SUMMARY
Planarians grow and regenerate organs by coordinating proliferation and differentiation of pluripotent stem cells with remodeling of post-mitotic tissues. Understanding how these processes are orchestrated requires characterizing cell type-specific gene expression programs and their regulation during regeneration and homeostasis. To this end, we analyzed the expression profile of planarian intestinal phagocytes, cells responsible for digestion and nutrient storage/distribution. Utilizing RNA interference, we identified cytoskeletal regulators required for intestinal branching morphogenesis, and a modulator of bioactive sphingolipid metabolism, ceramide synthase, required for the production of functional phagocytes. Additionally, we found that a gut-enriched homeobox transcription factor, nkx-2.2, is required for somatic stem cell proliferation, suggesting a niche-like role for phagocytes. Identification of evolutionarily conserved regulators of intestinal branching, differentiation, and stem cell dynamics demonstrates the utility of the planarian digestive system as a model for elucidating the mechanisms controlling post-embryonic organogenesis.
INTRODUCTION
Many animals replenish the cells of internal organs in response to physiological turnover, but only some can regenerate organs in response to traumatic injury. For example, in widely studied vertebrates and invertebrates, the epithelial lining of the digestive tract is renewed continuously, and some molecular pathways utilized in the context of tissue homeostasis are also employed in response to infection and cytotoxic damage (Faro et al., 2009; van der Flier and Clevers, 2009; Jiang and Edgar, 2012). By contrast, other animals are capable of de novo morphogenesis of gastrointestinal (GI) organs: sea cucumbers regenerate their digestive tract after spontaneous evisceration (Mashanov and Garcia-Arraras, 2011); several annelids regrow missing portions of their GI organs during asexual reproduction (Takeo et al., 2008; Zattara and Bely, 2011); and the ascidian Polyandrocarpa misakiensis regenerates its esophagus, stomach, and intestine after amputation (Kaneko et al., 2010). Among vertebrates, the ability to recover from intestinal transection has been identified only in amphibians (Goodchild, 1956; O’Steen, 1958). Because animals capable of GI regeneration have not been accessible to molecular genetic approaches, the mechanisms underlying GI regeneration remain obscure.
The planarian Schmidtea mediterranea has emerged as a useful model for studying organ regeneration. Planarians regenerate in response to nearly any type of amputation; even small tissue fragments are capable of regenerating into complete animals (Newmark and Sánchez Alvarado, 2002; Reddien and Sánchez Alvarado, 2004). Pluripotent somatic stem cells called neoblasts are distributed throughout most regions of the planarian body (Newmark and Sánchez Alvarado, 2000; Wagner et al., 2011). After amputation, neoblasts proliferate and differentiate, regenerating a variety of organs, including the nervous system (Cebrià, 2007; Agata and Umesono, 2008), excretory system (Rink et al., 2011; Scimone et al., 2011), and intestine (Forsthoefel et al., 2011). As the only dividing somatic cells, neoblasts also supply new tissue in response to cellular turnover, and during growth (Newmark and Sánchez Alvarado, 2000; Eisenhoffer et al., 2008; Wagner et al., 2011).
The planarian intestine provides an attractive system in which to examine mechanisms of stem cell-based organogenesis, including the regulation of differentiation, the maintenance and remodeling of organ morphology by post-mitotic cells, and the influence of differentiated tissue on stem cell dynamics. Neoblasts differentiate into enterocytes during both growth and regeneration (Forsthoefel et al., 2011; Wagner et al., 2011). Furthermore, post-mitotic enterocytes remodel during the addition of intestinal branches in growing animals and the re-establishment of gut morphology after amputation (Forsthoefel et al., 2011). However, aside from the potential influence of a number of axial polarity cues (Forsthoefel and Newmark, 2009; Reddien, 2011), the mechanisms that regulate differentiation and remodeling of enterocytes are unknown. Similarly, the role of differentiated cells in influencing neoblast dynamics is poorly understood. Because neoblast proliferation is upregulated after feeding (Baguñà, 1974, 1976a; Baguñà and Romero, 1981), and neoblasts associate with the severed ends of intestinal branches after amputation (Wenemoser and Reddien, 2010), the intestine could serve as a source of signals that regulate neoblast dynamics.
Expression of genes required for organ morphogenesis is often maintained by differentiated cells in fully developed organs (Cebrià, 2007; Zorn and Wells, 2009; Scimone et al., 2011; Lapan and Reddien, 2012). Thus, expression profiling of post-mitotic tissues is an important step in elucidating the regulation of organ growth and regeneration. Here, we have developed a protocol for isolating planarian intestinal phagocytes, enabling us (i) to characterize the gene expression profile of this cell type; and (ii) to perform a targeted RNAi screen to identify genes required for intestinal morphogenesis and function.
RESULTS
Isolation of intestinal phagocytes
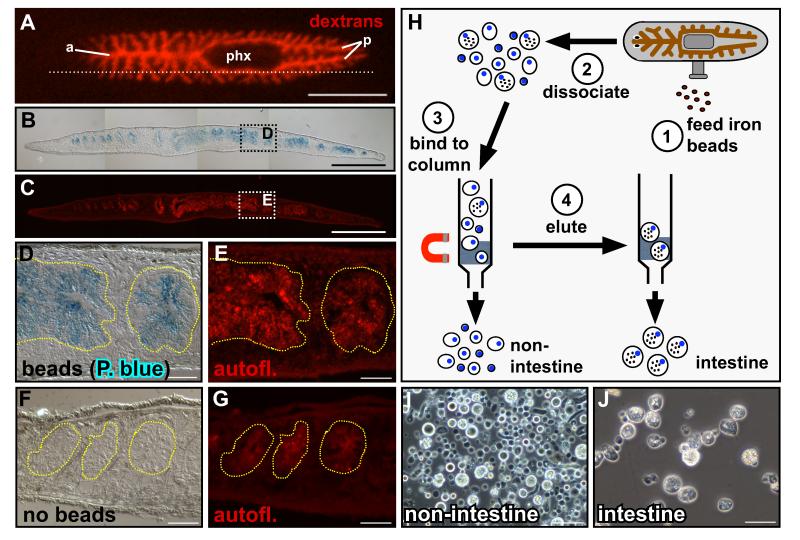
The planarian intestine is an extensively branched system of epithelial tubes (Figure 1A) comprised of a single layer of columnar cells resting on a basement membrane and encircled by enteric muscles (Willier et al., 1925; Ishii, 1965). As a “blind” gut, food enters the intestine and waste is excreted through a centrally located, muscular pharynx (Figure 1A) (Hyman, 1951). Two intestinal cell types have been identified histologically: absorptive phagocytic enterocytes that engulf food particles for intracellular digestion, and secretory goblet or “gland” cells that release enzymes (Ishii, 1965; Garcia-Corrales and Gamo, 1986, 1988).
Figure 1.
Isolation of planarian intestinal phagocytes. (A) Intestinal branches in a live planarian fed Alexa 568-conjugated dextrans. One primary anterior branch (a) extends to the head; two posterior branches (p) project around the pharynx (phx); secondary and tertiary branches extend laterally. Dotted line indicates sectioning plane in (B) and (C). (B) Prussian Blue-labeled parasagittal section of a planarian fed iron beads. (C) Correlation with intestinal autofluorescence (red) demonstrates intestine-specific iron uptake. (D and E) Magnification of boxed regions in (B) and (C); yellow dashed lines delineate gut branches. (F and G) Animals fed liver only do not label with Prussian Blue. (H) Schematic depicting intestinal phagocyte purification. (I and J) Non-intestinal (“flow-through,” I) and purified intestinal cells (J) obtained by magnetic sorting. (A) Anterior is to the left, dorsal view. (B-G) Anterior is to the left, dorsal is up. Scale bars: 1 mm (A); 500 μm (B and C); 50 μm (D-G, I and J).
Phagocytes retain fluorescent conjugates and other molecules after feeding and digestion (Figure 1A) (Morgan, 1900; Saló and Baguñà, 1985; Forsthoefel et al., 2011). The intestine also retains ‘Feridex,’ an aqueous colloid of superparamagnetic iron oxide associated with dextran (Figures 1B-1E). Prussian Blue stains intestinal branches in Feridex-fed animals (Figures 1B-1E), but not in control animals fed only liver (Figures 1F and 1G), demonstrating the specificity of iron uptake. We developed a purification strategy in which we dissociated Feridex-fed planarians into single cell suspensions, and separated phagocytes from other cell types via magnetic column (Figures 1H-1J).
