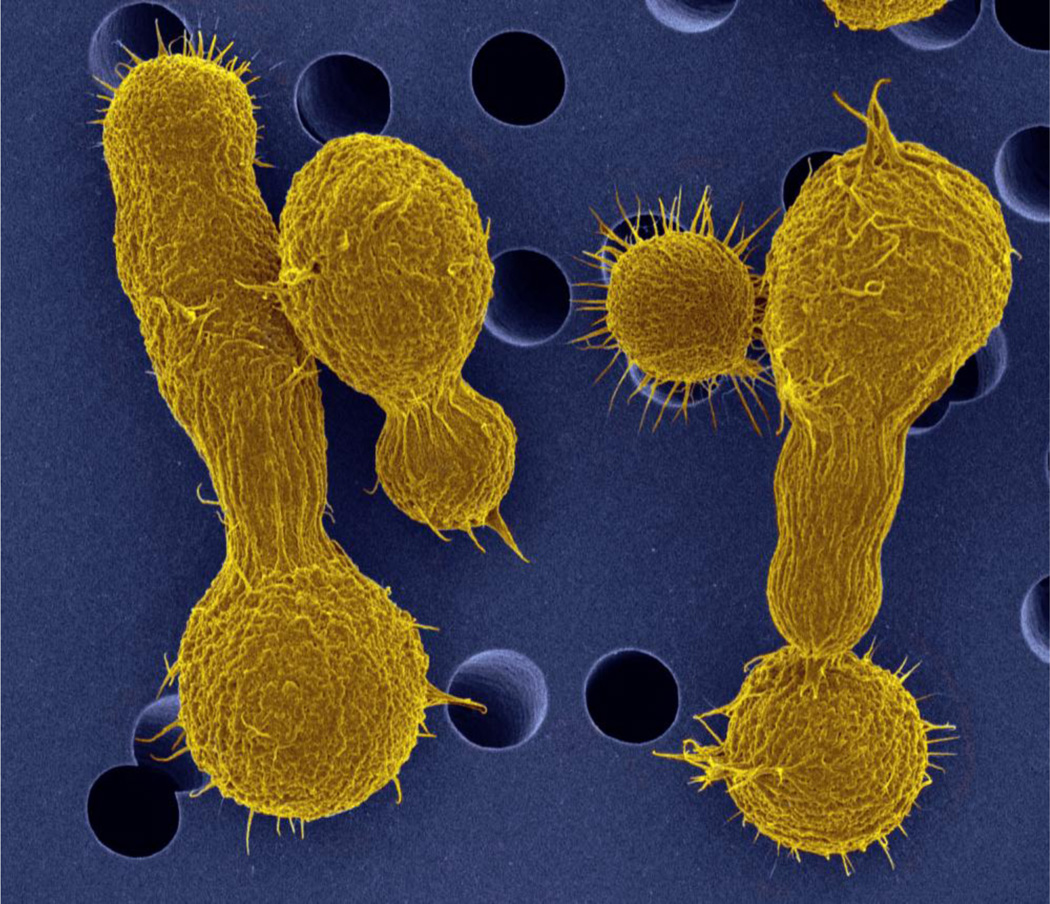
Cryptococcus gattii (formerly C. neoformans var. gattii) was first recognized by its unique cigar-shaped, elongated morphology (Figure 1) in the cerebral spinal fluid of an infected Congolese boy and later raised to species level due to differences from C. neoformans in morphology, biochemical attributes, and molecular sequences [1–5]. Cryptococcosis is a disease resulting from the inhalation of the infectious propagules of Cryptococcus from the environment. An initial lung infection can be rapidly cleared, progress to fulminant infection, or persist asymptomatically for long periods of time latently and then disseminate through the bloodstream into the brain, causing neurological symptoms, meningoencephalitis, and eventually death if untreated. Studies of virulence in C. gattii have benefitted from comparison with the well-studied sibling species, Cryptococcus neoformans. Many of the virulence factors of C. neoformans are conserved in C. gattii, including capsule, melanin, and the ability to grow at 37°C, the mechanisms of regulation are not always conserved [5–7].
Figure 1.
Pseudo-colored image of Cryptococcus gattii yeast cells demonstrating elongated-cell morphology. “Original black and white SEM image Copyright Dennis Kunkel Microscopy, Inc , pseudo colored image by Deb Springer.”
Historically, C. gattii was considered endemic in tropical and subtropical regions of the world. However, beginning in 1999, there has been a continuous and expanding outbreak in the temperate region in the Pacific Northwest. This outbreak was first recognized by the concurrent identification of humans and animals presenting with pneumonia and/or meningoencephalitis in 1999 on Vancouver Island [8, 9]. The infection rate of C. gattii on Vancouver Island between 1999 and 2003 was found to be substantially higher (~9 times) than in Australia, an endemic region for C. gattii [10]. The outbreak in the Pacific Northwest (PNW) was preceded many years earlier by a human infection with C. gattii (NIH444 =CBS6956/ATCC 32609), which was isolated in the 1970’s from the sputum of a patient in Seattle, WA, U.S.A., which lies in close proximity to Vancouver Island, B.C., Canada, and shares a similar climate [5, 11]. Furthermore, prior to the recognition of the outbreak, molecular type VGII was isolated from the environment in the San Francisco area in 1990 (CBS7750) [12]. In 2004, the outbreak expanded beyond Vancouver Island, with cases observed on the Canadian mainland in humans who had not traveled to Vancouver Island or other locations known to be high risk for C. gattii [13]. The outbreak progressed, and starting in 2005/2006, human and animal cases began to be reported in Oregon and Washington [14]. It is currently unknown how far it will eventually spread, and what barriers may exist to further spread. Monitoring the progression of this outbreak is important for public health officials as well as physicians and veterinarians.
Virulence
Under laboratory conditions, both yeast cells and spores can initiate infection and cause disease in animal models [15, 16]. Due to the smaller size of spores they may be more effectively aerosolized in nature and more efficiently enter the bronchial tubes and alveolar spaces within the lungs [15, 17]. Additionally, spores are implicated in promoting infection and disease development because, in contrast to yeast cells, they do not require opsonization for phagocytosis by macrophages [16]. The ability to persist inside of host cells is a key virulence attribute. In an evolutionary sense, this is likely linked to the interaction between Cryptococcus species, its environmental niche, and other organisms (amoeba, nematodes, insects, plants, and other fungi) [18–23]. However, virulence in the human host entails survival and non-lytic expulsion after phagocytosis by macrophages (vomocytosis) [22, 24]. Cryptococcus is a facultative intracellular pathogen, and proliferation within macrophages and the formation of tubular mitochondria has been correlated with virulence in the murine and Galleria model systems [25, 26]. Increased intracellular proliferation rates (IPR) were associated with upregulation in protein degradation and synthesis, carbohydrate metabolism, stress response, nucleotide metabolism, vesicular fusion, and transport [25]. Fusion of mitochondria resulted in morphological changes and the appearance of tubular mitochondria, which may protect against cell death and mutations by allowing complementation between mitochondria. Macrophages are implicated in both the dissemination of Cryptococcus from the lungs and the ferrying of Cryptococcus across the blood-brain barrier to the central nervous system [27]. The ability of spores to be more readily phagocytosed may dramatically impact latency and the course of infection.
