Abstract
By searching the GenBank database, we identified sequences encoding three new zebrafish cytosolic sulfotransferases (SULTs). These three new zebrafish SULTs, designated SULT1 ST9, SULT3 ST4, and SULT3 ST5, were cloned, expressed, purified, and characterized. SULT1 ST9 appeared to be mostly involved in the metabolism and detoxification of xenobiotics such as β-naphthol, β-naphthylamine, caffeic acid and gallic acid. SULT3 ST4 showed strong activity toward endogenous compound such as dehydroepiandrosterone (DHEA), pregnenolone, and 17β-estradiol. SULT3 ST5 showed weaker, but significant, activities toward endogenous compounds such as DHEA and corticosterone, as well as xenobiotics including mestranol, β-naphthylamine, β-naphthol, and butylated hydroxyl anisole (BHA). pH-dependency and kinetic constants of these three enzymes were determined with DHEA, β-naphthol, and 17β-estradiol as substrates. Reverse transcription-polymerase chain reaction (RT-PCR) was performed to examine the expression of these three new zebrafish SULTs at different developmental stages during embryogenesis, through larval development, and on to maturity.
Keywords: Cytosolic sulfotransferase, SULT, 17β-estradiol, dehydroepiandrosterone, molecular cloning, developmental expression, zebrafish
Introduction
Sulfation is a major Phase II metabolic pathway for the detoxification of a wide range of xenobiotics (Mulder and Jakoby, 1990; Falany and Roth, 1993; Weinshilboum and Otterness, 1994). Increasingly, it has been shown to be also involved in the biotransformation and homeostasis of endogenous steroid/thyroid hormones and catecholamine neurotransmitters (Falany, 1997; Weinshilboum et al., 1997; Strott, 2002). The responsible enzymes, the cytosolic sulfotransferases (SULTs), catalyze the transfer of a sulfonate group from the active sulfate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to acceptor substrate compounds containing hydroxyl or amino group (Lipmann, 1958).
Although considerable progress has been made in recent years in the study of the SULTs, some fundamental issues concerning, particularly, their ontogeny, regulation, and physiological involvement remain poorly understood. To help resolve these issues, a suitable animal model is needed. Zebrafish has in recent years emerged as a popular animal model for developmental and biomedical research (Briggs 2002; Ward and Lieschke, 2002). Its advantages, compared with mouse, rat, or other vertebrate animal models, include the small size, availability of a relatively large number of eggs, rapid external development of virtually transparent embryos, and short generation time. These unique characteristics make the zebrafish an excellent model for a systematic investigation of the developmental stage-dependent and cell type/tissue/organ-specific expression, as well as physiological involvement of the SULTs. A prerequisite for using the zebrafish in these studies is the identification of the various SULTs and their functional characterization. We have recently embarked on the molecular cloning of zebrafish SULTs. To date, fifteen zebrafish SULTs have been cloned, including 8 SULT1 STs, 3 SULT2 STs, 3 SULT3 STs, and a SULT6 ST (Sugahara et al., 2003a, 2003b, 2003c, 2003d, Ohkimoto et al., 2004; Liu et al., 2005, 2008; Yasuda et al., 2005a, 2005b, 2006b, 2008b, 2009).
We report here the identification of three new zebrafish SULTs. These three new SULTs, designated SULT1 ST9, SULT3 ST4, and SULT3 ST5, were cloned, expressed, and purified. The sulfating activities of the three enzymes toward a variety of endogenous compounds and xenobiotics were examined. Their pH-dependence and kinetic parameters in catalyzing the sulfation of representative endogenous compounds and xenobiotics were determined. Moreover, the developmental stage-dependent expression of these three new zebrafish SULTs was investigated.
Materials and Methods
Materials
Dehydroepiandrosterone (DHEA), 17β-estradiol, 17α-ethynylestradiol, estrone, triiodothyronin (T3), thyroxine (T4), butylated hydroxyanisole (BHA), caffeic acid, gallic acid, mestranol, cholorogenic acid, β-naphthylamine, β-naphthol, 3-(N-morpholino) propanesulfonic acid (MOPS), 2-morpholinoethanesulfonic acid (MES), N-2-hydroxylpiperazine-N2-ethanesulfonic (HEPES), 3-[N-tris-(hydroxymethyl) methylamino[-propanesulfonic acid (TAPS), 2-(cyclohexylamino) ethanesulfonic acid (CHES), 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), dimethyl sulfoxide (DMSO), dithiothreitol (DTT), and isopropyl-β-d-thiogalactopyranoside (IPTG) were products of Sigma Chemical Company (St. Louis, MO). Ecolume scintillation cocktail, progesterone, pregn enolone, corticosterone, androstene-3,17-dione, and carrier free sodium [35S]sulfate were purchased from MP Biomedical (Solon, OH). Cellulose thin-layer chromatography (TLC) plates were from EMD chemicals (Gibbstown, NJ). Recombinant human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase was prepared as previously described (Yanagisawa et al., 1998). TRI reagent was obtained from Molecular Research Center, Inc (Cincinnati, OH). Unfertilized zebrafish eggs, embryos, and larvae at different developmental stages were from Scientific Hatcheries (Huntington Beach, CA). Takara Ex Taq DNA polymerase was purchased from Fisher Scientific (Pittsburgh, PA). Oligonucleotide primers were synthesized by MWG Biotech (Huntsville, AL). All other reagents were of the highest grades commercially available.