Global analysis of phagocyte gene expression
We compared the gene expression profiles of purified phagocytes (Figure 1J) to other planarian cells (Figure 1I) using microarrays (Table S1). 1,514 of 11,521 transcripts represented on the arrays were upregulated in phagocytes at levels ≥1.5-fold than in other planarian cell types (Figures S1A-S1F, and Table S1). 1,152 genes (~76%) are predicted to encode homologs of proteins found in other organisms (Table S1). For example, the top BLASTX hits for 30 genes matched proteins from the parasitic flatworm Schistosoma japonicum (Table S1).
To characterize the phagocyte expression profile, we assigned gene ontology (GO) terms (Ashburner et al., 2000) and determined which biological process, molecular function, and cellular component categories were over-represented in intestinal cells (Table S2 and Figures S1G-S1I). Many enriched categories are consistent with a polarized cell type that remodels after injury and is responsible for intracellular digestion, nutrient distribution, and storage. For example, the most highly represented biological process categories include: regulation of cell shape; secretion; transport; tube development; and morphogenesis of an epithelium (Figure S1G). Within the cellular component hierarchy, gene products are predicted to localize to membrane-based organelles and the actin cytoskeleton (Figure S1I).
Validation reveals dynamic expression during regeneration and distinguishes subpopulations of intestinal phagocytes
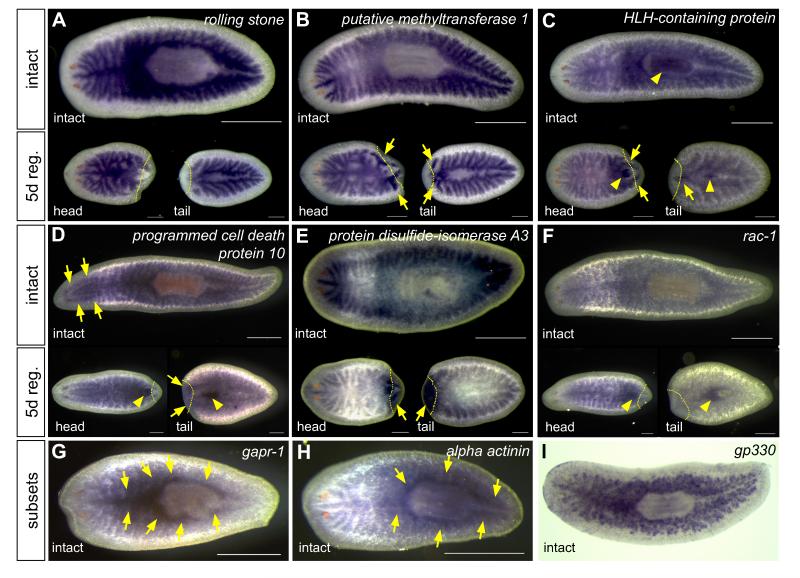
In order to validate the microarray results, we analyzed the expression of a subset of upregulated genes using whole-mount in situ hybridization. We focused on genes predicted to regulate several aspects of intestinal biology, including: remodeling or branching morphogenesis (cytoskeletal regulators); gene expression or differentiation (transcription factors, RNA binding proteins, and cell cycle modulators); and organ physiology (e.g., trafficking, transport, and digestive processes). We also randomly selected genes expressed over a range of levels and intestinal enrichment (Table S3). 216 of 218 genes analyzed (~99%) were expressed in the intestine in intact, uninjured animals (Figure 2 and Table S3), indicating the efficiency of phagocyte isolation and microarray analysis. For 42 genes (~19%), transcripts were detectable only in the intestine (Figures 2A and 2B), while other genes were also expressed in the pharynx, central nervous system, peripharyngeal secretory cells, and/or other tissues (Figure 2C and 2D; Figure S1J and S1K; Table S3). Approximately half of the genes we analyzed were expressed at elevated levels after amputation (Figures 2B and 2C, Figure S1K; Table S3), suggesting roles during regeneration of intestinal branches. Some genes were upregulated in only head or tail fragments, indicating that they may have region-specific functions, or that expression level varies with the plane of transection. Only one gene (Smed-protein disulfide-isomerase A3) was expressed ubiquitously in blastemas (Figure 2E). Intriguingly, although ~23% of genes were expressed in the pharynx of intact animals, over twice as many (~52%) were expressed in the regenerating pharynx (Figures 2C, 2D, 2F; Figure S1K), suggesting that some intestine-enriched genes may also function during pharynx regeneration. Finally, several genes were expressed in subsets of cells and/or regions of the intestine (Figures 2G-2I), hinting that phagocyte identity may be more diverse than ultrastructural studies suggest (Garcia-Corrales and Gamo, 1986, 1988).
Figure 2.
Validation of intestinal phagocyte gene expression by whole-mount in situ hybridization. (A) Example of a gene expressed in the intestine, but not upregulated during regeneration. (B-F) Genes upregulated in regenerating intestinal branches (arrows, B and C), pharynx (arrowheads, C, D, and F), central nervous system (arrows, D), or in the blastema (arrows, E). (G-I) Examples of genes upregulated in primary branches (G and H) or subsets of cells (I). In panels A-I, anterior is to the left. Gene identity is indicated in each panel. Yellow dotted lines indicate plane of amputation in five-day regenerates. Scale bars: 500 μm (intact animals); 250 μm (regenerates). See also Figure S1 and Tables S1, S2, and S3.
Gut-enriched genes are required for viability, feeding, blastema formation, intestinal regeneration, and branching morphogenesis
To identify genes required for growth, regeneration, and function of the intestine, we conducted an RNAi screen focused on a subset of 100 validated genes (Tables 1 and S4). Knockdown of 17 (17%) resulted in “gross” phenotypes, including cessation of feeding, the development of dorsal lesions followed by animal lysis and death, and the production of smaller blastemas after amputation (Tables 1 and S4). These phenotypes occurred over a range of time points during our dsRNA feeding regimen (Table 1). Examining a subset of the knockdowns by quantitative RT-PCR revealed that in 5/6 genes examined, >75% reduction in mRNA levels was achieved after 1-3 dsRNA feedings (Figure S2A). Although our RNAi targets were not chosen randomly, the observation that 15% were required for animal viability suggests that the intestine, like the protonephridial system (Rink et al., 2011; Scimone et al., 2011), plays essential physiological roles.
Table 1. Summary of genes with RNAi phenotypes in the primary screen.
| Gross (Live Animal) Phenotypes | |||||
|---|---|---|---|---|---|
| Gene ID |
Gene
Abbrev. |
Stop
Feeding |
Lysis * |
Small
Blastemas |
Putative Functions ** |
| egfp (control) | egfp | -- | -- | -- | -- |
|
engulfment and cell
motility protein 2 |
elmo-2 | + | -- | + | phagocytosis |
| tropomyosin-1 | tpm-1 | + | -- | -- | cytoskeletal regulation |
| early lysis (before 3rd feeding) | |||||
| bcl2-1 | bcl2-1 | n.a. | 1st | n.d. | anti-apoptosis 1 |
|
nuclear factor NF-kappa-
B p105 subunit |
NFkB-
p105 |
n.a. | 1st | n.d. | transcription; survival; inflammation/immunity |
| actin-1 | act-1 | + | 2nd | n.d. | cytoskeletal regulation |
|
capping protein subunit
a2 |
capz-a2 | + | 2nd | n.d. | cytoskeletal regulation |
| arp-2 | arp-2 | + | 2nd | n.d. | cytoskeletal regulation |
| nkx-2.2 | nkx-2.2 | + | 2nd | +c | transcription; differentiation |
| clathrin heavy chain | chc | + | 2nd | n.d. | endocytosis; vesicular trafficking 2 |
| cdc42 | cdc42 | + | 2nd | n.d. | cytoskeletal regulation |
| late lysis (after 3rd feeding) | |||||
|
Neural Wiskott-Aldrich
syndrome protein-like |
N-WASP | + | 3rd | + | cytoskeletal regulation |
|
partitioning defective 6
homolog beta |
par6-b | + | 3rd +CURL a | + | cell polarity; asymmetric division; adhesion |
|
coiled-coil domain
containing protein 93 |
ccdc-93 | + | 3rd | -- | intracellular trafficking; unknown |
|
putative homeodomain
transcription factor 2 |
phtf-2 | + | 3rd +BLOAT b | -- | transcription |
| rho-A | rho-A | + | 3rd | -- | cytoskeletal regulation |
| ceramide synthase 1 | CerS1 | + | 4th | -- | sphingolipid metabolism |
|
mediator of RNA
polymerase II complex subunit 21 |
med-21 | + | 5th | + | transcription |
For lysis, intact animals begin to lyse after the RNAi feeding listed.
Putative functions based on gene ontology assignments and homologs' roles in other organisms. n.a., not applicable; n.d., not done.
animals also curl ventrally after the 3rd RNAi feeding
animals also bloat after the 3rd RNAi feeding
nkx-2.2(RNAi) animals were only fed twice to assess regeneration.