Molecular types
Historical perspectives on cryptococcosis due specifically to C. gattii in AIDS patients are restricted due to a previous lack of differentiating C. gattii from C. neoformans. Cryptococcal serotypes were first readily distinguished by agglutination assays into Serotypes A, B, C, and D and later serotypes were associated with C. neoformans (Serotype A, D, and AD hybrids) and C. gattii (Serotype B and C) [28–31]. Canavanine-glycine-bromothymol blue (CGB) agar is now widely used for initial identification of C. gattii from clinical samples [30, 32, 33]. Genotyping of Cryptococcus species has resulted in major advances in differentiating C. gattii isolates. PCR fingerprinting and amplified fragment length polymorphisms (AFLP) have redefined our understanding of C. gattii into 4 molecular types: VGI, VGII, VGIII, and VGIV [31, 34–38]. C. gattii-C. neoformans hybrids are rare, obtained only from clinical samples, and identified as a result of Amplified fragment length polymorphism (AFLP), and multi-locus sequencing typing (MLST) analysis (AFLP8 and AFLP9) [39–41]. Only VGI and VGII have been associated thus far with forming hybrids with C. neoformans [40, 42].
C. gattii VGI is the most prevalent type of veterinary, clinical, and environmental isolate sampled world-wide. Clinical and veterinary cases associated with C. gattii VGI in the Netherlands, China, PNW, Eastern USA, combined with the recent documented environmental isolation of VGI from the Netherlands, Spain, Italy, and PNW strongly suggest a greatly expanded ecological niche of VGI outside the historically endemic regions [43–49]. VGII has a more restricted distribution and is most prominently associated with disease in the PNW and Australia although it has also been associated with disease in Central and South America, India, Korea, and China [50–55]. VGI and VGII primarily infect immunocompetent or non-AIDS immunocompromised hosts. C. gattii VGIII and VGIV are associated with immunocompromised AIDS patients but can also occur in patients with no apparent immune dysfunction [56–58]. The clinical presence of VGIII is prominent in California but recent reports from New Mexico suggest that VGIII has established an environmental niche in the Southwestern USA. In stark contrast to the clinical presence of VGI, VGII, and VGIII only two isolates of C. gattii, CBS7750 (VGII, 1990, San Francisco) and WM161 (VGIII, San Diego), have ever been recovered environmentally from California [31, 59]. In the Southwestern USA, identification of the ecological reservoir(s) of C. gattii appears elusive even with increased environmental sampling [50, 60, 61]. Furthermore VGIII has been associated with sporadic infections in Mexico, Colombia, Brazil, and Korea [31, 50, 57, 62]. Isolation of VGIV is much more sporadic outside of Africa but has been associated with clinical cases in Mexico, Puerto Rico, Brazil, South America, and India [31, 56, 58, 62–64]. The sporadic cases of VGIII and VGIV appear to suggest unknown environmental reservoirs because many of these patients lacked travel history to known regions of endemic populations of VGIII or VGIV [57, 63, 65].
AFLP and PCR fingerprinting readily became the methods of choice over serotyping due to higher resolution differences observed in C. gattii. The major limitation of PCR-fingerprinting and AFLP analysis is that reference strains must always be included because the results are often inconsistent which can prevent direct comparisons between research groups. Technological advances and the reduced cost of DNA sequencing have led to widespread sequencing and phylogenetic analysis of Cryptococcus. Comparisons between strains are in some cases limited because different labs routinely utilize different primer sets. The widespread implementation of MLST analysis of C. gattii has expanded our understanding of its diversity [31, 66, 67]. The proposed consensus MLST scheme for C. neoformans and C. gattii put forth by the International Society for Human and Animal Mycology (ISHAM) recommends the use of 7 unlinked loci (CAP59, GPD1, LAC1, PLB1, SOD1, URA5 and the IGS1) for all global genotyping [68]. At each locus an allele number is assigned for each unique genetic sequence observed and sequence type (ST) is then assigned based on the combination of alleles at each locus. The allele and sequence type database for C. gattii is maintained and publicly accessible at http://mlst.mycologylab.org/. The MLST approach allows for the direct comparison between global isolates of Cryptococcus and further substantiates the recognition of the 4 major molecular types of C. gattii previously recognized by PCR-fingerprinting and AFLP analysis. Three additional subclades (potential varieties) within VGII/AFLP6 (VGIIa/AFLP6a, VGIIb/AFLP6b, VGIIc/AFLP6c), and VGIII (VGIIIa and VGIIIb) [26, 39, 61, 66, 69] have been observed due to MLST analysis. Global analysis of the major MLST loci has revealed no evidence for nuclear exchange between the four VG types and these may therefore constitute four species within C. gattii [26, 31, 61, 66, 69]. One limitation of the currently recommended MLST scheme is that gene(s) within the mating type locus are not included, necessitating further effort to assign mating type.
The mitochondrial genomes between Cryptococcus species differ drastically in size and may play a role in pathogenesis [25, 26, 70]. Although gene synteny appears to be conserved, the mitochondrial genome of C. gattii (34.7 kb) is larger in comparison to C. neoformans var. neoformans (32 kb) and C. neoformans var. grubii (24 kb) [70–72]. A recent mitochondrial hybridization event was observed in the VGI lineage that may reflect hybridization and mitochondrial DNA exchange from VGII to VGI [70, 72]. Mitochondrial genomes of highly virulent C. gattii strains may be under selection because increased genetic variation was detected in coding regions versus highly conserved intergenic regions as made evident by mitochondrial recombination in C. gattii [70, 72].
Isolates of VGII and VGIII have been demonstrated to mate in laboratory settings but viable and fertile progeny are limited [3, 61]. Genetic rearrangements between the VG types may serve as reproductive barriers and limit productive recombination.
Molecular types, virulence, and the Pacific Northwest outbreak
MLST (multi-locus sequence typing) and VNTR (variable number of tandem repeats) analysis has distinguished C. gattii VGII (VGIIa and VGIIb) genotypes that are present in the Pacific Northwest [26, 61, 73]. VGIIa and VGIIb are the major and minor isolates implicated in the ongoing outbreak. The minor VGIIb genotype shows diminished virulence in comparison to VGIIa, causes a substantially smaller fraction of the outbreak-associated infections, and occurs in Australia. VGIIa and VGIIb share approximately 50% of the MLST markers analyzed, suggesting that they are related [69]. Key virulence factors like capsule, melanin, and urease are maintained between C. gattii molecular types, but the regulation of these factors can differ [6]. CAS3 in C. gattii was not linked to capsule size in vitro, in contrast to C. neoformans, but was important for virulence of both species in vivo [6]. Within C. gattii, CAS3 was found to be up-regulated in VGIIa in contrast to VGIIb. Microarray data identified several key virulence factors up-regulated in VGIIa compared to VGIIb that included Cas3, the Map kinase Mpk1 (involved in growth at 37°C), and the laccases Lac1 and Lac2 (involved in melanin synthesis) [6]. The original description of VGIIa in the PNW suggested this genotype may be restricted to the ongoing outbreak, but recently a VGIIa strain identical to R265 at 11 MLST loci has been reported from a cerebral cryptoccoma case in Japan. The patient reportedly traveled to Guam and Saipan and had no recent travel history to the USA [74]. The reservoir and occurrence of VGIIa outside the USA has not been determined but its isolation from an otherwise healthy Japanese patient suggests that VGIIa may be spreading, or have unknown geographic and environmental reservoirs outside of North America.