Cloning, bacterial expression, and purification of recombinant zebrafish SULTs
By searching the GenBank database, three zebrafish sequences (GenBank Accession #XM_001919250 (SULT1 ST9), XM_6950520 (SULT3 ST4), and CR_936460 (SULT3 ST5)) encoding putative SULTs were identified. To generate corresponding cDNAs for cloning into the pGEX-2T or pMAL-c5x prokaryotic expression vector, sense and antisense oligonucleotide primers designed based on 5′- and 3′- regions of the respective coding sequences were synthesized with Bam HI restriction site incorporated at the end (Table 1). Using these primer sets, PCRs were carried out under the action of EX Taq DNA polymerase, with the first-strand cDNA reverse-transcribed from the total RNA of a 3-month-old zebrafish as template. Amplification conditions were 2 min at 94°C and 20 cycles of 94°C for 35 sec, 60°C for 40 s, and 72°C for 1 min. The final reaction mixtures were applied onto a 0.9% agarose gel, separated by electrophoresis, and visualized by ethidium bromide staining. The PCR product bands detected were excised from the gel, and the DNAs therein were isolated by spin filtration. Purified PCR products were subjected to Bam HI restriction and cloned into Bam HI-restricted pMAL-c5x (for SULT1 ST9 and SULT3 ST5) or pGEX-2T (for SULT3 ST4) vector, and verified for authenticity by nucleotide sequencing (Sanger et al., 1977). Recombinant protein expression using pMAL-c5x or pGEX-2T expression system allows for the production of maltose-binding protein (MBP) or glutathione S-transferase (GST) fusion protein which can be conveniently purified by affinity chromatography using amylose resin or glutathione-Sepharose. To express recombinant zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5, competent Escherichia coli BL21 (DE3) cells transformed with prokaryotic expression vector harboring the cDNA encoding SULT1 ST9, SULT3 ST4, or SULT3 ST5 were grown in 1 L LB medium supplemented with 60 μg/ml ampicillin. After the cell density reached 0.6 OD600 nm, IPTG was added to induce the production of recombinant MBP- or GST-SULT fusion protein. After induction, the cells were collected by centrifugation and homogenized in 25 ml ice-cold lysis buffer using an Aminco French Press. Twenty μl of 10 mg/ml aprotinin (a protease inhibitor) were added to the crude homogenate. The crude homogenate was subjected to centrifugation at 10,000 × g for 15 min at 4°C. The supernatant collected was fractionated using 2.5 ml of affinity resin. Upon washing with lysis buffer to remove unbound proteins, the MBP or GST fusion protein was eluted or cleaved off from the resin using either a stepwise gradient of maltose in 50 mM Tris-HCl, pH 8.0 or bovine thrombin in a thrombin digestion buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 2.5 mM CaCl2). The information for the preparation of the recombinant SULTs is summarized in Table 2. MBP-SULT1 ST9 and MBP-SULT3 ST5 fusion proteins or SULT3 ST4 released upon thrombin digestion were analyzed for purity by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and subjected to enzymatic characterization.
Table 1. Oligonucleotide primers used in the cloning and RT-PCR analysis of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5.
| Target Sequence | Sense and Antisense Oligonucleotide Primers | |
|---|---|---|
| SULT1 ST9 | Sense: | 5′-CGCGGATCCATGGAAATCCAAGGCAAATCCTCTACTGATTTA-3′ |
| Antisense: | 5′-CGCGGATCCTCACTCAGTAGGGAACTTGAGAGTGGAATTCT-3′ | |
| SULT3 ST4 | Sense: | 5′-CGCGGATCCATGGCTCAAGAGGAATGCAAAATGATTAGTGAC-3′ |
| Antisense: | 5′-CGCGGATCCTCAACTATGCAGTTCTGTGATGTCCCAGACCAG-3′ | |
| SULT3 ST5 | Sense: | 5′-CGCGGATCCATGGCTCAGGAGGAATGCAAAATGATTAGTGAC-3′ |
| Antisense: | 5′-CGCGGATCCTCAGCCACGCAGTTCTGTGATGTCCCAGACCAG-3′ | |
| β-Actin | Sense: | 5′-ATGGATGAGGAAATCGCTGCCCTGGTC-3′ |
| Antisense: | 5′- TTAGAAGCACTTCCTGTGAACGATGGA-3′ |
Recognition sites of Bam HI restriction endonuclease in the oligonucleotides are underlined. Initiation and termination codons for translation are in bold type.
The sense and antisense oligonucleotide primer sets listed were verified by BLAST Search to be specific for the corresponding zebrafish SULT or β-actin nucleotide sequence.
Table 2.
Summary of the procedures for the expression and purification of recombinant zebrafish SULT1 ST9, SULT3 ST4 and SULT3 ST5.
| Zebrafish SULT | Vector | Induction Condition | Affinity Resin | Elution/Digestion |
|---|---|---|---|---|
| SULT1 ST9 | pMAL-c5x | 0.5 mM IPTG; | Amylose Resin | 1 mM to 50 mM maltose; |
| SULT3 ST5 | 5 hours at 37°C | 15 min at RT*) | ||
| SULT3 ST4 | pGEX-2T | 0.1 mM IPTG; | Glutathione -Sepharose | 5 U/ml bovine thrombin; |
| Overnight at RT | -Sepharose | 15 min at RT |
RT refers to room temperature.
Enzymatic assay
The assay for the sulfating were also included. The reaction was started by the addition of the enzyme, allowed to continue at 28° activity of zebrafish SULT enzymes was performed using radioactive PAP[35S] as the sulfate donor. The reaction mixture for the standard enzymatic assay, prepared in a final volume of 25 μl, contained, 50 mM MOPS at pH 7.0, 14 μM of PAP[35S], 1 mM DTT, and 50 μM substrate. Controls with water or DMSO replacing substrate;C for 5 minutes, and terminated by placing the tube containing the reaction mixture on a heating block at 100°C for 1 minute. Heated reaction mixture was subjected to centrifugation to pellet down the precipitates formed. Afterwards, 2 μl of the reaction mixture was spotted on a cellulose TLC plate and the spotted TLC plate was subjected to TLC analysis using a solvent system containing n-butanol, isopropanol, 88% formic acid and H2O in a ratio of 3:1:1:1 (by volume) (Liu and Lipmann, 1984). Upon completion of TLC, the TLC plate was air-dried and autoradiographed by using an X-ray film. The radioactive spot corresponding to the sulfated product was located, cut out, and eluted in 0.5 ml water in a glass vial. 4.5 ml of Ecolume scintillation liquid was added to each vial, mixed thoroughly, and the radioactivity therein was counted by using a liquid scintillation counter. Controls were installed to ensure that the [35S]sulfated product was not destructed during heat treatment, lost to precipitates upon centrifugation at the end of the enzymatic assay, or incompletely eluted from TLC plate into water. The cpm count obtained was used to calculate the specific activity in the unit of nmol of sulfated product/minute/mg enzyme. Protein determination of purified SULT enzymes was based on the method of Bradford (1976) with bovine serum albumin as a standard. The detection limit for the specific activity was ∼0.01 nmol/min/mg enzyme. To examine the pH-dependence of the sulfation of 17β-estradiol, DHEA or β-naphthol, different buffers (sodium acetate at pH 4.5, 5.0, or 5.5; Mes at pH 5.5, 6.0, or 6.5; Mops at pH 6.5, 7.0, or 7.5; Hepes at pH 7.0, 7.5 or 8.0; Taps at pH 8.0, 8.5 or 9.0; Ches at pH 9.0, 9.5, or 10.0; and Caps at pH 10.0, 10.5, 11.0 or 11.5), instead of 50 mM Mops (pH 7.0), were used in the reactions with 50 μM of each substrate. For the kinetic studies on the sulfation of 17β-estradiol and β-naphthol by SULT1 ST9 and the sulfation of DHEA and p-naphthol by SULT3 ST4 or SULT3 ST5, varying concentrations of these substrate compounds and 50 mM Mops buffer at pH 7.0 were used.