(Inoue et al., 2007). See also Supplemental Table S4.
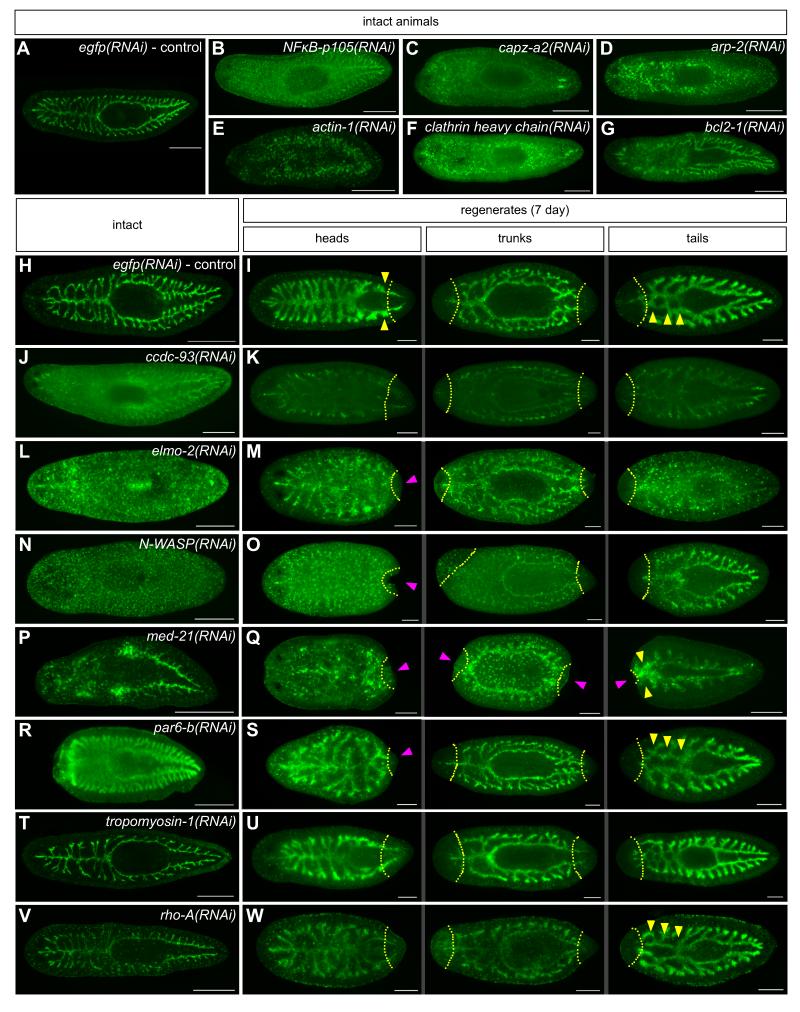
To assess gene function during intestinal morphogenesis, we generated a monoclonal antibody (3G9) that specifically recognizes the intestine (Figures 3A, 3H, and 3I) (Forsthoefel et al., in preparation). RNAi knockdowns that did not cause feeding cessation or lysis showed normal labeling with this antibody (not shown). Knockdowns resulting in gross phenotypes, however, displayed a range of phenotypes in uninjured animals, including reduced intestinal labeling, increased diffuse background, and elevated punctate background (Figure 3B, 3C, 3J, 3L, 3N, 3P, and Table 1). In some cases, the integrity of the intestine was severely disrupted and intestinal cells appeared to dissociate from one another or even lyse (Figures 3D-G, 3L, 3P, and Table 1). In regenerates, in addition to the phenotypes observed in intact animals, some knockdowns resulted in a delay or block in intestinal regeneration (Figures 3K, 3M, 3O, 3Q, 3S, and Table 1).
Figure 3.
RNAi phenotypes visualized by an intestine-specific MAb. (A) egfp(RNAi) control: primary, secondary, and tertiary intestinal branches are labeled by MAb 3G9 (green). (B-G) Phenotypes of early-lysing RNAi knockdowns in intact animals, including loss of intestinal labeling (B-C), increased diffuse background (B-D, F), increased punctate background (B-D), dissociation of intestinal cells from each other (D-F), and intestinal disintegration (F-G). (H-W) Phenotypes of late- and non-lysing RNAi knockdowns in intact and regenerating animals. (H and I) egfp(RNAi) controls: regenerating branches indicated by yellow arrowheads (I). RNAi phenotypes include decreased labeling (J and K), loss of integrity, dissocation and/or lysis of intestinal cells (L-Q), small blastemas (magenta arrowheads in M, O, Q, and S), and stalled or delayed intestinal regeneration (yellow arrowheads in Q and S). (T and U) tpm-1 knockdown causes decreased branching in uninjured animals (T); regenerates are unaffected (U). (V and W) In rho-A(RNAi) tail regenerates, branches do not fuse at the midline (W, yellow arrowheads, compare to I and U); intact animals and anterior fragments are unaffected, aside from decreased signal. Dashed yellow lines indicate plane of amputation. Gene identity is indicated in each panel. Anterior is to the left in all panels. Scale bars: 500 μm (A-G, H, J, L, N, P, R, T, and V); 200 μm (I, K, M, O, Q, S, U, and W). See also Figure S2 and Table S4.
Knockdown of two putative cytoskeletal regulators, Smed-tropomyosin-1 (tpm-1) and Smed-rho-A, altered intestinal morphology more specifically, causing distinct defects in branching morphogenesis. tpm-1 knockdown resulted in fewer secondary and tertiary branches in intact animals (Figure 3T, Figure S2B-D). However, tpm-1(RNAi) head, trunk, and tail fragments regenerated normally (Figure 3U), suggesting that TPM-1 is not required for the re-establishment of anteroposterior intestinal morphology after injury. Conversely, rho-A was required for fusion of the anterior regions of intestinal branches in tail fragments (Figure 3W) while intestinal morphology in uninjured animals, head fragments, and trunk fragments was normal (Figure 3V, 3W). These phenotypes were specific to rho-A, since knockdown of several closely related rac-like GTPases did not cause a similar phenotype (Table S4). Thus, cytoskeletal regulators can function in specific contexts to maintain or re-establish intestinal branching morphology in intact or regenerating animals.
ceramide synthase is required for the differentiation of functional phagocytes
Knockdown of several genes resulted in a reduction in intestinal labeling by MAb 3G9 (Figure 3, Figure 4E, and Table 1). We further characterized Smed-ceramide synthase 1 (CerS1) (Table 1, Figure S3A), which encodes a planarian member of a family of enzymes that catalyze the production of ceramide and dihydroceramide, precursors of a variety of sphingolipids important for differentiation, lipid metabolism, and other cellular processes (Spassieva et al., 2006; Hannun and Obeid, 2008; Teufel et al., 2009).
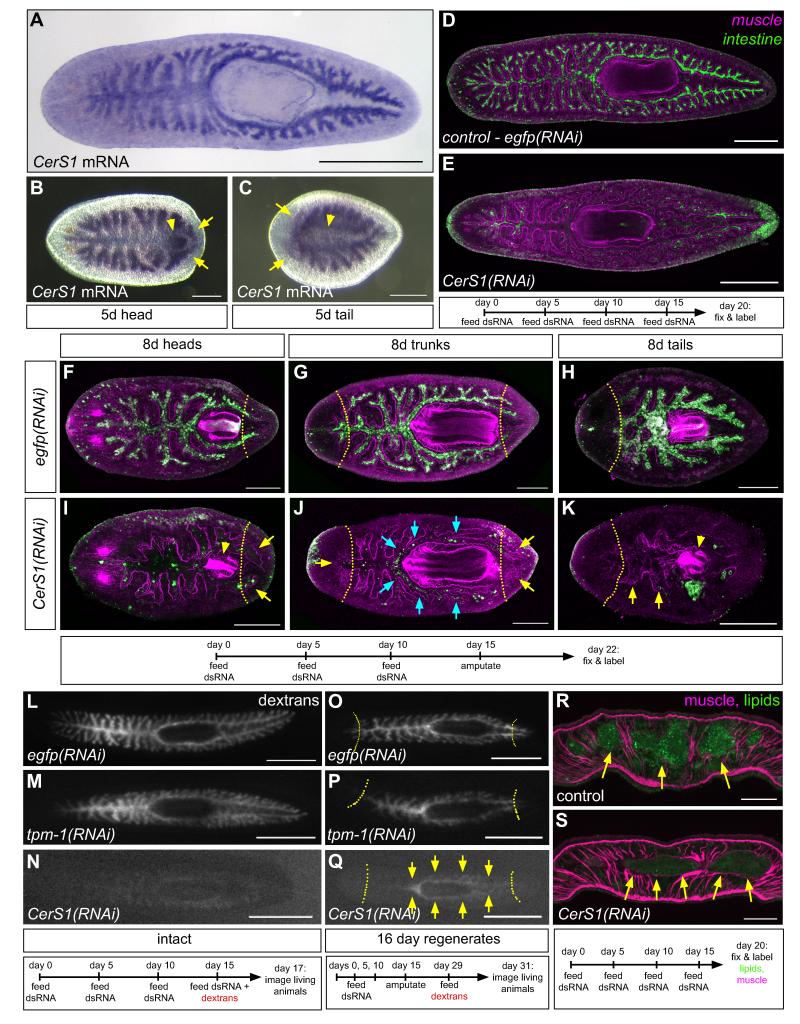
Figure 4.