Byrnes et al. (2010) identified a new genotype, VGIIc, as a component of the outbreak expansion in Oregon. VGIIc demonstrates higher virulence similar to VGIIa in a murine model [26]. How the VGIIa genotype evolved is still unknown but two hypotheses have been advanced. The two divergent genotypes could be siblings from unknown parents, or one isolate (VGIIa) is the progeny of a mating occurring between VGIIb and another unknown isolate of the same mating type [69]. The second hypothesis for the origin of the VGIIa major genotype is that it resulted from opposite-sex mating, possibly in South America where MATa strains are more prevalent. There appears to be substantial evidence for recombination occurring in natural populations that suggests the possibility of same-sex mating, opposite-sex mating, or both [26, 75]. This is further substantiated by the collection of spore-sized particles using air samplers in the Vancouver Island region, which may be produced by mating because formation of spores requires sexual reproduction [73, 76]. The apparent absence of MATa strains in the outbreak region suggests that the source of the spores, and of the recombination, is likely α-α same-sex mating. As a result of extensive environmental sampling in the PNW, one VGIIa α/α diploid strain was recovered, which supports the hypothesis that same sex mating among C. gattii is contributing to the outbreak [69]. Evidence of recombination occurring within the natural population also supports ongoing sexual reproduction [26, 75]. The origin of VGIIc is not yet understood. Thus far, VGIIc appears restricted and has not been isolated from patients outside Oregon or from any environmental sampling. The origin of VGIIc is unknown but could have resulted from a locally derived mutation and recombination event or from a recent introduction event. The ongoing clinical and environmental sampling and the low cost of whole genome sequencing will facilitate the sequencing of multiple strains as an alternative to MLST analysis. This approach will likely yield further understanding of the relationships between genotype, origin, and virulence of C. gattii [77].
Environmental distribution of C. gattii
Cryptococcosis is caused by the pathogenic Cryptococcus species including C. neoformans and C. gattii. Although attention is drawn to Cryptococcus as mammalian pathogens, natural populations occur as part of the environmental flora associated with trees, soil, and bird droppings. These environmental niches serve as avenues of exposure and disease acquisition because they are essentially reservoirs of the infectious yeast and spore particles. Thus, studying the distribution and pathogenicity of naturally occurring, non-clinical isolates will advance our understanding of Cryptococcus.
Environmental sampling involves gathering debris, decaying wood, and bird droppings, as well as swabbing various plants including, but not limited to, Eucalyptus, Almond, Douglas fir, Laurel, and Cacti [10, 63, 78–81]. Using conventional methods involving selective, defined nutrient media in addition to molecular analyses including MLST and AFLP markers, natural populations of C. gattii have been identified in both tropical and temperate regions. The global distribution of the four VG groups within C. gattii is summarized in Table 1. VGIV is relatively rare in non-clinical environments and most commonly found in sub-Saharan African AIDS patients, but also has been reported from Mexico, Brazil, and Puerto Rico [58, 62, 63]. VGIII is clinically prevalent in California, Mexico, was recently found as the cause of a fatal clinical case in New Mexico, and isolated from cacti in Cuba [60, 61, 82]. Although C. gattii is traditionally thought to be restricted to the tropics, not all tropical regions have reported natural populations. VGIII has not been isolated from indigenous trees (mopane trees) or feral pigeon droppings in South Africa [83].
Table 1.
Cryptococcus molecular types, geographic and clinical distribution
| Species/Hybrids* | Geographic distribution | Group | Source | Patient risk | Reference Strains |
|---|---|---|---|---|---|
| C. neoformans var. grubii | Most common worldwide | VNI | Clinical, Veterinary, & Environmental | world-wide in compromised, immunocompetent China and Korea | H99(α); 125.91 (MATa) |
| Aftrica | VNB | Clinical | compromised, rare in immunocompetent | BT148(α); BT63 (MATa) | |
| Australia | VNII | Clinical, Veterinary, & Environmental | compromised, rare in immunocompetent | 8-1(α) | |
| C. n. var. grubii -C. n. var. neoformans* | Europe, South America | VNIII | Clinical | compromised, rare in immunocompetent | KW5αADa; CDC228 |
| C. neoformans var. neoformans | Europe, South America | VNIV | Clinical, Veterinary, & Environmental | compromised, rare in immunocompetent | JEC21(α); JEC20 (MATa) |
| C. gattii | Most comon C. gattii , Europe, less common in Africa, South America, North America, Australia | VGI | Clinical, Veterinary, & Environmental | Immunocompetent, rare in compromised | WM276(α); E566 (MATa) |
| North and South America, Australia, less common in Oceania | VGII | Clinical | Imunocompetent | WM178(α); CBS1930 (MATa) | |
| North America, reported from Asia, Australia, Europe, South America | VGIIa | Clinical, Veterinary, & Environmental | R265(α) | ||
| Northwestern USA, Australia, Southwestern Canada | VGIIb | Clinical, Veterinary, & Environmental | R272(α) | ||
| Oregon, USA | VGIIc | Clinical & Veterinary | EJB18(α) | ||
| South America less common in Noth America, Australia, Europe, Africa | VGIIIa | Clinical, Veterinary, & Environmental | Immunocompetent, HIV+ | WM161(α); CA1872 (MATa) | |
| VGIIIb | Clinical | NIH312(α); B4546 (MATa) | |||
| Africa, India, South America | VGIV | Clinical, Veterinary, & Environmental | Immunocompetent, HIV+ | BT26; WM779(α) | |
| C. n. var. neoformans(VNIII) -C. gattii (VGI)* | Africa, Netherlands, South America | NA | Clinical | Compromised | CBS10488, CBS10489, CBS10490 |
| C. n. var. grubii (VNI)-C. gattii (VGI)* | Europe, North America | CBS10496 |
Recent identification of the clinical presence of VGI in the Netherlands, Spain, Italy, and the USA in combination with successful environmental isolation from the Netherlands, Spain, Italy, India, and China is beginning to mirror the documented outbreak and expansion of the ecological niche observed for VGII in Canada and the USA [44, 46–48, 84–86]. These observations suggest an ongoing outbreak and expansion of VGI. VGI has been historically common in Australia and was found predominantly in environmental sampling across India [48, 51, 84, 85, 87]. Very little gene flow was detected between the populations of different geographic locations or between different host tree species in India, questioning the dissemination of the strains beyond their local niches. Overall, in both Australia and India, a strong clonality was found in the population structure of VGI as compared to VGII, which shows moderate signs of sexual recombination in Australia [14, 88]. VGII is very common in the PNW, Australia, and India and has recently been found in clinical and environmental isolates from Brazil [55, 89]. In Brazil, more isolates (94%) in the clinical environment were found to be VGII than VGI (6%); but, the proportion of VGI (25%) increased in the non-clinical isolates collected in soil and bird droppings, suggesting a higher virulence of VGII in mammalian hosts [90].
Taken together, the distribution patterns of VG lineages within C. gattii do not fit what is expected under the “everything is everywhere” hypothesis but rather provide evidence that the VG types are endemic to specific regions of the world. Such population structure may not allow frequent gene flow, promoting speciation within C. gattii in nature. Nevertheless, the frequent occurrence of all VG groups except VGIII and VGIV in non-clinical environments warrants further research into the dissemination of these populations as possible sources of infection of mammalian hosts.