RT-PCR Analysis for the developmental stage-dependent expression of zebrafish SULT1ST9, SULT3 ST4, and SULT3 ST5
All experiments and animal care procedures were approved by the Institutional Animal Care and Use Committee of the University of Toledo under Protocol No. 105414. The developmental stage-dependent expression of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 was investigated by employing reverse transcriptase-polymerase chain reaction (RT-PCR). Using the TRI Reagent, the total RNA from unfertilized zebrafish eggs, zebrafish embryos and larvae at various developmental stages and adult (male/female) fish were isolated. The first-strand cDNAs were prepared using the total RNAs isolated as templates for the subsequent PCR amplification. The PCR amplification conditions were 2 min at 94°C for initial denaturation, followed by 35 cycles of 30 sec at 94°C for denaturation, 35 sec at 56°C for annealing, and 1 min at 72°C for extension. The final PCR reaction mixtures were subjected to 0.9% agarose gel electrophoresis and visualized by ethidium bromide staining. PCR amplification of the sequence encoding zebrafish β-actin was concomitantly performed as a control using the above-described first-strand cDNAs as templates.
Miscellaneous methods
The sulfate donor, PAP [35S], was synthesized from ATP and carrier-free [35S]sulfate using the bifunctional human ATP sulfurylase/APS kinase (Yanagisawa et al., 1998). The synthesized PAP[35S] was adjusted to the desired concentration and specific activity by the addition of nonradioactive (cold) PAPS. SDS-PAGE was performed on a 12% polyacrylamide gel using the method of Laemmli (1970).
Results
Molecular cloning of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5
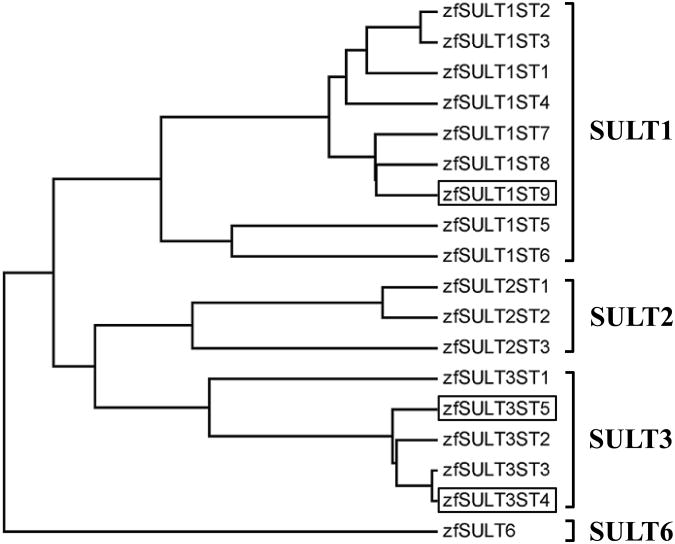
The cDNAs encoding zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 were amplified by RT-PCR, cloned into the pMAL-c5x or pGEX-2T vector, and subjected to nucleotide sequencing for verifying their authenticity. The nucleotide sequences obtained were submitted to the GenBank database under the Accession number JN896760, JN896762, and JN896761 for SULT1 ST9, SULT3 ST4, and SULT3 ST5, respectively. Figure 1 shows the aligned amino acid sequences of the three newly cloned zebrafish SULTs. The open reading frames of SULT1 ST9, SULT3 ST4, and SULT3 ST5 encompass, respectively, 903, 900, and 900 nucleotides and code for 300-, 299-, and 299-amino acid polypeptides. Similar to other SULTs, these three new zebrafish SULTs contain sequences resembling the so-called “signature sequences” (YPKSGTxW in the N-terminal region and RKGxxGDWKNxFT in the C-terminal region) characteristic of known SULT enzymes (Weinshilboum et al., 1997). Of these two sequences, YPKSGTxW has been demonstrated by X-ray crystallography to be involved in the binding to the 5′-phosphosulfate group of the co-substrate for SULT-catalyzed sulfation reactions, PAPS (Lipmann, 1958), and has been named the “5′-phosphosulfate binding (5′-PSB) motif” (Negishi et al., 2001). Sequence analysis based on BLAST pairwise search revealed that the deduced amino acid sequence of zebrafish SULT1 ST9 displays 52 and 49% amino acid sequence identity to mouse SULT1D1 and human SULT1A3, and up to 84% amino acid sequence identity to other zebrafish SULT1 STs. Similar BLAST search revealed that zebrafish SULT3 ST4 and SULT3 ST5 display, respectively, 46 and 46% amino acid sequence identity to mouse SULT3A1, and 49 and 48% identity to rabbit SULT3A1. Moreover, zebrafish SULT3 ST4 and SULT3 ST5 display only 33-37% amino acid sequence identity to the previously identified zebrafish SULT1 STs, and 39-40% amino acid sequence identity to the previously reported zebrafish SULT2 STs. Between zebrafish SULT3 ST4 and SULT3 ST5, 89% amino acid sequence identity was observed. And, compared with the three zebrafish SULT3 STs previously identified (Yasuda et al., 2008; Yasuda et al, 2009), zebrafish SULT3 ST4 and SULT3 ST5 exhibited 50 and 50% amino acid identity to zebrafish SULT3 ST1, 88 and 86% identity to zebrafish SULT3 ST2, and 98 and 86% identity to zebrafish SULT3 ST3. Figure 2 shows a dendrogram depicting the classification of the three new SULTs together with the fifteen SULTs previously identified.