Expression and functional analysis of Smed-CerS1. (A-C) Whole-mount in situ hybridization to detect CerS1 transcripts in uninjured animals (A), and head (B) and tail (C) regenerates (arrows, gut branches; arrowheads, regenerating pharynx). (D and E) MAb 3G9 (green) and enteric muscle labeling (magenta) of uninjured control (D) and CerS1(RNAi) animals (E). (F-K) MAb 3G9 (green) and enteric muscle labeling (magenta) of regenerates. Blue arrows, partial retention of labeling in branches surrounding the pharynx in CerS1(RNAi) fragments. Yellow dashed lines (F-K), plane of amputation; yellow arrows (I-K), regenerating intestinal branches; arrowheads (I and K), regenerating pharynges. Confocal projections (D-K). (L-N) Visualization of fluorescent dextran retention after feeding in intact control (L and M) and CerS1(RNAi) animals (N). (O-Q) Fluorescent dextran retention in trunk regenerates of control (O and P) and CerS1(RNAi) (Q) animals. Arrows (Q) indicate dextran retention only in branches around the pharynx, but not in anterior or posterior branches. Images of living animals (L-Q). (R and S) Visualization of intestinal lipid storage in control (R) and CerS1(RNAi) (S) animals. Epifluorescent images of fixed cryosections labeled with phalloidin (magenta, muscles) and BODIPY (green, lipids). (A-Q) Anterior, to the left. (R-S) Dorsal, to the top. Time courses are indicated below each experiment. Scale bars: 500 μm (A); 250 μm (B-E); 200 μm (F-K); 1 mm (L-Q); 100 μm (R-S). See also Figure S3.
CerS1 expression was largely restricted to intestinal branches in intact animals; after amputation, CerS1 was moderately upregulated in regenerating branches and in the regenerating pharynx (Figures 4A-4C). In uninjured CerS1(RNAi) animals, the intestine no longer labeled with MAb 3G9, but the layer of surrounding enteric muscles remained intact (Figures 4D and 4E). In regenerates, reorganization of gut branches occurred normally, indicated by enteric muscle labeling (Forsthoefel et al., 2011) (Figures 4F-4K). 3G9 labeling was lost in head and tail fragments, and in the elongating anterior and posterior branches of trunk fragments, a region where new intestinal cells normally differentiate (Forsthoefel et al., 2011) (Figures 4I-4K). However, 3G9 labeling was retained in branches around the pharynx (Figure 4J). Despite the loss of 3G9 labeling, the number and localization of goblet cells appeared to be unaffected in either intact or regenerating CerS1(RNAi) animals (Figures S3B-S3I). Furthermore, CerS1 knockdown did not inhibit the birth of BrdU-positive enterocytes in intact animals (Figures S3J and S3K), blastemas were normal, and heads and pharynges regenerated properly (Figures 4I and 4K). Thus, CerS1 knockdown did not skew fate selection in favor of goblet cells, block the initial generation of intestinal cells, or affect regeneration of other tissues, suggesting that loss of 3G9 labeling might reflect a more specific defect, during later stages of phagocyte differentiation.
To test this idea, we examined whether functional enterocytes were produced in CerS1(RNAi) animals. First, we found that although CerS1(RNAi) animals ingested food, they did not retain fluorescent dextrans, a lipid-binding dye, or a lysosome marker after feeding (Figures 4L-4N) (Figures S3L-S3Q), suggesting that phagocytosis, metabolic processing, and/or nutrient storage is disrupted. Extending this assay to regenerates, we reasoned that if CerS1 is required for the maturation of new phagocytes, and not merely the maintenance of phagocyte function, then functional defects would be most pronounced in regions where differentiation occurs preferentially after amputation; older cells that existed prior to injury would still be able to retain and engulf food. Indeed, in CerS1(RNAi) trunk fragments fed after regeneration, dextran retention was minimal in newly regenerated anterior and posterior branches, and was confined to intestinal branches around the pharynx (Figures 4O-4Q), consistent with the partial retention of 3G9 labeling (Figure 4J).
Planarian phagocytes are also the main cell type in which lipid droplets are stored (Jennings, 1957; Fried and Grigo, 1977). In most CerS1(RNAi) animals (7/10), enteric lipid content was dramatically reduced (Figure 4R and 4S), reinforcing the idea that CerS1 is required for the differentiation of functional phagocytes. Finally, analyzing CerS1(RNAi) animals by electron microscopy revealed, in addition to lower lipid content, frequent long intercellular gaps between the lateral surfaces of enterocytes that were often filled with amorphous material (Figures S3R-S3U). Apical cell junctions (Pedersen, 1961), autophagosomes (Bowen, 1980; Garcia-Corrales and Gamo, 1987), and goblet cell morphology (Pascolini and Gargiulo, 1975; Garcia-Corrales and Gamo, 1986), however, appeared normal. Based on these results we conclude that CerS1 is required for later stages of differentiation of functional phagocytes.
nkx-2.2 is required for neoblast proliferation
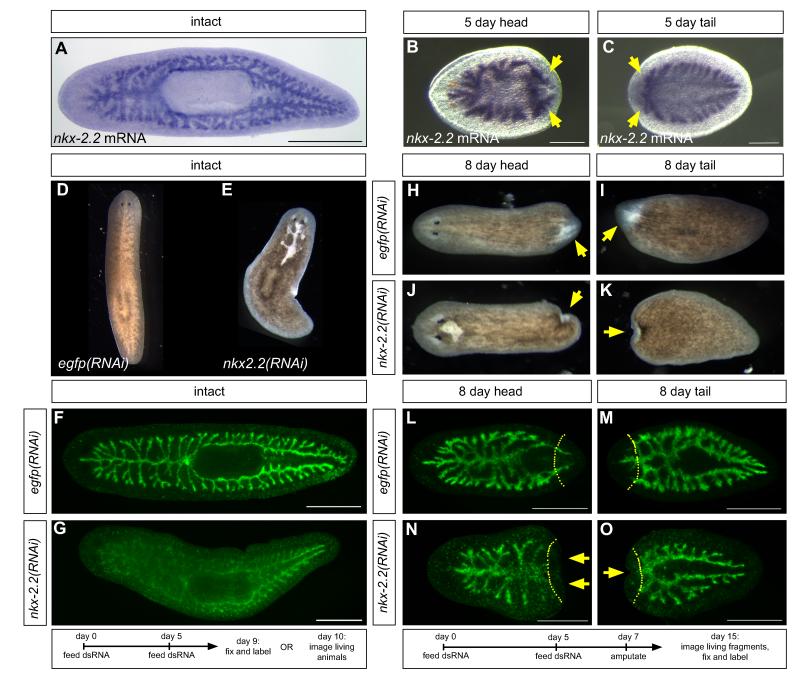
We further characterized Smed-nkx-2.2 (Figure S4A) because it was one of the most differentially expressed genes to yield an RNAi phenotype (Table S4), and because nkx-2.2 orthologs regulate transcription and differentiation in the mammalian intestine and pancreas (Sussel et al., 1998; Desai et al., 2008; Wang et al., 2009b). nkx-2.2 is expressed throughout the intestine in intact animals (Figure 5A); in amputated fragments, nkx-2.2 is upregulated in regenerating branches (Figures 5B and 5C). nkx-2.2(RNAi) animals lysed in a pattern over gut branches within five days after the second RNAi feeding (Figures 5D and 5E). In knockdown animals fixed just prior to the formation of lesions, the intestine of uninjured animals labeled less intensely, but intestinal morphology was otherwise normal (Figure 5F and 5G). In regenerates, blastemas were severely reduced in size (Figures 5H-5K). In head fragments, posterior intestine branches did not form (Figures 5L and 5N), while in tail fragments, anterior branches fused at the midline, but did not elongate as in controls (Figures 5M and 5O).
Figure 5.