AIDS and Cryptococcus gattii
Prior to the 1950’s, the incidence of cryptococcosis was relatively low and most frequently encountered in the tropics. The emergence of acquired immunodeficiency syndrome (AIDS) dramatically altered the prevalence of Cryptococcus. In Africa, cryptococcosis is associated with an increased occurrence of meningitis in an unusually younger cohort of patients. The increased awareness of cryptococcosis in the Congo River Basin led to the initial identification of cryptococcosis caused by an atypical strain with elongated-morphology later recognized as C. gattii [1, 91–93]. Today, infections caused by Cryptococcus are recognized as a significant cause of morbidity and mortality in HIV/AIDS patients and are annually responsible for >1,000,000 infections, >620,000 deaths, and up to one-third of all AIDS associated deaths [94, 95]. Recent estimates indicate that Cryptococcus is a more prevalent cause of HIV related mortality in sub-Saharan Africa than tuberculosis [95]. In other developing countries, Cryptococcus infections are second only to tuberculosis, and the two frequently co-occur in the AIDS population [96, 97]. The endemic, world-wide, environmental prevalence of the closely related varieties or species, C. neoformans var. neoformans and C. neoformans var. grubii, has resulted in their frequent association with AIDS patients. The proportion of Cryptococcus/AIDS related infections that are inherently due to C. gattii has only recently been addressed because reporting of Cryptococcus infections and the determination of species is not prevalent world-wide. Retrospective and more recent clinical studies indicate C. gattii associated disease in AIDS patients is less common than C. neoformans, but the actual incidence may be dependent upon geographical origin, travel, and environmental exposure to the pathogen. C. gattii causes many fewer infections in AIDS patients compared to C. neoformans. However C. gattii is more prevalent than C. neoformans in immunocompetent and apparently healthy individuals. Serotype C, VGIII and VGIV are the most prolific C. gattii types associated with AIDS patients, but Serotype B, VGI, VGII and C. gattii-C. neoformans (VGI/VNI) hybrids have been sporadically isolated from AIDS patients [40, 41, 61, 98–101]. In the United States C. gattii molecular type VGIII (12%) is most prevalent in Californian AIDS patients. In South Africa VGIII (1%) and VGIV (1%) were found in AIDS patients [58, 61, 98, 101, 102]. In Sub-Saharan Africa, Botswana, and Malawi, VGIV was prevalent in AIDS patients (13.3%) [64]. Case reports from other parts of the world (UK, Mexico, and India) have also described C. gattii in HIV+ individuals [41, 103–105]. Long term outcomes of patients with AIDS and Cryptococcus have improved due to the decreased cost and continual development of antifungal drugs, highly active antiretroviral therapy (HAART), and combination therapy regimens, but unfortunately cryptococcal infections are still common, frequently recur, and remain difficult to cure. Key issues in undeveloped nations include difficulties in managing treatment with amphotericin B, the fact that flucytosine is not available, and that most patients receive only fluconazole, which is fungistatic and not fungicidal and can lead to drug resistance.
Sexual cycle of C. gattii
Cryptococcus gattii can reproduce asexually through budding and sexually through mating between the two mating types (a-α, opposite-sex) (Figure 1). However, natural populations of C. gattii predominantly contain strains of α mating type, limiting the opportunities for opposite-sex mating. This mating type is ubiquitous in all geographic regions, in both clinical and environmental isolates, and in every molecular type within C. gattii (VGI, VGII VGIII, and VGIV). For example, all VGI strains in India and Europe (Spain, Italy, Netherlands) and VGII strains in the PNW have the mating type α, which is also common in VGIII strains in southern California and Mexico [26, 48, 61, 87]. These observations suggest that C. gattii, like its sibling species C. neoformans, may undergo unisexual mating (α-α). In C. neoformans, similar environmental cues trigger opposite-sex and unisexual mating. Morphological features that distinguish the two types of mating are also known and include fused vs. unfused clamp connections and dikaryotic vs. monokaryotic hyphae (Table 2). Unisexual mating in C. gattii has not yet been described under laboratory conditions; however, indirect evidence has been adduced. For instance, phylogenetic incompatibilities, evidence of clonality, and limited but unambiguous evidence of recombination is found in unisexual populations of C. gattii, suggesting that unisexual mating may contribute to generating genotypic diversity in natural populations [106]. Here, we review the recent advancements in our understanding of the significance and frequency of opposite-sex and unisexual mating in natural populations and its impact on the pathogenicity of C. gattii.
Table 2.
Features distinguishing unisexual and opposite-sex reproduction in C. neoformans (Lin et al 2005)
| Feature | Unisexual reproduction |
Opposite-sex reproduction |
|---|---|---|
| Mating types involved | α | a and α |
| Hyphae | Monokaryotic | Dikaryotic |
| Clamp connections | Unfused | Fused |
| Blastospores | Haploid or diploid, always α | Haploid or diploid, could be a or α |
| Spores | All α | a and α in a 1:1 ratio |
Frequency of sex
The recent discovery of a sexual cycle in C. amylolentus indicates that virtually all species of the Cryptococcus sensu stricto clade can undergo sexual reproduction [107]. Studies on C. gattii indicate that some strains within a species could experience more sexual reproduction in nature than others. For example, strains in the VGIII lineage of C. gattii are highly fertile and produce more abundant spores than VGII strains. VGIII also has a higher genetic diversity, with 13 unique genotypes in southern CA, and more instances of recombination than VGII, which has a largely clonal population in the PNW. This discrepancy may reflect either a high frequency of sex in natural populations of VGIII, that VGIII was introduced to the southern California region a long time ago, or both [61]. Moreover, the VGIIIb subgroup mates more avidly under laboratory conditions and shows more signs of greater recombination than VGIIIa. The largely clonal structure of many populations of C. gattii may suggest either less instances of opposite-sex mating relative to asexual reproduction or frequent instances of unisexual mating between closely related or identical isolates in nature. Direct demonstration of unisexual mating in the laboratory would verify the latter possibility.
Lack of allelic exchange between the VG groups, based on MLST analyses on ~300 global isolates, is indicative of speciation [14]. VGI, VGII, and VGIII are molecularly distinct, geographically separated, which may limit opportunities for mating. Although hybridization events among C. gattii VG types have been demonstrated in the laboratory, limited evidence exists from nature [3, 108]. Recent MLST analysis suggests introgression events may have occurred between the VGIIIa and VGIIIb subgroups as indicated by the non-ancestral shared allele of PLB1 [61]. C. neoformans hybrids with C. gattii VGI, VGII and VGIII also have been reported from clinical isolates, and mating has been demonstrated under laboratory conditions [40, 41, 61, 109]. It is unclear whether all VG groups are cross-fertile or mate with closely related species since fertility varies widely between isolates.
Consequences of sex
Among the pathogenic Cryptococcus complex, C. gattii is a robust pathogen of immunocompetent individuals in contrast to C. neoformans, which is primarily restricted to immunocompromised patients. However, less commonly infections in apparently healthy individuals by C. neoformans have been reported and some molecular types of C. gattii are more frequently associated with disease in immunocompromised individuals [61, 110–112]. The changes underlying these host shifts may include rewiring of already existing virulence machinery through sexual recombination. Hence, studying the sexual cycle in C. gattii may be fundamental to understanding the evolution of its pathogenicity. Sexual reproduction could contribute to the pathogenicity of C. gattii by generating genetically diverse progeny (basidiospores) that are also infectious propagules. In addition, spores are resistant to desiccation, are readily airborne, easily inhaled, and capable of penetrating the alveoli [113]. In Southern California, both mating types are present in VGIII clinical populations, suggesting the possibility for opposite-sex mating in nature [14]. Several of these isolates were indeed found to be highly fertile in laboratory tests, producing abundant spores. In populations where only one mating type predominates, unisexual mating may generate similar infectious propagules in C. gattii.
The process of meiotic recombination during mating may bring together useful gene combinations, producing progeny better suited to expand the host range including immunocompetent individuals and temperate regions as implicated in VGII isolates found in the PNW. Clinical isolates of the VGII lineage also show higher minimum inhibitory concentrations for azole drugs and may require more aggressive and longer term treatment [14, 114]. Sex may also help generate greater variance in virulence characteristics. For example, in the VGIII lineage, which is generally very fertile, frequent recombination may have created larger variation in virulence with some progeny emerging as more virulent and others less virulent. Similarly, the subgroup VGIIIb is more fertile and less virulent as compared to VGIIIa, both in murine models and IPR assays [61]. The VGII and VGIIIa genotypes may represent highly virulent sexual progeny whose virulence could be maintained through clonal reproduction. Both VGII and VGIIIa lineages contain isolates almost exclusively of α mating type. Consequently, unisexual mating may also contribute to maintaining the gene combinations responsible for increased virulence.