Figure 1.
Alignment of deduced amino acid sequences of the zebrafish SULT1 ST9, SULT3 ST4 and SULT3 ST5. Residues conserved among these three SULT enzymes are in shaded boxes. Two “signature sequences” located, respectively, in the N-terminal and C-terminal regions, as well as a conserved sequence in the middle region are underlined.
Figure 2.
Classification of the zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5, together with previously identified zebrafish SULTs, on the basis of their amino acid sequences. The dendrogram, generated based on Greedy algorithm (Brodsky et al., 1995; Nikolaev et al., 1997), shows the degree of amino acid sequence homology among different zebrafish SULTs.
Expression, purification, and characterization of recombinant zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5
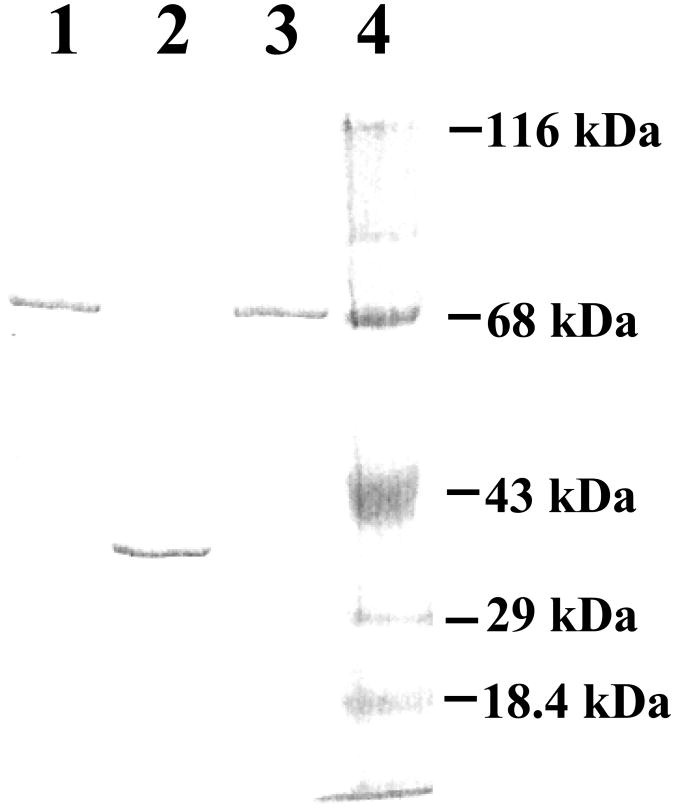
A pilot experiment showed that of the three new zebrafish SULTs, SULT1 ST9 and SULT3 ST5 could only be expressed using the pMAL-c5x expression system, and SULT3 ST4 could only be expressed using the pGEX expression system. Based on these information, pMAL-c5x and pGEX-2T harboring zebrafish SULT1 ST9, SULT3 ST4, or SULT3 ST5 cDNA were transformed into E. coli BL21 (DE3) cells for the expression of recombinant enzyme. Recombinant zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 were purified from the E. coli cell extract as MBP-fusion protein (for SULT1 ST9 and SULT3 ST5) or thrombin-digested tag-free enzyme (for SULT3 ST4). As shown in Figure 3, upon SDS-PAGE, purified SULT1 ST9, SULT3 ST4, and SULT3 ST5 migrated at approximately 75, 35, and 75 kDa positions. Taking into consideration of the 40 kDa molecular mass of the MBP portion in the MBP fusion proteins, these results are in agreement with the predicted molecular weight (34,731, 35,217, and 34780) of SULT1 ST9, SULT3 ST4, and SULT3 ST5, based on their deduced amino acid sequences. Purified zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 were subjected to functional characterization with respect to their enzymatic properties. It should be pointed out that, while the free form of SULT1 ST9 and SULT3 ST5 generated upon digestion with Factor Xa was also prepared, the tag-free SULT1 ST9 and SULT3 ST5 exhibited dramatically lower and unstable sulfating activity in comparison with their MBP-fusion protein counterparts (data not shown). The MBP-SULT1 ST9 (or SULT3 ST5) fusion proteins therefore, were used for the enzymatic characterization. (The specific activities determined in the following studies were corrected for the molecular mass (40 kDa) of the MBP moiety in the fusion protein form of the enzymes.)
Figure 3.
SDS gel electrophoretic pattern of the purified recombinant zebrafish SULTs. SDS-PAGE was performed on a 12% gel, followed by Coomassie blue staining. Samples analyzed in lanes 1, 2, and 3 were SULT1 ST9, SULT3 ST4 and SULT3 ST5. Positions of protein molecular weight markers co-electrophoresed are marked on the right, β-lactoglobulin (Mr = 18,400), carbonic anhydrase (Mr = 29,000), ovalbumin (Mr = 43,000), bovine serum albumin (Mr = 68,000), and β-galactosidase (Mr = 116,000).
Substrate specificity
An initial study revealed that SULT1 ST9, SULT3 ST4, and SULT3 ST5 exhibited differential sulfating activities toward the different substrates tested (Table 3). SULT1 ST9 displayed low, but significant (compared with the detection limit of the assay), activities toward 17β-estradiol, estrone, and T4, among the endogenous compounds tested as substrates. Among the xenobiotic compounds tested, SULT1 ST9 showed sulfating activities toward all of them except mestranol, with the strongest activity being toward β-naphthol (at 21.32 nmol/min/mg enzyme). In contrast, SULT3 ST4 showed sulfating activities towards all of the endogenous compounds tested, with strongest activities being found with DHEA and pregnenolone (at 13.83 and 9.04 nmol/min/mg enzyme, respectively). In addition, SULT3 ST4 also exhibited significant sulfating activity toward some xenobiotics including mestranol, butylated hydroxyanisole, 17α-ethynylestadiol, β-naphthylamine, and β-naphthol. SULT3 ST5 exhibited weaker, but significant, activities toward many of the endogenous and xenobiotic compounds tested as substrates. SULT3 ST5 showed sulfating activities toward endogenous compounds including DHEA, corticosterone, androstene-3,17-dione, 17β-estradiol, pregnenolone, T4, but no detectable activity towards estrone and progesterone. Among the xenobiotics tested, SULT3 ST5 displayed activities toward xenobiotics including butylated hydroxyanisole, mestranol, 17α-ethynylestadiol, β-naphthylamine and β-naphthol.