Expression and functional analysis of Smed-nkx-2.2. (A-C) In situ hybridization to detect nkx-2.2 expression in uninjured animals (A), and regenerating head (B) and tail (C) fragments (arrows, upregulation in regenerating branches). (D and E) Lysis of uninjured nkx-2.2(RNAi) planarians, dorsal to intestinal branches. (F and G) MAb 3G9 labeling of control (F) and nkx-2.2(RNAi) animals (G). (H-K) Blastemas (arrows) in control (H and I) and nkx-2.2(RNAi) (J and K) regenerates. (L-O) MAb 3G9 labeling of control (L and M) and nkx-2.2(RNAi) (N and O) regenerates. Arrows indicate lack of intestinal branches in nkx-2.2(RNAi) fragments. Dashed lines, planes of amputation. (D and E, H-K) Images of live planarians. (D and E) Anterior to the top; all other panels, anterior to the left. Scale bars: 250 μm (B and C); 500 μm (A, F, G, L-O).
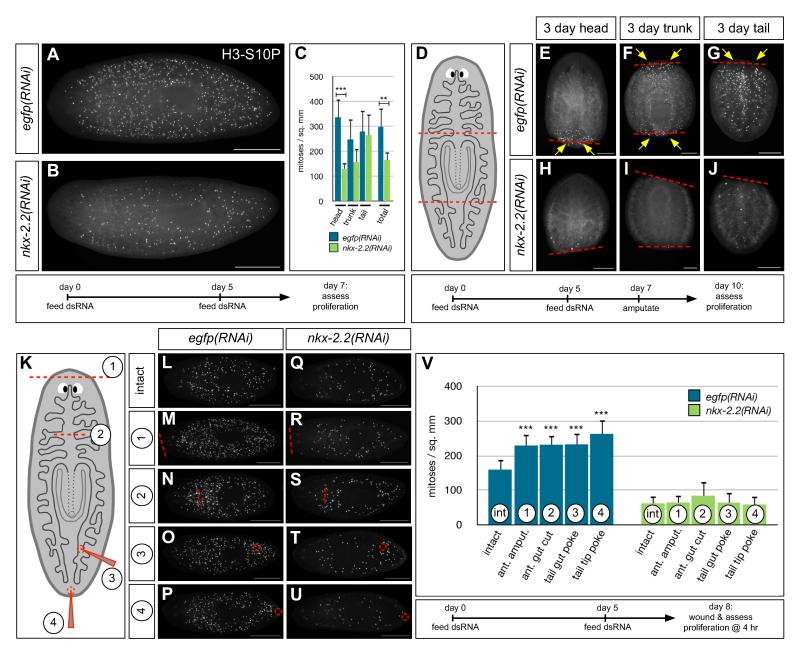
Reduced blastema growth is often associated with knockdown of genes required for stem cell maintenance or proliferation, since new cells are no longer generated in response to injury (Reddien et al., 2005; Guo et al., 2006; Rouhana et al., 2010). Therefore, we next asked whether neoblasts were able to proliferate in various contexts. In uninjured animals, neoblast proliferation increases up to five-fold after feeding, to supply new cells for growth (Figure 6A) (Baguñà, 1974, 1976a; Baguñà and Romero, 1981). However, proliferation in nkx-2.2(RNAi) animals after feeding was reduced by ~45% (Figures 6A-6C). The failure in neoblast proliferation was accompanied by only a moderate loss of stem cells anteriorly, suggesting that nkx-2.2(RNAi) affects neoblast proliferation, not survival (Figure S4B).
Figure 6.
Smed-nkx-2.2 is required for neoblast proliferation. (A and B) Phosphohistone H3-S10 (H3-S10P) labeling of uninjured control (A) and nkx-2.2(RNAi) (B) planarians two days after feeding. (C) Proliferation is most dramatically reduced anterior to the pharynx, and by ~45% overall. Error bars, standard deviation. ***p<0.001, **p<0.01, Student’s t-test. n≥5 animals per condition. Regions were delineated as in the schematic in (D). (D-J) H3-S10P labeling of three-day regenerates; proliferation is nearly abolished in nkx-2.2(RNAi) fragments (arrows). Red dashed lines, plane of amputation. (K-U) H3-S10P labeling four hours after various minor injuries, including amputation of the anterior head tip (“1”), mid-anterior incision through an intestine-containing region (“2”), posterior tail poke through an intestine-containing region (“3”), and a poke injury to the tail tip (“4”). (V) Quantification of experiments in (L-U). Error bars, standard deviation. ***p<0.0005, Student’s t-test (injuries were compared to intact samples within each RNAi treatment group); n≥8 animals per condition. Orientation: anterior to the left (A, B, L-U); to the top (E-J). H3P-S10-labeled animals in all panels were imaged under epifluorescence. Scale bars: 500 μm (A, B, L-U); 250 μm (E-J). See also Figure S4.
Neoblasts also respond to tissue loss: several days after amputation, proliferation increases near the site of injury to provide new cells for the regeneration of missing structures (Baguñà, 1976b; Saló and Baguñà, 1984). In nkx-2.2(RNAi) fragments three days after amputation, regenerative proliferation in head, trunk, and tail fragments was almost entirely abolished (Figures 6D-6J). Because at this time point, intestinal regeneration is in its earliest stages (Forsthoefel et al., 2011), it is likely that the failure of intestinal regeneration in nkx-2.2(RNAi) animals is secondary to the proliferation defect.
Neoblasts also proliferate in an earlier, animal-wide mitotic burst that peaks at 4-10 hr after injury, even in response to wounds that do not involve tissue loss (Wenemoser and Reddien, 2010). nkx-2.2 is required for this earlier mitotic peak after a variety of minor wounds (Figures 6K-6V), indicating that the effect of nkx-2.2 knockdown on proliferation is not dependent exclusively on either tissue loss or intestinal damage. Intriguingly, after minor injuries such as anterior amputation or tail tip incisions, wound healing occurred normally, and some nkx-2.2 knockdown animals partially regenerated, even when lysis had begun (Figure S4C). These observations imply that neoblasts remaining after nkx-2.2 knockdown retain the ability to migrate and differentiate, further supporting the idea that nkx-2.2(RNAi) specifically affects proliferation.
We examined whether the effect of nkx-2.2(RNAi) on proliferation was due to general defects in phagocyte viability, function, or integrity of the intestinal epithelium. We did not observe increased apoptosis in the intestine or elsewhere (Figure S4D). Furthermore, in contrast to CerS1 knockdown experiments (Figure S3D), most nkx-2.2(RNAi) animals retained a lipid-binding dye (Figure S4E) and fluorescent dextrans (not shown), in some cases even after lysis had begun (Figure S4E). Similarly, we were able to isolate equivalent numbers of viable phagocytes by magnetic sorting from control and nkx-2.2(RNAi) animals fed iron particles concurrently with the second RNAi feeding (not shown).
Finally, we examined whether nkx-2.2 might act autonomously in neoblasts. Colorimetric in situ hybridizations suggested only weak, diffuse nkx-2.2 expression outside the intestine (Figure 6A-6C), but fluorescent detection revealed scattered, punctate nkx-2.2 expression in mesenchymal cells (Figure S4F). These cells persisted for several days after gamma irradiation, a treatment that selectively ablates neoblasts (Dubois, 1949; Hayashi et al., 2006), demonstrated by the loss of smedwi-1-mRNA- positive cells (Figure S4F). Non-intestinal expression of Smed-hepatocyte nuclear factor 4 (hnf4), another putative endodermal marker (Wagner et al., 2011), was similarly maintained for several days after irradiation (Figure S4F). Delayed loss of gene expression in mesenchymal cells after irradiation is an indicator of fate-restricted stem cell progeny that are differentiating (Eisenhoffer et al., 2008). These observations imply that the role of NKX-2.2 in neoblasts, if any, is restricted to differentiation, and that the observed proliferation defects are due to the loss of nkx-2.2 in postmitotic intestinal phagocytes.
DISCUSSION
The planarian intestine is a fascinating system for studying post-embryonic organogenesis: during both growth and regeneration, new intestinal branches are generated by the coordinated remodeling of differentiated tissues with the birth of new enterocytes (Forsthoefel et al., 2011). In this study, we have begun to elucidate the molecular mechanisms regulating these processes by purifying a single intestinal cell type, the phagocyte, and identifying over 1500 genes that are expressed differentially in the intestine. Functional characterization of several of these genes demonstrates that planarians utilize evolutionarily conserved molecules to regulate branching morphogenesis and the generation of functional enterocytes during growth and regeneration.