Lastly, a direct link between mating and virulence pathway is possible because the mating type locus (MAT) may directly contribute to regulation of virulence. The MAT locus regulates the response to mating pheromones and subsequent hyphae and basidiospore formation. Previous studies in C. neoformans have shown that congenic strains, which differ only at the MAT locus, differ in virulence, indicating that MAT contributes to pathogenicity of the species [109, 115–117]. Additional investigations observed no difference in virulence between strains of opposite mating types indicating that the influence of MAT depends on the genetic background [117–119]. Virulence is a polygenic trait and the MAT locus governs virulence in concert with other genes, stressing the importance of genetic background in regulation of virulence. The MAT locus in the VGIII lineage exhibits genomic plasticity including gene rearrangements, truncation, and gene loss [61]. The observed plasticity may be a direct cause or a consequence of the associated high fertility of VGIII. C. gattii has a bipolar MAT locus similar to its pathogenic sister species, C. neoformans. Studies on closely related non-pathogenic species of Cryptococcus suggest that the transition to bipolarity occurred recently and concomitantly with the emergence of the pathogenic C. neoformans/C. gattii species cluster [107]. Studying the structure and function of the MAT loci of various clinical and non-clinical isolates and its association with virulence is necessary to establish the role of mating in the regulation of pathogenicity in C. gattii.
Conclusion
Research has demonstrated that C. gattii is more than just an opportunistic pathogen because it harbors distinct mechanisms geared to evade, subvert, and manipulate the host immune system, while maintaining intracellular growth. Cryptococcus gattii is likely to have a wider geographical distribution than presently appreciated based on the ongoing expansion of clinical and environmental reports worldwide. Historically, the prevalence of C. gattii has been overlooked because most clinical laboratories did not distinguish C. gattii from C. neoformans. Due to the increasing documentation of C. gattii in previously undocumented regions, observed differences in drug sensitivity, and disease outcome, clinicians should pursue species-specific diagnoses for all cases presenting with cryptococcosis. CGB agar and MLST analyses are quick, inexpensive, and reliable methods of distinguishing C. gattii from C. neoformans and should be incorporated into standard clinical testing procedures for cryptococcosis.
Acknowledgements
We thank Jo Kingsbury, Anna Averette, Marianna Feretzaki, and Virginia Lehman for critical review of the manuscript. This research is supported by R37 award AI39115 and RO1 award AI50113 from the NIH/NIAID.
Footnotes
Disclosure
D. J. Springer: none; S. Phadke: none; B. Billmyre: grant from the NIH; J. Heitman: employed by Duke, grants from NIH, Merck and Astellas
References
Papers of particular interest, published recently have been highlighted as:
• Of importance
•• Of major importance
- 1.Vanbreuseghem R, Takashio M. An atypical strain of Cryptococcus neoformans (San Felice) Vuillemin 1894. II. Cryptococcus neoformans var. gattii var. nov. Ann Soc Belges Med Trop Parasitol Mycol. 1970;50:695–702. [PubMed] [Google Scholar]
- 2.Sorrell TC. Cryptococcus neoformans variety gattii. Medical Mycology. 2001;39:155–168. [PubMed] [Google Scholar]
- 3.Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- 4.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae) Taxon. 2002;51:804–806. [Google Scholar]
- 5. Cryptococcus: from human pathogen to model yeast. Washington, DC: ASM Press; 2011. •• Comprehensive book on Cryptococcus. Assembled chapters from a contingent of international experts on the up to date assembly of research on Cryptococcus.
- 6.Ngamskulrungroj P, Price J, Sorrell T, Perfect JR, Meyer W. Cryptococcus gattii virulence composite: candidate genes revealed by microarray analysis of high and less virulent Vancouver island outbreak strains. PLoS ONE. 2011;6:e16076. doi: 10.1371/journal.pone.0016076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngamskulrungroj P, Chang Y, Roh J, Kwon-Chung KJ. Differences in nitrogen metabolism between Cryptococcus neoformans and C. gattii the two etiologic agents of Cryptococcosis. PLoS ONE. 2012;7:e34258. doi: 10.1371/journal.pone.0034258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoang LMN, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997–2002): epidemiology, microbiology and histopathology. J. Med. Microbiol. 2004;53:935–940. doi: 10.1099/jmm.0.05427-0. [DOI] [PubMed] [Google Scholar]
- 9.Stephen C, Lester S, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43:792–794. [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc. Natl. Acad. Sci. U.S.A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryotic Cell. 2005;4:1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeiffer T, Ellis D. Environmental isolation of Cryptococcus neoformans var. gattii from California. J Infect Dis. 1991;163:929–930. doi: 10.1093/infdis/163.4.929. [DOI] [PubMed] [Google Scholar]
- 13.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Byrnes EJ, 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J Infect Dis. 2009;199:1081–1086. doi: 10.1086/597306. •• Demonstrated C. gattii expansion outside of Vancouver, BC into the USA.
- 15.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun. 2009;77:3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botts MR, Giles SS, Gates MA, Kozel TR, Hull CM. Isolation and characterization of Cryptococcus neoformans spores reveal a critical role for capsule biosynthesis genes in spore biogenesis. Eukaryot Cell. 2009;8:595–605. doi: 10.1128/EC.00352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz A, Neilson JB, Bulmer GS. Control of Cryptococcus neoformans in nature by biotic factors. Sabouraudia. 1982;20:21–29. [PubMed] [Google Scholar]
- 19.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A. 2001;98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malliaris SD, Steenbergen JN, Casadevall A. Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. Medical Mycology. 2004;42:149–158. doi: 10.1080/13693786310001616500. [DOI] [PubMed] [Google Scholar]
- 21.Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect. Immun. 2003;71:4862–4872. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Current Biology. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 23. Springer DJ, Ren P, Raina R, Dong Y, Behr MJ, McEwen BF, et al. Extracellular fibrils of pathogenic yeast Cryptococcus gattii are important for ecological niche, murine virulence and human neutrophil interactions. PLoS ONE. 2010;5:e10978. doi: 10.1371/journal.pone.0010978. • First to demonstrate increased virulence of C. gattii from growth on plant material.
- 24.Alvarez M, Burn T, Luo Y, Pirofski LA, Casadevall A. The outcome of Cryptococcus neoformans intracellular pathogenesis in human monocytes. BMC Microbiol. 2009;9:51. doi: 10.1186/1471-2180-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A. 2009;106:12980–12985. doi: 10.1073/pnas.0902963106. •• A comprehensive study linking tubular mitochondria, mitochondrial regulation and enhanced intracellular proliferation of C. gattii
- 26. Byrnes EJ, 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000850. e1000850. •• MLST study including many isolates that provides evidence of clonal expansion and the emergence of novel genotypes. Also distinguishes VGIIc as a highly virulent type by murine virulence and intracellular proliferation.