Table 3. Specific activities of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 with endogenous and xenobiotic compounds as substratesa.
| Endogenous compounds | SULT1 ST9 | SULT3 ST4 | SULT3 ST5 | Xenobiotic compounds | SULT1 ST9 | SULT3 ST4 | SULT3 ST5 |
|---|---|---|---|---|---|---|---|
| DHEA | NDb | 13.83 ± 0.20 | 0.58 ± 0.02 | 17α-Ethynylestradiol | 0.08 ± 0.01 | 0.58 ± 0.02 | 1.04 ± 0.02 |
| Androstene-3,17-dione | ND | 0.40 ± 0.02 | 0.10 ± 0.01 | Caffeic acid | 7.53 ± 0.07 | ND | ND |
| 17β-Estradiol | 0.08 ± 0.01 | 3.45 ± 0.07 | 0.08 ± 0.01 | Gallic acid | 0.77 ± 0.04 | ND | ND |
| Progesteron e | ND | 0.28 ± 0.01 | ND | Mestranol | ND | 0.98 ± 0.07 | 0.16 ± 0.01 |
| Estrone | 0.22 ± 0.01 | 0.21 ± 0.01 | ND | Chrologenic acid | 15.29 ± 0.44 | ND | ND |
| Pregnenolone | ND | 9.04 ± 0.06 | 0.06 ± 0.01 | β-Naphthylamine | 0.38 ± 0.01 | 0.31 ± 0.01 | 0.03 ± 0.01 |
| Corticosterone | ND | 0.80 ± 0.02 | 0.14 ± 0.01 | β-Naphthol | 21.32 ± 0.25 | 0.14 ± 0.01 | 0.06 ± 0.01 |
| T3 | ND | ND | ND | Butylated hydroxyl anisole | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.11 ± 0.01 |
| T4 | 0.51 ± 0.02 | 0.05 ± 0.01 | 0.10 ± 0.01 |
Specific activity refers to nmol substrate sulfated/min/mg purified enzyme. Data represent mean ± S.D. derived from three determinations. The concentration of the substrate used in the assay mixture was 50 μM.
ND refers to activity not detected. Specific activity determined was lower than the detection limit (estimated to be ∼0.01 nmol/min/mg protein).
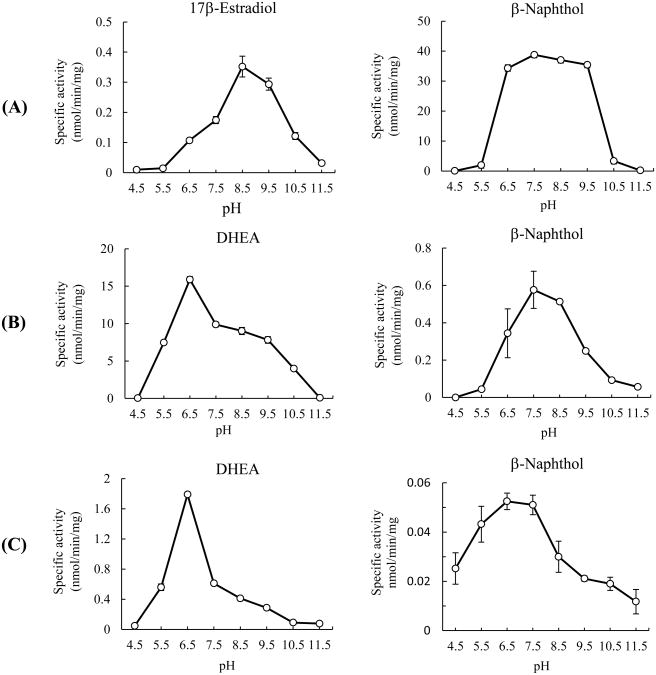
To investigate further the enzymatic characteristics of the three newly identified zebrafish SULTs in catalyzing the sulfation of endogenous and xenobiotic compounds, the pH-dependency and kinetic parameters were examined. For SULT1 ST9, 17β-estradiol, a representative estrogen substrate of zebrafish SULT1 STs (Yasuda et al., 2005b), and β-naphthol, with which SULT1 ST9 showed the strongest activity among xenobiotics tested, were selected as representative endogenous and xenobiotic substrate, respectively. For SULT3 ST4 and SULT3 ST5, DHEA, with which both SULT3 STs displayed the strongest activity among the endogenous compounds, and β-naphthol, for use as a xenobiotic substrate for comparison with SULT1 ST9, were selected.
pH-dependency
SULT1 ST9 with 17β-estradiol as the substrate displayed strong sulfating activity between pH 7.5 and 9.5, with maximum activity being at pH 8.5, (Figure 4A). With β-naphthol as the substrate, SULT1 ST9 showed a broad pH optimum spanning pH 6.5 to 9.5, with a maximum activity at pH 7.5 (Figure 4A). SULT3 ST4 with DHEA as a substrate showed a broad pH optimum spanning pH 5.5 to 9.5, with a maximum sulfating activity at pH 6.5 (Figure 4B). SULT3 ST4 with β-naphthol as a substrate also showed a broad pH optimum spanning pH 6.5 to 9.5, with a maximum sulfating activity at pH 7.5 (Figure 4B). In contrast, SULT3 ST5 with DHEA as a substrate exhibited a narrower pH optimum between pH 5.5 and 7.5, with a maximum sulfating activity at pH 6.5 (Figure 4C). SULT3 ST5 with β-naphthol as a substrate exhibited a broad pH optimum spanning pH 4.5 to 8.5, with a maximum sulfating activity at pH 6.5 (Figure 4C).