Phagocytes express a complement of intestine-specific cytoskeletal regulators, including homologs of actin, myosin, tropomyosin, cofilin, Arp2/3 complex proteins, Rho-family GTPases, EVL (Ena/VASP-like), and F-actin capping protein (Table S1). Knockdown of several of these genes caused rapid animal lysis and death after only one or two dsRNA feedings, indicating that they may play critical roles in adhesion, polarity, or other processes required for intestinal integrity. Two genes, however, regulated branching morphogenesis specifically in either uninjured (tpm-1) or regenerating (rho-A) animals. Although best known for their role in actomyosin contractility in skeletal and cardiac muscle, many mammalian tropomyosin isoforms are expressed in specific spatiotemporal patterns during development (Gunning et al., 2008), and also regulate cell migration and motility (Bach et al., 2010). To the best of our knowledge, however, tropomyosins have not been implicated in the morphogenesis of epithelial tubes. Rho-family GTPases control the dynamics of the actin and microtubule cytoskeletons, regulating polarity, adhesion, and motility in response to Wnts and other signals (Heasman and Ridley, 2008; Schlessinger et al., 2009). Knockdown of rho-A affected only tail fragment regeneration, suggesting a role downstream of axial polarity cues that influence the maintenance and regeneration of the intestine’s anteroposterior morphology (Forsthoefel and Newmark, 2009; Reddien, 2011). Future experiments will be required to determine whether TPM-1 and RHO-A regulate migration of differentiating enterocytes into the intestinal epithelium, and/or coordination of cell shape changes and collective migration during remodeling.
We also identified a requirement for CerS1 during phagocyte differentiation into functional cells capable of food uptake and lipid storage. CerS1 encodes a planarian member of the LASS (longevity assurance homolog) family of ceramide synthases, enzymes that catalyze the production of ceramide and dihydroceramide, precursors for a variety of sphingolipids (Figure S3A) (Spassieva et al., 2006; Teufel et al., 2009; Mullen et al., 2012). Sphingolipid biosynthetic pathways are found in all eukaryotes, and control the production of structural components of membranes (Grassme et al., 2007; Silva et al., 2012). Additionally, ceramide is the precursor for ceramide-1-phosphate, sphingosine-1-phosphate, and other bioactive lipids (Hannun and Obeid, 2008; Mullen et al., 2012). These metabolites regulate diverse processes including cell junction formation (Wang et al., 2009a), cell polarity (Bieberich, 2011), the response of epithelial sheets to dying cells (Gu et al., 2011; Eisenhoffer et al., 2012), and differentiation of neural stem cells and adipocytes (Xu et al., 2010; Bieberich, 2011, 2012). Furthermore, ceramide and other sphingolipids also regulate lipid storage and mobilization (Bauer et al., 2009; Kohyama-Koganeya et al., 2011; Worgall, 2011). Thus, further exploration of sphingolipid metabolic pathways in planarians may illuminate novel role(s) for bioactive lipids, and the coordination of energy stores during growth and regeneration.
Finally, knockdown of the homeodomain transcription factor nkx-2.2 blocked intestinal regeneration and neoblast proliferation after feeding, amputation, and minor injuries. nkx-2.2 is expressed predominantly throughout the intestine, in addition to a small number of mesenchymal cells outside the intestine. We found that nkx-2.2-positive mesenchymal cells persist after irradiation, and others have noted that mesenchymal expression only increases during later stages of regeneration, after initial proliferative peaks have occurred (Garcia-Fernàndez et al., 1993). These data suggest that mesenchymal nkx-2.2-positive cells are likely neoblast progeny in the process of differentiation. Consistent with this idea, nkx-2.2 is required in mice for the specification of hormone-secreting pancreatic islet cells and enteroendocrine cells of the small intestine (Sussel et al., 1998; Wang et al., 2004; Desai et al., 2008).
Despite a potential role in differentiation, the primary defect we observe in nkx-2.2(RNAi) animals, attenuation of proliferation, occurs prior to early stages of intestinal regeneration. nkx-2.2 does not appear to be required for neoblast survival or migration, nor is it required for phagocyte viability, food uptake, or nutrient storage. Therefore, we suggest that the major cause of decreased neoblast proliferation during the early phase of the nkx-2.2(RNAi) phenotype is not abrogated differentiation or phagocyte death, but the dysregulation of NKX-2.2 targets in terminally differentiated intestinal phagocytes. Intriguingly, in addition to the role of NKX-2.2 in the differentiation of endocrine cell types, in the mouse pancreas NKX-2.2 is required for the expression of several secreted molecules, including peptide hormones such as insulin and glucagon (Anderson et al., 2009).
In planarians, the mechanisms by which differentiated tissues communicate with stem cells to regulate their numbers and differentiation into specific cell types are poorly understood. Mitotically active neoblasts are closely associated with the intestine in uninjured animals, and cluster around the severed tips of intestinal branches near the plane of amputation after injury (Newmark and Sánchez Alvarado, 2000; Wenemoser and Reddien, 2010). Furthermore, after feeding (Baguñà, 1974, 1976a) or tissue loss (Baguñà, 1976b; Saló and Baguñà, 1984; Wenemoser and Reddien, 2010), neoblast proliferation increases dramatically, but the molecular cues that stimulate cell division in these contexts have not been identified.
Attenuation of neoblast proliferation by knockdown of an intestinally expressed transcription factor provides evidence that intestinal phagocytes may play a niche-like role in the regulation of neoblast dynamics. In planarians, phagocytes are the main cell type in which lipid droplets and glycogen granules are found, and the only cell type responsible for food digestion (Willier et al., 1925; Garcia-Corrales and Gamo, 1987, 1988). In addition, planarian body size is extremely plastic: planarians grow when food is available, and degrow through cell loss during starvation (Baguñà and Romero, 1981), processes that are regulated in part by insulin-like peptide signaling (Miller and Newmark, 2012). Intestinal phagocytes are uniquely positioned to coordinate these processes by playing an endocrine-like role in the communication of metabolic state to other cells, including neoblasts. Because some proliferative responses are affected even in the absence of gut damage and tissue loss, future experiments must determine how NKX-2.2 and/or its targets regulate proliferation, and whether they respond differently to stimuli such as feeding or various kinds of injury.
Ongoing investigations in a range of organisms have begun to elucidate the molecular mechanisms by which digestive organs respond to damage (Amcheslavsky et al., 2009; Chatterjee and Ip, 2009; Jiang et al., 2009; Ashton et al., 2010) and nutrient availability (Park and Takeda, 2008; Choi et al., 2011; O’Brien et al., 2011). Furthermore, recent studies have emphasized the importance of understanding the role of post-mitotic cells in regulating proliferation and differentiation in the gastrointestinal stem cell niche (Sato et al., 2011; Durand et al., 2012; Kim et al., 2012; Yilmaz et al., 2012), and after traumatic injury to organs in other regenerating animals such as salamander, newt, and zebrafish (Kumar et al., 2007; Berg et al., 2011; Choi and Poss, 2012). Our work illustrates the utility of the planarian intestine as a complementary system in which to unravel the mechanisms that coordinate cellular responses during stem cell-based organ growth and regeneration.
EXPERIMENTAL PROCEDURES
Animal care and maintenance
Asexual Schmidtea mediterranea (clonal line ClW4) were maintained as described (Cebrià and Newmark, 2005).
Purification of intestinal phagocytes
Large planarians (>8 mm in length) were fed a mixture of 135 μL liver homogenate (1:3, liver paste:H2O), 0.7 μL food coloring (Durkee), 60 μL 2% ultra-low melting point agarose, and 4 μL of a 1:4 dilution of Feridex (AMAG Pharmaceuticals). Forty-eight hours later, animals were dissociated into single cell suspensions (Reddien et al., 2005) using dispase (0.6 U/mL, Worthington Biochemical) instead of trypsin. Dissociated cells were filtered sequentially through 160 μm, 53 μm, and 30 μm meshes, resuspended in de-gassed CMF with 0.5 mM EDTA, and purified on MACS LS separation columns (Miltenyi).
RNA isolation, amplification, labeling, and microarray analysis
Total RNA was isolated from intestinal and non-intestinal cells using Trizol (Invitrogen). RNA was amplified with the Amino Allyl MessageAmp II aRNA Amplification Kit (Ambion), coupled to Alexa Fluor 555 or Alexa Fluor 647 succinimidyl ester dyes (Molecular Probes), and hybridized to CombiMatrix oligonucleotide arrays according to manufacturer protocols. Arrays were imaged using a GenePix 4000B Scanner and GenePix Pro 6.0 software (Axon Instruments/Molecular Devices). Data were processed in Bioconductor (Gentleman et al., 2004; Smyth, 2004; Ritchie et al., 2007). Detailed protocols in Extended Experimental Procedures.