- 27.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans. Infect Immun. 2009;77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans EE. The antigenic composition of Cryptococcus neoformans. I. A serologic classification by means of the capsular and agglutination reactions. J Immunol. 1950;64:423–430. [PubMed] [Google Scholar]
- 29.Wilson DE, Bennett JE, Bailey JW. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127:820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]
- 30.Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C) J. Clin. Microbiol. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel RA. The indirect fluorescent antibody test for the detection of antibody in human cryptococcal disease. The Journal of Infectious Diseases. 1966;116:573–580. doi: 10.1093/infdis/116.5.573. [DOI] [PubMed] [Google Scholar]
- 34.Boekhout T, van Belkum A, Leenders AC, Verbrugh HA, Mukamurangwa P, Swinne D, et al. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int. J. Syst. Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 35.Meyer W, Mitchell TG, Freedman EZ, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J. Clin. Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chowdhary A, Randhawa HS, Sundar G, Kathuria S, Prakash A, Khan Z, et al. In vitro antifungal susceptibility profiles and genotypes of 308 clinical and environmental isolates of Cryptococcus neoformans var. grubii and Cryptococcus gattii serotype B from northwestern India. Journal of Medical Microbiology. 2011;60:961–967. doi: 10.1099/jmm.0.029025-0. • A detailed analysis and comparison of different species and molecular types demonstrating differences in antifungal susceptibility.
- 37.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WCJ, Abeln ECA, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 38.Marra RE, Huang JC, Fung E, Nielsen K, Heitman J, Vilgalys R, et al. A genetic linkage map of Cryptococcus neoformans variety neoformans serotype D (Filobasidiella neoformans) Genetics. 2004;167:619–631. doi: 10.1534/genetics.103.023408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hagen F, Illnait-Zaragozi M-T, Bartlett KH, Swinne D, Geertsen E, Klaassen CHW, et al. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 2010;54:5139–5145. doi: 10.1128/AAC.00746-10. • A detailed analysis demonstrating differences in antifungal susceptibility of large collection of C. gattii isolates.
- 40.Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, et al. AIDS patient death caused by novel Cryptococcus neoformans x C. gattii hybrid. Emerg Infect Dis. 2008;14:1105–1108. doi: 10.3201/eid1407.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aminnejad M, Diaz M, Arabatzis M, Castaneda E, Lazera M, Velegraki A, et al. Identification of novel hybrids between Cryptococcus neoformans var. grubii VNI and Cryptococcus gattii VGII. Mycopathologia. 2012;173:337–346. doi: 10.1007/s11046-011-9491-x. [DOI] [PubMed] [Google Scholar]
- 42.Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 43. Romeo O, Scordino F, Criseo G. Environmental isolation of Cryptococcus gattii serotype B, VGI/MATalpha strains in southern Italy. Mycopathologia. 2011;171:423–430. doi: 10.1007/s11046-010-9389-z. • Environmental isolation of C. gattii from Italy.
- 44. Chowdhary A, Randhawa HS, Boekhout T, Hagen F, Klaassen CH, Meis JF. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerging Infectious Diseases. 2012;18:172–174. doi: 10.3201/eid1801.111190. • First environmental isolation of C. gattii VGI from the Netherlands.
- 45. Sellers B, Hall P, Cine-Gowdie S, Hays AL, Patel K, Lockhart SR, et al. Cryptococcus gattii: An emerging fungal pathogen in the Southeastern United States. The American Journal of the Medical Sciences. 2012;343:510–511. doi: 10.1097/MAJ.0b013e3182464bc7. • Expanded incidence of C. gattii VGI outside thw western USA and Canada.
- 46.Byrnes EJ, 3rd, Li W, Lewit Y, Perfect JR, Carter DA, Cox GM, et al. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS ONE. 2009;4:e5851. doi: 10.1371/journal.pone.0005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCulloh RJ, Phillips R, Perfect JR, Byrnes EJ, 3rd, Heitman J, Dufort E. Cryptococcus gattii genotype VGI infection in New England. The Pediatric Infectious Disease Journal. 2011;30:1111–1114. doi: 10.1097/INF.0b013e31822d14fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Colom MF, Hagen F, Gonzalez A, Mellado A, Morera N, Linares C, et al. Ceratonia siliqua (carob) trees as natural habitat and source of infection by Cryptococcus gattii in the Mediterranean environment. Medical Mycology. 2012;50:67–73. doi: 10.3109/13693786.2011.574239. • First environmental isolation of C. gattii VGI from Spain.
- 49.Chen M, Liao WQ, Wu SX, Yao ZR, Pan WH, Liao Y. Taxonomic analysis of Cryptococcus species complex strain S8012 revealed Cryptococcus gattii with high heterogeneity on the genetics. Chinese Medical Journal. 2011;124:2051–2056. [PubMed] [Google Scholar]
- 50.Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, Carriconde F, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Res. 2010;10:769–778. doi: 10.1111/j.1567-1364.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng X, Yao Z, Ren D, Liao W, Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 2008;8:930–938. doi: 10.1111/j.1567-1364.2008.00422.x. [DOI] [PubMed] [Google Scholar]
- 52.Matos CS, de Souza Andrade A, Oliveira NS, Barros TF. Microbiological characteristics of clinical isolates of Cryptococcus spp. in Bahia, Brazil: molecular types and antifungal susceptibilities. European Journal of Clinical Microbiology & Infectious Diseases. 2012;31:1647–1652. doi: 10.1007/s10096-011-1488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debourgogne A, Hagen F, Elenga N, Long L, Blanchet D, Veron V, et al. Successful treatment of Cryptococcus gattii neurocryptococcosis in a 5-year-old immunocompetent child from the French Guiana Amazon region. Revista Iberoamericana de Micologia. 2012 doi: 10.1016/j.riam.2012.01.008. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 54.Pasa CR, Chang MR, Hans-Filho G. Post-trauma primary cutaneous cryptococcosis in an immunocompetent host by Cryptococcus gattii VGII. Mycoses. 2012;55:e1–e3. doi: 10.1111/j.1439-0507.2011.02058.x. [DOI] [PubMed] [Google Scholar]
- 55.Martins LM, Wanke B, Lazera Mdos S, Trilles L, Barbosa GG, de Macedo RC, et al. Genotypes of Cryptococcus neoformans and Cryptococcus gattii as agents of endemic cryptococcosis in Teresina, Piaui (northeastern Brazil) Memorias do Instituto Oswaldo Cruz. 2011;106:725–730. doi: 10.1590/s0074-02762011000600012. [DOI] [PubMed] [Google Scholar]
- 56.Cogliati M, Chandrashekar N, Esposto MC, Chandramuki A, Petrini B, Viviani MA. Cryptococcus gattii serotype-C strains isolated in Bangalore, Karnataka, India. Mycoses. 2012;55:262–268. doi: 10.1111/j.1439-0507.2011.02082.x. [DOI] [PubMed] [Google Scholar]
- 57.Firacative C, Torres G, Rodriguez MC, Escandon P. First environmental isolation of Cryptococcus gattii serotype B, from Cucuta, Colombia. Biomedica. 2011;31:118–123. doi: 10.1590/S0120-41572011000100014. [DOI] [PubMed] [Google Scholar]
- 58.Olivares LR, Martinez KM, Cruz RM, Rivera MA, Meyer W, Espinosa RA, et al. Genotyping of Mexican Cryptococcus neoformans and C. gattii isolates by PCR-fingerprinting. Med Mycol. 2009;47:713–721. doi: 10.3109/13693780802559031. [DOI] [PubMed] [Google Scholar]
- 59.Byrnes EJ, 3rd, Marr KA. The outbreak of Cryptococcus gattii in western North America: epidemiology and clinical issues. Current Infectious Disease Reports. 2011;13:256–261. doi: 10.1007/s11908-011-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Walraven CJ, Gerstein W, Hardison SE, Wormley F, Lockhart SR, Harris JR, et al. Fatal disseminated Cryptococcus gattii infection in New Mexico. PLoS ONE. 2011;6:e28625. doi: 10.1371/journal.pone.0028625. • First clinical report of C. gattii in New Mexico. Evidence for a wider distribution of C. gattii.