Figure 4.
pH-dependency of the sulfating activity of the zebrafish SULT1 ST9 (A), SULT3 ST4 (B) and SULT3 ST5 (C) with endogeneous (17β-estradiol or DHEA) and xenobiotic (β-naphthol) compounds as substrates. The enzymatic assays were carried out under standard assay conditions as described in Materials and Methods, using different buffer systems as indicated. The data represent calculated mean ± SD derived from three experiments.
Kinetic parameters
Enzymatic assays were performed based on the same procedure as described in the Materials and Methods, except that different substrate concentrations were used. Data obtained were used to generate Lineweaver-Burk double-reciprocal plots in order to calculate the Km, Vmax, and Vmax/Km for each of the three enzymes in catalyzing the sulfation of indicated substrates. The calculated values of Km, Vmax, and Vmax/Km for the three enzymes are compiled in Table 4. For SULT1 ST9, while the Km values with 17β-etsradiol and β-naphthol were comparable (53.27 and 68.11 μM, respectively), the Vmax was 280 times higher for p-naphthol than for 17β-etsradiol. Based on calculated Vmax/Km, it can be concluded that SULT1 ST9 is likely more important for the sulfation of xenobiotics such as β-naphthol than for endogenous compounds such as 17β-etsradiol. In contrast, both SULT3 ST4 and SULT3 ST5 showed lower Km values with DHEA than with β-naphthol and higher Vmax values with DHEA than with p-naphthol. Compared with SULT1 ST9, the catalytic efficiency, as reflected by calculated Vmax/Km, with β-naphthol as substrate was much lower for both SULT3 ST4 and SULT3 ST5.
Table 4. Kinetic constants of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5 with 17β-estradiol, DHEA, or β-naphthol as substratesa.
| Vmax (nmol/min/mg) | Km (μM) | Vmax/Km | |
|---|---|---|---|
| SULT1 ST9 | |||
| with 17β-estradiol | 0.53 | 53.27 | 0.002 |
| with β-naphthol | 148.9 | 68.11 | 2.19 |
| SULT3 ST4 | |||
| with DHEA | 75.76 | 37.88 | 1.99 |
| with β-naphthol | 1.04 | 213.6 | 0.005 |
| SULT3 ST5 | |||
| with DHEA | 19.12 | 6.23 | 3.02 |
| with β-naphthol | 2.02 | 699.1 | 0.003 |
Results shown represent means ± S.D. derived from three determinations.
Developmental stage-dependent expression of zebrafish SULT1 ST9, SULT3 ST4, and SULT3 ST5
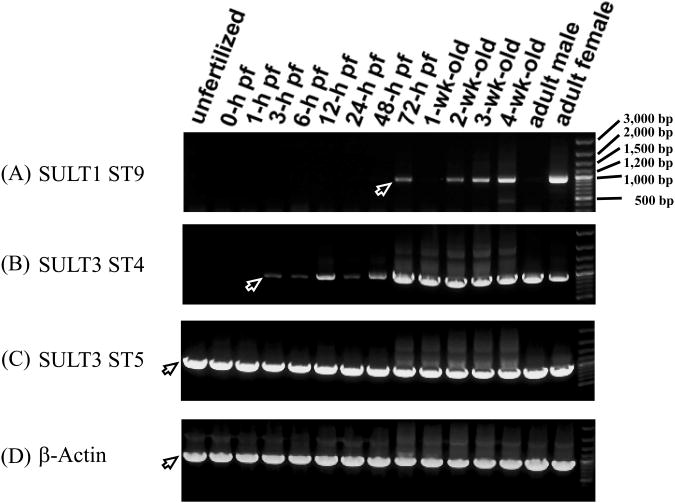
The expression of mRNAs encoding SULT1 ST9, SULT3 ST4, and SULT3 ST5 from embryogenesis to maturity was examined using RT-PCR in order to obtain clues to their physiological involvement. mRNA encoding SULT1 ST9 was undetectable in unfertilized eggs or embryos up to the phyaryngula period, and started appearing at a low level at the hatching period, which then disappeared in the first week of larval development (Figure 5A). Afterwards, increasing expression of SULT1 ST9 mRNA was detected from the second week of larval development on to maturity to adult fish. Interestingly, the expression of the mRNA encoding SULT1 ST9 was detected only in female, but not in male adult fish. For SULT3 ST4, there was no detectable level of its coding mRNA in unfertilized egg (Figure 5B). Upon fertilization, a low level of SULT3 ST4 mRNA was detected during the zygote period, which then disappeared during the cleavage period. Afterwards, a significant level of SULT3 ST4 mRNA was detected during the blastula, gastrula, segmentation, pharyngula and hatching periods. It then started to increase in the larval stages and on to maturity in both male and female adult fish. In contrast, the expression of the mRNA encoding SULT3 ST5 was detected at all developmental stages from embryogenesis on to maturity (Figure 5C). As a control, β-actin, a housekeeping protein, was found to be constantly expressed throughout all developmental stages (Figure 5D).
Figure 5.
Developmental stage-dependent expression of the zebrafish SULT1 ST9, SULT3 ST4 and SULT3 ST5. (A) - (C) RT-PCR analysis of the expression of mRNAs encoding SULT1 ST9, SULT3 ST4 and SULT3 ST5 at different stages during embryogenesis and larval development onto maturity. Final PCR mixtures were subjected to 0.9% agarose electrophoresis. Samples analyzed correspond to unfertilized zebrafish eggs, zebrafish embryos during the zygote period (0-hour post-fertilization (pf), cleavage period (1-hour pf), blastula period (3-hour pf), gastrula period (6-hour pf), neurula/segmentaion period (12-hour pf), pharyngula period (24-hour pf), and hatching period (48- and 72-hour pf), 1, 2, 3, 4-week-old zebrafish larvae, and 3-month-old adult male or female zebrafish. The positions of PCR products corresponding to different zebrafish SULT1 ST9, SULT3 ST4 or SULT3 ST5 cDNAs, visualized by ethydium bromide staining, are marked by arrows. (D) RT-PCR analysis of the expression of the zebrafish β-actin at the same developmental stages as those described above. The figure is representative of three independent repetitions with different samples.