GO annotation
Gene ontology terms (Ashburner et al., 2000) were assigned using Annot8r (http://www.nematodes.org/bioinformatics/annot8r/index.shtml) (Schmid and Blaxter, 2008), and mapped to custom slim ontologies with “map2slim” in GO-Perl. Detailed methods in Extended Experimental Procedures.
Whole mount and fluorescent in situ hybridization (WISH, FISH)
Validation of microarray results by WISH was conducted on Carnoy’s fixed samples as described (Forsthoefel et al., 2011). For Smed-CerS1 and Smed-nkx-2.2 intact (uninjured) animals, WISH was conducted as in Pearson et al. (2009). FISH was also performed as described (Pearson et al., 2009) except that 1% casein (Sigma) was added to maleic acid blocking buffer and anti-DIG antibody solution; detection was performed using Cy3-tyramide (Perkin Elmer).
RNAi experiments
ESTs (Zayas et al., 2005) were amplified from pBluescript, and then TA-cloned into a derivative of the vector “p2T7TAblue” (Alibu et al., 2005) (http://trypanofan.path.cam.ac.uk/trypanofan/vector/). (Detailed vector modifications are in Extended Experimental Procedures.) Constructs were transformed into E. coli strain HT115(DE3); dsRNA expression was induced and bacteria were mixed with 30-60 μL of liver homogenate and fed to planarians as described (Gurley et al., 2008). In the primary screen, planarians were fed five times, every five days; for regenerates, animals were fed three times, then amputated and allowed to regenerate for seven days. For immunofluorescent characterization of knockdowns that caused lysis, intact animals were fixed when animals first began to develop lesions. 3G9 phenotypes and blastema phenotypes were scored if present in >25% of undamaged/non-lysing animals. A secondary screen was performed to verify all phenotypes; genes with non-reproducible phenotypes were not considered further. For further gene characterizations, dsRNA was sometimes fed fewer times prior to lysis to enable fixation and amputation (see Table S4), and non-eating animals were removed after each round of feeding.
Immunofluorescence
For MAb 3G9 labeling, animals were relaxed and killed in 0.66 M MgCl2 (diluted in planarian salts) for 15-30 sec, rinsed in PBS, shaken in ice-cold 2% HCl (3 min), fixed in methacarn (15 min), rinsed in methanol (15 min), then bleached O/N in 6% H2O2. For proliferation analysis, animals were fixed in Carnoy’s (Umesono et al., 1997). After rehydration, standard methods (Forsthoefel et al., 2011) were utilized to label animals with MAb 3G9 supernatant (Forsthoefel et al., in preparation), rabbit anti-muscle serum, rhodamine-LCA (Vector Labs) (Zayas et al., 2010), and anti-phosphohistone-H3-S10. Detailed protocols in Extended Experimental Procedures.
Feeding of fluorescent compounds
Dextran feeding was conducted as described (Forsthoefel et al., 2011). Alternatively, BODIPY 493/503 (1 mg/mL in DMSO, Molecular Probes) or Neutral Red (4.5 mg/mL in PBS, Sigma) were diluted 1:10 into liver homogenate. In experiments to assess phagocytosis/storage in CerS1 and nkx-2.2 knockdowns, dextrans or BODIPY were diluted as above into liver mixed with dsRNA-expressing E. coli. Animals that did not ingest food initially were removed so as not to confound analysis of dextran/BODIPY retention two days later.
BODIPY and Prussian Blue labeling
dsRNA-fed animals were relaxed and killed individually in 0.66 M MgCl2 as above, rinsed quickly in PBS, and fixed overnight in 4% formaldehyde/PBS. Following two rinses in PBS, animals were incubated for 10 min each in 15% sucrose/PBS and 30% sucrose/PBS, mounted in Tissue Freezing Medium (TBS), then cryosectioned at 12 μm. Sections were adhered to Superfrost Plus slides (Fisher) coated with 0.5% gelatin and 0.05% chromium potassium sulfate. Slides were rehydrated in deionized water, rinsed in PBS-Tween-20 (0.01%), then incubated for 15 min at room temperature in BODIPY 493/503 (0.2 μg/mL), Alexa 568-phalloidin (2 U/ml, Molecular Probes), and DAPI at 1 μg/mL for 15 min at RT. Following three washes in PBS-Tween-20 (0.01%) (5 min each), slides were mounted in Vectashield.
For Prussian Blue labeling, cryosections from animals fed Feridex were prepared as above (fixed two days after feeding), then incubated for 20 minutes in a 1:1 solution of 10% potassium ferrocyanide and 20% concentrated HCl (7.2% final). Slides were rinsed in 95% ethanol, 100% ethanol, and xylenes, then mounted in Permount.
Imaging
Imaging and image processing were conducted as described (Forsthoefel et al., 2011). Anterior amputations, tail incisions, and other minor injuries were performed as described (Wenemoser and Reddien, 2010); living animals were imaged on a Leica MZ125 with a MicroFIRE camera (Optronics) and PictureFrame 3.00.03. Confocal imaging was conducted on a Zeiss LSM 710 running Zen.
Supplementary Material
ACKNOWLEDGMENTS
We thank: John Brubacher, Jim Collins, Tingxia Guo, Ryan King, Melanie Issigonis, Rachel Roberts-Galbraith, Labib Rouhana, James Sikes, Yuying Wang, Ricardo Zayas, and other Newmark lab members for their insights, comments, and discussions; Tracy Chong for collaboration on dispase dissociation; Ryan King for advice and collaboration on FISH optimization; David Horn and Doug LaCount for kindly sharing p2T7TAblue and other trypanosome dsRNA expression vectors; Liping Wang, Rachel Breitenfeld, and Xiaoxia Wang of the Immunological Resources Center at Illinois for monoclonal antibody production; Jenny Drnevich Zadeh for guidance in microarray analysis; Barbara Pilas for suggesting magnetic sorting; and Diana Thomas for helping with magnetic sorting. We apologize to authors whose work we were unable to cite due to space constraints. This work was supported by a Ruth L. Kirschstein National Research Service Award from NIH (F32-DK077469) to DJF and R01-HD043403 to PAN. PAN is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
CombiMatrix microarray data are deposited in GEO (accession number GSE35565).
SUPPLEMENTAL INFORMATION
Supplemental Information incudes Extended Experimental Procedures, four figures, and four tables and can be found within this article online at doi: .