- 61. Byrnes EJ, 3rd, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, et al. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathogens. 2011;7 doi: 10.1371/journal.ppat.1002205. e1002205. •• A primary report providing a detailed account of the population structure and endemicity of C. gattii in California. Crucial in highlighting the prevalence of C. gattii VGIII in HIV+ individuals of California.
- 62.Trilles L, Lazera Mdos S, Wanke B, Oliveira RV, Barbosa GG, Nishikawa MM, et al. Regional pattern of the molecular types of Cryptococcus neoformans and Cryptococcus gattii in Brazil. Mem Inst Oswaldo Cruz. 2008;103:455–462. doi: 10.1590/s0074-02762008000500008. [DOI] [PubMed] [Google Scholar]
- 63. Loperena-Alvarez Y, Ren P, Li X, Schoonmaker-Bopp DJ, Ruiz A, Chaturvedi V, et al. Genotypic characterization of environmental isolates of C. gattii from Puerto Rico. Mycopathologia. 2010;170:279–285. doi: 10.1007/s11046-010-9296-3. • First report expanding C. gattii niche to cacti.
- 64.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 65. Souza LKH, Souza Junior AH, Costa CR, Faganello J, Vainstein MH, Chagas ALB, et al. Molecular typing and antifungal susceptibility of clinical and environmental Cryptococcus neoformans species complex isolates in Goiania, Brazil. Mycoses. 2010;53:62–67. doi: 10.1111/j.1439-0507.2008.01662.x. • Demonstrates differences in sensitivity to antifungals between C. gattii molecular types.
- 66.Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, et al. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS ONE. 2009;4:e5862. doi: 10.1371/journal.pone.0005862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katsu M, Kidd S, Ando A, Moretti-Branchini ML, Mikami Y, Nishimura K, et al. The internal transcribed spacers and 5.8S rRNA gene show extensive diversity among isolates of the Cryptococcus neoformans species complex. FEMS Yeast Research. 2004;4:377–388. doi: 10.1016/S1567-1356(03)00176-4. [DOI] [PubMed] [Google Scholar]
- 68.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:1–14. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. •• First report to demonstrate the unique genetics and clonal spread of C. gattii strains associated with the Vancouver Island outbreak.
- 70.Bovers M, Hagen F, Kuramae EE, Boekhout T. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 2009;9:489–503. doi: 10.1111/j.1567-1364.2009.00494.x. [DOI] [PubMed] [Google Scholar]
- 71.Litter J, Keszthelyi A, Hamari Z, Pfeiffer I, Kucsera J. Differences in mitochondrial genome organization of Cryptococcus neoformans strains. Antonie Van Leeuwenhoek. 2005;88:249–255. doi: 10.1007/s10482-005-8544-x. [DOI] [PubMed] [Google Scholar]
- 72. Xu J, Yan Z, Guo H. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol Ecol. 2009;18:2628–2642. doi: 10.1111/j.1365-294X.2009.04227.x. • Demonstrates historical mitochondrial genome divergence within C. gattii with recent evidence for hybridization and recombination in the mitochondrial genome.
- 73.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Envir. Microbiol. 2007;73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, et al. Cryptococcus gattii genotype VGIIa infection in man, Japan, 2007. Emerging Infectious Diseases. 2010;16:1155–1157. doi: 10.3201/eid1607.100106. • First report of VGIIa outside of its previously described region of occurrence in the the USA.
- 75. Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, et al. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population--an emerging outbreak in Australia. PLoS ONE. 2011;6:e16936. doi: 10.1371/journal.pone.0016936. • Important analysis of the population genetics of VGII in Australia. Discusses and contrasts same-sex and opposite-sex hypotheses for the outbreak in the Pacific Northwest and Australia.
- 76. Kidd SE, Bach PJ, Hingston AO, Mak S, Chow Y, MacDougall L, et al. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg Infect Dis. 2007;13:51–57. doi: 10.3201/eid1301.060823. • Important reference for the natural and human-driven dispersal mechanisms of C. gattii in the Pacific Northwest, with worldwide implications.
- 77. Gillece JD, Schupp JM, Balajee SA, Harris J, Pearson T, Yan Y, et al. Whole genome sequence analysis of Cryptococcus gattii from the Pacific Northwest reveals unexpected diversity. PLoS ONE. 2011;6:e28550. doi: 10.1371/journal.pone.0028550. • Whole genome analysis of C. gattii drawing attention to similarities and differences between molecular types.
- 78.Callejas A, Ordonez N, Rodriguez MC, Castaneda E. First isolation of Cryptococcus neoformans var. gattii serotype C, from the environment in Colombia. Med Mycol. 1998;36:341–344. [PubMed] [Google Scholar]
- 79.Mseddi F, Sellami A, Jarboui MA, Sellami H, Makni F, Ayadi A. First environmental isolations of Cryptococcus neoformans and Cryptococcus gattii in Tunisia and review of published studies on environmental isolations in Africa. Mycopathologia. 2011;171:355–360. doi: 10.1007/s11046-010-9381-7. [DOI] [PubMed] [Google Scholar]
- 80.Pfeiffer TJ, Ellis DH. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. Medical Mycology. 1992;30:407–408. [PubMed] [Google Scholar]
- 81.Randhawa HS, Kowshik T, Chowdhary A, Preeti Sinha K, Khan ZU, Sun S, et al. The expanding host tree species spectrum of Cryptococcus gattii and Cryptococcus neoformans and their isolations from surrounding soil in India. Med Mycol. 2008;46:1–11. doi: 10.1080/13693780802124026. [DOI] [PubMed] [Google Scholar]
- 82.Illnait-Zaragozi MT, Martinez-Machin GF, Fernandez-Andreu CM, Perurena-Lancha MR, Theelen B, Boekhout T, et al. Environmental isolation and characterisation of Cryptococcus species from living trees in Havana city, Cuba. Mycoses. 2012;55:e138–e144. doi: 10.1111/j.1439-0507.2012.02168.x. [DOI] [PubMed] [Google Scholar]
- 83. Litvintseva AP, Mitchell TG. Population genetic analyses reveal the African origin and strain variation of Cryptococcus neoformans var. grubii. PLoS Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002495. e1002495. • A primary reference article revealing the ancestry and mode of transmission of the closely related species C. n. var. grubii. Important example of population genetics and analysis of diversity leading to insights on the geographical origin of a Cryptococcus species.
- 84.Romeo O, Scordino F, Chillemi V, Criseo G. Cryptococcus neoformans/Cryptococcus gattii species complex in southern Italy: an overview on the environmental diffusion of serotypes, genotypes and mating-types. Mycopathologia. 2012 doi: 10.1007/s11046-012-9547-6. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 85.Iatta R, Hagen F, Fico C, Lopatriello N, Boekhout T, Montagna MT. Cryptococcus gattii Infection in an immunocompetent patient from Southern Italy. Mycopathologia. 2011;174:87–92. doi: 10.1007/s11046-011-9493-8. [DOI] [PubMed] [Google Scholar]
- 86. Viviani MA, Cogliati M, Esposto MC, Lemmer K, Tintelnot K, Colom Valiente MF, et al. Molecular analysis of 311 Cryptococcus neoformans isolates from a 30-month ECMM survey of cryptococcosis in Europe. FEMS Yeast Res. 2006;6:614–619. doi: 10.1111/j.1567-1364.2006.00081.x. • Demonstrates the presence of C. gattii cases in Europe, outside its typical tropical niche.