Discussion
To develop a zebrafish model for systematically investigating how the SULTs function during the developmental process, we have embarked on the molecular cloning and characterization of the zebrafish SULTs, which have so far led to the identification of fifteen distinct SULT enzymes (Sugahara et al., 2003a, 2003b, 2003c, 2003d, Ohkimoto et al., 2004; Liu et al., 2005; Yasuda et al., 2005a, 2005b, 2006b, 2008b, 2009). In the current study, we cloned, expressed, purified, and characterized three additional zebrafish SULTs. As shown in an updated dendrogram (Figure 2), SULT1 ST9 was classified into a small cluster previously consisted of SULT1 ST7 and SULT1 ST8 (Liu et al., 2008). SULT3 ST4 and SULT3 ST5 paired with, respectively, SULT3 ST2 and SULT3 ST3 (Figure 2) in two sub-clusters, which together with SULT3 ST1 constitute the SULT3 family.
Substrate specificity analysis revealed that SULT1 ST9 displayed strong sulfating activities toward phenolic xenobiotic compounds, particularly β-naphthol and chrologenic acid (Table 3). In contrast, SULT3 ST4 showed strong sulfating activities toward hydroxysteroids and estrogens, particularly DHEA and pregnenolone. It should be noted that SULT3 ST4 and previously identified SULT3 ST3 (Yasuda, et al., 2009), which share the highest amino acid sequence identity (98%; cf. Figure 2), exhibited a high degree of overlapping substrate specificity toward hydroxysteroids and estrogens. SULT3 ST5 also exhibited sulfating activities toward hydroxysteroids and xenobiotics, although the activities detected were much lower than those of, respectively, SULT3 ST4 and SULT1 ST9. Further characterization of the enzymatic properties showed that the pH-dependency of SULT3 ST4 and SULT3 ST5 with DHEA and β-naphthol were comparable, whereas SULT1 ST9 displayed a different pH-dependency from the two SULT3 STs (Figure 4). The different pH optima and ranges within which the three SULTs were shown to be active likely correlate with the chemical properties of the endogenous and/or xenobiotic compounds which they may use as substrates. Based on the kinetic parameters, particularly Vmax/Km which reflects the catalytic efficiency, of the three newly identified SULTs (Table 4), SULT1 ST9 is likely more important for the sulfation of xenobiotics than for endogenous compounds. Both SULT3 ST4 and SULT3 ST5 appear more important for the sulfation of DHEA and other hydroxysteroids. These activity data suggest that SULT1 ST9, like other zebrafish SULT1 enzymes (Sugahara et al., 2003a, 2003b; Liu et al., 2005; Yasuda et al., 2005a, 2005b), may be primarily involved in the metabolism of xenobiotics, whereas SULT3 ST4 and SULT3 ST5 are more important for the metabolism and homeostasis of hydroxysteroids. It is also interesting to point out that the two newly identified zebrafish SULT3 STs and the previously reported zebrafish SULT2 and SULT3 STs exhibited differential but overlapping substrate specificity toward hydroxysteroids and estrogens (Sugahara et al., 2003d; Yasuda et al., 2006, 2008, 2009), which collectively may serve to regulate a wide range of levels and chemical entities of hydroxysteroids and estrogens in zebrafish at different developmental stages. Whether the sulfation of xenobiotics and hydroxy steroid compounds by the newly identified SULT1 ST9, SULT3 ST4, and SULT3 ST5 is physiologically relevant, nevertheless, may depend on the local expression and levels of these enzymes and the concentrations of target molecules in their local environment inside the cell.
RT-PCR analysis showed that the mRNAs encoding the three newly identified zebrafish SULTs exhibited distinct patterns of expression during embryogenesis, through larval development, and on to maturity (Figure 5). Similar to mRNAs encoding SULT1 ST5 and SULT1 ST7 (which catalyze the sulfation of primarily xenobiotics including β-naphthol and chlorogenic acid) (Liu, et al., 2008; Yasuda, et al., 2005), SULT1 ST9 mRNA was not detected until the hatching period (72-h pf). The appearance of SULT1 ST9 during the hatching period may imply its involvement in xenobiotic detoxification in hatched zebrafish larvae. It is noted that SULT3 ST4 and the previously identified SULT3 ST3 (Yasuda et al., 2009), which share the highest degree of amino acid sequence identity (98%) among the five zebrafish SULT3 STs, showed comparable developmental expression patterns, except that SULT3 ST3, but not SULT3 ST4, mRNA was detected in unfertilized zebrafish eggs (Yasuda, et al., 2009). Interestingly, SULT3 ST5 mRNA, similar to SULT3 ST1 mRNA (Yasuda, et al., 2008), was present throughout the embryonic/larval development and on to maturity. Further studies are needed in order to clarify the physiological relevance of the distinct developmental expression of the three newly identified zebrafish SULTs.
To summarize, we have cloned, expressed, purified, and characterized three new zebrafish SULT enzymes. This study represents part of an overall effort to obtain a complete repertoire of the SULT enzymes present in zebrafish. The identification of the various SULTs and their biochemical characterization is a prerequisite for using the zebrafish as a model for investigating the mechanism underlying the regulation and homeostasis of hydroxysteroids and other key endogenous compounds, as well as the detoxification of xenobiotics, during the developmental process. Further work is warranted to achieve this goal.
Highlights.
>Three zebrafish SULTs, SULT1 ST9, SULT3 ST4, and SULT3 ST5, catalyze the sulfation of xenobiotic and endogenous compounds.>pH-dependence results imply differential substrate recognitions among these three SULTs.>Kinetic data indicate that the three zebrafish SULTs prefer different xenobiotic and endogenous compounds as substrates.>RT-PCR analysis revealed distinct patters of developmental expression of the three zebrafish SULTs.
Acknowledgments
This work was supported in part by a National Institutes of Health grant GM085756 and a startup fund from College of Pharmacy, The University of Toledo.
Abbreviations
- SULT
sulfotransferase
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- RT-PCR
reverse transcription-polymerase chain reaction
- DHEA
dehydroepiandrosterone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication .As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantitiesof protein utilizing the principle of protein–dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briggs JP. The zebrafish: a new model organism for integrative physiology. Am J Physiol Regul Integr Comp Physiol. 2002;282:R3–R9. doi: 10.1152/ajpregu.00589.2001. [DOI] [PubMed] [Google Scholar]
- Brodsky LI, Ivanov VV, Kalaidzidis YL, Leontovich AM, Nikolaev VK, Feranchuk SI, Drachev VA. GeneBee-NET: Internet-based server for analyzing biopolymers structure. Biochemistry. 1995;60:923–928. [PubMed] [Google Scholar]
- Falany C, Roth JA. Properties of human cytosolic sulfotransferases involved in drug metabolism. In: Jeffery EH, editor. Human Drug Metabolism: From Molecular Biology to Man. CRC Press; Boca Raton, FL: 1993. pp. 101–115. [Google Scholar]
- Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- Liu MC, Lipmann F. Decrease of tyrosine-O-sulfate-containing proteins found in rat fibroblasts infected with Rous sarcoma virus or Fujinami sarcoma virus. Proc Natl Acad Sci USA. 1984;81:3695–3698. doi: 10.1073/pnas.81.12.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MY, Yang YS, Sugahara T, Yasuda S, Liu MC. Identification of a novel zebrafish SULT1 cytosolic sulfotransferase: cloning, expression, characterization, and developmental expression study. Arch Biochem Biophys. 2005;437:10–19. doi: 10.1016/j.abb.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Liu TA, Bhuiyan S, Snow R, Yasuda S, Yasuda T, Yang SY, Williams FE, Liu MY, Suiko M, Carter G, Liu MC. Identification and characterization of two novel cytosolic sulfotransferases, SULT1 ST7 and SULT1 ST8, from zebrafish. Aquat Toxicol. 2008;89:94–102. doi: 10.1016/j.aquatox.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Mulder GJ, Jakoby WB. Sulfation in Conjugation Reactions. In: Mulder GJ, editor. Drug Metabolism. Taylor and Francis; London: 1990. pp. 107–161. [Google Scholar]
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch Biochem Biophys. 2001;390:149–157. doi: 10.1006/abbi.2001.2368. [DOI] [PubMed] [Google Scholar]
- Nikolaev VK, Leontovich AM, Drachev VA, Brodsky LI. Building multiple alignment using iterative analyzing biopolymers structure dynamic improvement of the initial motif alignment. Biochemistry. 1997;62:578–582. [Google Scholar]
- Ohkimoto K, Liu MY, Suiko M, Sakakibara Y, Liu MC. Characterization of azebrafish estrogen-sulfating cytosolic sulfotransferase: inhibitory effects and mechanism of action of phytoestrogens. Chem Biol Interact. 2004;147:1–7. doi: 10.1016/j.cbi.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strott CA. Sulfonation and molecular action. Endocr Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Carter G, Pai TG, Liu MC. cDNA cloning, expression, and functional characterization of a zebrafish SULT1 cytosolic sulfotransferase. Arch Biochem Biophys. 2003a;414:67–73. doi: 10.1016/s0003-9861(03)00172-3. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Pai TG, Collodi P, Suiko M, Sakakibara Y, Nishiyama K, Liu MC. Sulfation of hydroxychlorobiphenyls. Molecular cloning, expression, and functional characterization of zebrafish SULT1 sulfotransferases. Eur J Biochem. 2003b;270:2404–2411. doi: 10.1046/j.1432-1033.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Liu CC, Pai TG, Liu MC. Molecular cloning, expression, andfunctional characterization of a novel zebrafish cytosolic sulfotransferase. Biochem Biophys Res Commun. 2003c;300:725–730. doi: 10.1016/s0006-291x(02)02915-7. [DOI] [PubMed] [Google Scholar]
- Sugahara T, Yang YS, Liu CC, Pai TG, Liu MC. Sulphonation of dehydroepiandrosterone and neurosteroids: molecular cloning, expression, and functional characterization of a novel zebrafish SULT2 cytosolic sulphotransferase. Biochem J. 2003d;375:785–791. doi: 10.1042/BJ20031050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AC, Lieschke GJ. The zebrafish as a model system for human disease. Front Biosci. 2002;7:827–833. doi: 10.2741/A814. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM. Sulfotransferase enzymes in conjugation–deconjugation reactions. In: Kaufmann FC, editor. Drug Metabolism and Toxicity. Springer-Verlag, Berlin; 1994. pp. 45–78. [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her CT, Raftogianis RB. Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11:3–14. [PubMed] [Google Scholar]
- Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Kumar AP, Liu MY, Sakakibara Y, Suiko M, Chen L, Liu MC. Identification of a novel thyroid hormone-sulfating cytosolic sulfotransferase, SULT1 ST5,from zebrafish. FEBS J. 2005a;272:3828–3837. doi: 10.1111/j.1742-4658.2005.04791.x. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu CC, Takahashi S, Suiko M, Chen L, Snow R, Liu MC. Identification of a novel estrogen-sulfating cytosolic SULT from zebrafish: molecular cloning, expression, characterization, and ontogeny study. Biochem Biophys Res Commun. 2005b;330:219–225. doi: 10.1016/j.bbrc.2005.02.152. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Liu MY, Yang YS, Snow R, Takahashi S, Liu MC. Identification of novel hydroxysteroid-sulfating cytosolic SULTs, SULT2 ST2 and SULT2 ST3, from zebrafish: cloning, expression, characterization, and developmental expression. Arch Biochem Biophys. 2006;455:1–9. doi: 10.1016/j.abb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Yasuda T, Yasuda S, Williams FE, Liu MY, Sakakibara Y, Bhuiyan S, Snow R, Carter G, Liu MC. Characterization and ontogenic study of novel steroid-sulfating SULT3 sulfotransferases from zebrafish. Mol Cell Endocrinol. 2008;294:29–36. doi: 10.1016/j.mce.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Burgess M, Yasuda T, Liu MY, Bhuiyan S, Williams FE, Kurogi K, Sakakibara Y, Suiko M, Liu MC. A novel hydroxysteroid-sulfating cytosolic sulfotransferase, SULT3 ST3, from zebrafish: identification, characterization, and ontogenic study. Drug Metab Lett. 2009;3:217–227. doi: 10.2174/187231209790218154. [DOI] [PubMed] [Google Scholar]