- 87.Chowdhary A, Rhandhawa HS, Prakash A, Meis JF. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: an update. Critical Reviews in Microbiology. 2012;38:1–16. doi: 10.3109/1040841X.2011.606426. [DOI] [PubMed] [Google Scholar]
- 88.Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, Xu J. Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Environmental Microbiology. 2011;13:1875–1888. doi: 10.1111/j.1462-2920.2011.02510.x. [DOI] [PubMed] [Google Scholar]
- 89.Da Silva BK, Freire AK, Bentes Ados S, Sampaio Ide L, Santos LO, Dos Santos MS, et al. Characterization of clinical isolates of the Cryptococcus neoformans-Cryptococcus gattii species complex from the Amazonas State in Brazil. Revista Iberoamericana de Micologia. 2012;29:40–43. doi: 10.1016/j.riam.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 90.Silva DC, Martins MA, Szeszs MW, Bonfietti LX, Matos D, Melhem MS. Susceptibility to antifungal agents and genotypes of Brazilian clinical and environmental Cryptococcus gattii strains. Diagnostic Microbiology and Infectious Disease. 2012;72:332–339. doi: 10.1016/j.diagmicrobio.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 91.Molez JF. The historical question of acquired immunodeficiency syndrome in the 1960s in the Congo River basin area in relation to cryptococcal meningitis. Am J Trop Med Hyg. 1998;58:273–276. doi: 10.4269/ajtmh.1998.58.273. [DOI] [PubMed] [Google Scholar]
- 92.Swinne D, Nkurikiyinfura JB, Muyembe TL. Clinical isolates of Cryptococcus neoformans from Zaire. Eur J Clin Microbiol. 1986;5:50–51. doi: 10.1007/BF02013464. [DOI] [PubMed] [Google Scholar]
- 93.Kwon-Chung KJ, Bennett JE. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralbl Bakteriol Mikrobiol Hyg [A] 1984;257:213–218. [PubMed] [Google Scholar]
- 94.Lewden C, Salmon D, Morlat P, Bevilacqua S, Jougla E, Bonnet F, et al. Causes of death among human immunodeficiency virus (HIV)-infected adults in the era of potent antiretroviral therapy: emerging role of hepatitis and cancers, persistent role of AIDS. Int J Epidemiol. 2005;34:121–130. doi: 10.1093/ije/dyh307. [DOI] [PubMed] [Google Scholar]
- 95. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. •• Important reference discussing the increasing incidence of Cryptococcosis worldwide.
- 96.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khanna N, Chandramuki A, Desai A, Ravi V, Santosh V, Shankar SK, et al. Cryptococcosis in the immunocompromised host with special reference to AIDS. Indian J Chest Dis Allied Sci. 2000;42:311–315. [PubMed] [Google Scholar]
- 98.Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. Cryptococcus gattii in AIDS patients, southern California. Emerg Infect Dis. 2005;11:1686–1692. doi: 10.3201/eid1111.040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, et al. Epidemiology and host-and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clinical Infectious Diseases. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 100.Karstaedt AS, Crewe-Brown HH, Dromer F. Cryptococcal meningitis caused by Cryptococcus neoformans var. gattii serotype C, in AIDS patients in Soweto, South Africa. Medical Mycology. 2002;40:7–11. doi: 10.1080/mmy.40.1.7.11. [DOI] [PubMed] [Google Scholar]
- 101.Morgan J, McCarthy KM, Gould S, Fan K, Arthington-Skaggs B, Iqbal N, et al. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002–2004. Clin Infect Dis. 2006;43:1077–1080. doi: 10.1086/507897. [DOI] [PubMed] [Google Scholar]
- 102.Tay ST, Rohani MY, Soo Hoo TS, Hamimah H. Epidemiology of cryptococcosis in Malaysia. Mycoses. 2009;53:509–514. doi: 10.1111/j.1439-0507.2009.01750.x. [DOI] [PubMed] [Google Scholar]
- 103.Bodasing N, Seaton RA, Shankland GS, Kennedy D. Cryptococcus neoformans var. gattii meningitis in an HIV-positive patient: first observation in the United Kingdom. Journal of Infection. 2004;49:253–255. doi: 10.1016/j.jinf.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 104.Castanon-Olivares LR, Lopez-Martinez R, Barriga-Angulo G, Rios-Rosas C. Crytococcus neoformans var. gattii in an AIDS patient: first observation in Mexico. J Med Vet Mycol. 1997;35:57–59. [PubMed] [Google Scholar]
- 105.Castanon-Olivares LR, Arreguin-Espinosa R, Ruiz-Palacios y Santos G, Lopez-Martinez R. Frequency of Cryptococcus species and varieties in Mexico and their comparison with some Latin American countries. Rev Latinoam Microbiol. 2000;42:35–40. [PubMed] [Google Scholar]
- 106.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Findley K, Sun S, Fraser JA, Hsueh YP, Averette AF, Li W, et al. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002528. e1002528. •• A primary research article that describes the sexual cycle and sex determinants in non-pathogenic sister species of Cryptococcus . Fulfills a critical step in understanding of the evolution and plasticity of the sexual machinery in fungi.
- 108.Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, Heitman J. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryotic Cell. 2005;4:1410–1419. doi: 10.1128/EC.4.8.1410-1419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chaturvedi V, Fan J, Stein B, Behr MJ, Samsonoff WA, Wickes BL, et al. Molecular genetic analyses of mating pheromones reveal intervariety mating or hybridization in Cryptococcus neoformans. Infect. Immun. 2002;70:5225–5235. doi: 10.1128/IAI.70.9.5225-5235.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Carniato A, Scotton PG, Miotti AM, Mengoli C. Cryptococcus neoformans meningoencephalitis among apparently immunocompetent patients: description of two cases. Infez Med. 2009;17:41–45. [PubMed] [Google Scholar]
- 111.Ma AL, Fong NC, Leung CW. Cryptococcal meningitis in an immunocompetent adolescent. Ann Trop Paediatr. 2008;28:231–234. doi: 10.1179/146532808X335697. [DOI] [PubMed] [Google Scholar]
- 112.Kang Y, Tanaka H, Moretti ML, Mikami Y. New ITS genotype of Cryptococcus gattii isolated from an AIDS patient in Brazil. Microbiol Immunol. 2009;53:112–116. doi: 10.1111/j.1348-0421.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 113.Kozubowski L, Heitman J. Profiling a killer, the development of Cryptococcus neoformans. FEMS Microbiology Reviews. 2012;36:78–94. doi: 10.1111/j.1574-6976.2011.00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lester SJ, Malik R, Bartlett KH, Duncan CG. Cryptococcosis: update and emergence of Cryptococcus gattii. Veterinary Clinical Pathology. 2011;40:4–17. doi: 10.1111/j.1939-165X.2010.00281.x. [DOI] [PubMed] [Google Scholar]
- 115.Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wickes BL. The role of mating type and morphology in Cryptococcus neoformans pathogenesis. International Journal of Medical Microbiology. 2002;292:313–329. doi: 10.1078/1438-4221-00216. [DOI] [PubMed] [Google Scholar]
- 117.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and a Isolates. Infect. Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Toffaletti DL, Nielsen K, Dietrich F, Heitman J, Perfect JR. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr Genet. 2004;46:193–204. doi: 10.1007/s00294-004-0521-9. [DOI] [PubMed] [Google Scholar]
- 119.Nielsen K, Marra RE, Hagen F, Boekhout T, Mitchell TG, Cox GM, et al. Interaction between genetic background and the mating-type locus in Cryptococcus neoformans virulence potential. Genetics. 2005;171:975–983. doi: 10.1534/genetics.105.045039. [DOI] [PMC free article] [PubMed] [Google Scholar]