REFERENCES
- Agata K, Umesono Y. Brain regeneration from pluripotent stem cells in planarian. Philos Trans R Soc Lond B Biol Sci. 2008;363:2071–2078. doi: 10.1098/rstb.2008.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alibu VP, Storm L, Haile S, Clayton C, Horn D. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2005;139:75–82. doi: 10.1016/j.molbiopara.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KR, White P, Kaestner KH, Sussel L. Identification of known and novel pancreas genes expressed downstream of Nkx2.2 during development. BMC Dev. Biol. 2009;9:65. doi: 10.1186/1471-213X-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, et al. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev. Cell. 2010;19:259–269. doi: 10.1016/j.devcel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach CT, Schevzov G, Bryce NS, Gunning PW, O’Neill GM. Tropomyosin isoform modulation of focal adhesion structure and cell migration. Cell. Adh. Migr. 2010;4:226–234. doi: 10.4161/cam.4.2.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J. Dramatic mitotic response in planarians after feeding, and a hypothesis for the control mechanism. J. Exp. Zool. 1974;190:117–122. doi: 10.1002/jez.1401900111. [DOI] [PubMed] [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. J. Exp. Zool. 1976a;195:53–64. [Google Scholar]
- Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. II. Mitotic studies during regeneration, and a possible mechanism of blastema formation. J. Exp. Zool. 1976b;195:65–80. [Google Scholar]
- Baguñà J, Romero R. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–194. [Google Scholar]
- Bauer R, Voelzmann A, Breiden B, Schepers U, Farwanah H, Hahn I, Eckardt F, Sandhoff K, Hoch M. Schlank, a member of the ceramide synthase family controls growth and body fat in Drosophila. EMBO J. 2009;28:3706–3716. doi: 10.1038/emboj.2009.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg DA, Kirkham M, Wang H, Frisen J, Simon A. Dopamine controls neurogenesis in the adult salamander midbrain in homeostasis and during regeneration of dopamine neurons. Cell Stem Cell. 2011;8:426–433. doi: 10.1016/j.stem.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Bieberich E. Ceramide in stem cell differentiation and embryo development: novel functions of a topological cell-signaling lipid and the concept of ceramide compartments. J. Lipids. 2011;2011:610306. doi: 10.1155/2011/610306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich E. It’s a lipid’s world: bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochem. Res. 2012;37:1208–1229. doi: 10.1007/s11064-011-0698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen ID. Phagocytosis in Polycelis tenuis. In: Smith DC, Tiffon Y, editors. Nutrition in the Lower Metazoa. Pergamon Press; Oxford: 1980. pp. 1–14. [Google Scholar]
- Cebrià F. Regenerating the central nervous system: how easy for planarians. Dev Genes Evol. 2007;217:733–748. doi: 10.1007/s00427-007-0188-6. [DOI] [PubMed] [Google Scholar]
- Cebrià F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development. 2005;132:3691–3703. doi: 10.1242/dev.01941. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Ip YT. Pathogenic stimulation of intestinal stem cell response in Drosophila. J. Cell. Physiol. 2009;220:664–671. doi: 10.1002/jcp.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl. Acad. Sci. USA. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Poss KD. Cardiac regeneration. Curr. Topics Dev. Biol. 2012;100:319–344. doi: 10.1016/B978-0-12-387786-4.00010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Loomis Z, Pugh-Bernard A, Schrunk J, Doyle MJ, Minic A, McCoy E, Sussel L. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev. Biol. 2008;313:58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F. Contribution á l ’ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull. Biol. Fr. Belg. 1949;83:213–283. [Google Scholar]
- Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, Perret C, Shroyer NF, Romagnolo B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc. Natl. Acad. Sci. U S A. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell. 2008;3:327–339. doi: 10.1016/j.stem.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA, Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro A, Boj SF, Clevers H. Fishing for intestinal cancer models: unraveling gastrointestinal homeostasis and tumorigenesis in zebrafish. Zebrafish. 2009;6:361–376. doi: 10.1089/zeb.2009.0617. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Newmark PA. Emerging patterns in planarian regeneration. Curr. Opin. Genet. Dev. 2009;19:412–420. doi: 10.1016/j.gde.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ, Park AE, Newmark PA. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev. Biol. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried B, Grigo KL. Histochemical, thin-layer chromatographic, and histochromatographic analyses of neutral lipids in the planarian Dugesia dorotocephala (Turbellaria) Trans. Am. Microsc. Soc. 1977;96:530–532. [Google Scholar]
- Garcia-Corrales P, Gamo J. The ultrastructure of the gastrodermal gland cells in the freshwater planarian Dugesia gonocephala s.l. Acta Zool. 1986;67:43–51. [Google Scholar]
- Garcia-Corrales P, Gamo J. Ultrastructural changes in the gastrodermal phagocytic cells of the freshwater planarian Dugesia gonocephala s.l. during starvation. J. Submicrosc. Cytol. 1987;19:291–302. [Google Scholar]
- Garcia-Corrales P, Gamo J. Ultrastructural changes in the gastrodermal phagocytic cells of the planarian Dugesia gonocephala s.l. during food digestion (Plathelminthes) Zoomorphol. 1988;108:109–117. [Google Scholar]
- Garcia-Fernàndez J, Baguñà J, Salò E. Genomic organization and expression of the planarian homeobox genes Dth-1 and Dth-2. Development. 1993;118:241–253. doi: 10.1242/dev.118.1.241. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild CG. Reconstitution of the intestinal tract in the adult leopard frog, Rana pipiens Schreber. J. Exp. Zool. 1956;131:301–327. [Google Scholar]
- Grassme H, Riethmuller J, Gulbins E. Biological aspects of ceramide-enriched membrane domains. Prog. Lipid. Res. 2007;46:161–170. doi: 10.1016/j.plipres.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Gu Y, Forostyan T, Sabbadini R, Rosenblatt J. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J. Cell. Biol. 2011;193:667–676. doi: 10.1083/jcb.201010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O’Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Guo T, Peters AHFM, Newmark PA. A bruno-like gene is required for stem cell maintenance in planarians. Dev. Cell. 2006;11:159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A. β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 2008;319:323–327. doi: 10.1126/science.1150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 2006;48:371–380. doi: 10.1111/j.1440-169X.2006.00876.x. [DOI] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Hyman LH. The Acoelomate Bilateria. II. McGraw-Hill; New York: 1951. The Invertebrates: Platyhelminthes and Rhynchoceola. [Google Scholar]
- Inoue T, Hayashi T, Takechi K, Agata K. Clathrin-mediated endocytic signals are required for the regeneration of, as well as homeostasis in, the planarian CNS. Development. 2007;134:1679–1689. doi: 10.1242/dev.02835. [DOI] [PubMed] [Google Scholar]
- Ishii S. Electron microscopic observations on the Planarian tissues II. The intestine. Fukushima J. Med. Sci. 1965;12:67–87. [PubMed] [Google Scholar]
- Jennings JB. Studies on feeding, digestion, and food storage in free-living flatworms (Platyhelminthes: Turbellaria) Biol. Bull. 1957;112:63–80. [Google Scholar]
- Jiang H, Edgar BA. Intestinal stem cell function in Drosophila and mice. Curr. Opin. Genet. Dev. 2012;22:354–360. doi: 10.1016/j.gde.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko N, Katsuyama Y, Kawamura K, Fujiwara S. Regeneration of the gut requires retinoic acid in the budding ascidian Polyandrocarpa misakiensis. Dev. Growth Differ. 2010;52:457–468. doi: 10.1111/j.1440-169X.2010.01184.x. [DOI] [PubMed] [Google Scholar]
- Kim TH, Escudero S, Shivdasani RA. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. U S A. 2012;109:3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama-Koganeya A, Nabetani T, Miura M, Hirabayashi Y. Glucosylceramide synthase in the fat body controls energy metabolism in Drosophila. J. Lipid. Res. 2011;52:1392–1399. doi: 10.1194/jlr.M014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Reports. 2012;2:294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashanov VS, Garcia-Arraras JE. Gut regeneration in holothurians: a snapshot of recent developments. Biol. Bull. 2011;221:93–109. doi: 10.1086/BBLv221n1p93. [DOI] [PubMed] [Google Scholar]
- Miller CM, Newmark PA. An insulin-like peptide regulates size and adult stem cells in planarians. Int. J. Dev. Biol. 2012;56:75–82. doi: 10.1387/ijdb.113443cm. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Regeneration in planarians. Archiv. Entwick. Mech. Org. 1900;10:58–119. [Google Scholar]
- Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 2012;441:789–802. doi: 10.1042/BJ20111626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev. Biol. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A. Not your father’s planarian: a classic model enters the era of functional genomics. Nat. Rev. Genet. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- O’Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Steen WK. Regeneration of the intestine in adult urodeles. J. Morphol. 1958;103:435–477. [Google Scholar]
- Park MS, Takeda M. Starvation suppresses cell proliferation that rebounds after refeeding in the midgut of the American cockroach, Periplaneta americana. J. Insect Physiol. 2008;54:386–392. doi: 10.1016/j.jinsphys.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Pascolini R, Gargiulo AM. Ultrastructural modifications of cells in the intestine of Dugesia lugubris. Boll. Zool. 1975;42:123–131. [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 2009;238:443–450. doi: 10.1002/dvdy.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen KJ. Some observations on the fine structure of planarian protonephridia and gastrodermal phagocytes. Z. Zellforsch. 1961;53:609–628. [Google Scholar]
- Pellettieri J, Fitzgerald P, Watanabe S, Mancuso J, Green DR, Sánchez Alvarado A. Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 2009;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Rink JC, Vu HT, Alvarado AS. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–3780. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev. Biol. 2010;341:429–443. doi: 10.1016/j.ydbio.2010.02.037. [DOI] [PubMed] [Google Scholar]
- Saló E, Baguñà J. Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J. Embryol. Exp. Morphol. 1984;83:63–80. [PubMed] [Google Scholar]
- Saló E, Baguñà J. Cell movement in intact and regenerating planarians. Quantitation using chromosomal, nuclear and cytoplasmic markers. J. Embryol. Exp. Morphol. 1985;89:57–70. [PubMed] [Google Scholar]
- Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 2009;23:265–277. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- Schmid R, Blaxter ML. annot8r: GO, EC and KEGG annotation of EST datasets. BMC Bioinformatics. 2008;9:180. doi: 10.1186/1471-2105-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LC, Ben David O, Pewzner-Jung Y, Laviad EL, Stiban J, Bandyopadhyay S, Merrill AH, Jr., Prieto M, Futerman AH. Ablation of ceramide synthase 2 strongly affects biophysical properties of membranes. J. Lipid Res. 2012;53:430–436. doi: 10.1194/jlr.M022715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- Takeo M, Yoshida-Noro C, Tochinai S. Morphallactic regeneration as revealed by region-specific gene expression in the digestive tract of Enchytraeus japonensis (Oligochaeta, Annelida) Dev. Dyn. 2008;237:1284–1294. doi: 10.1002/dvdy.21518. [DOI] [PubMed] [Google Scholar]
- Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast lag1 (Lass) gene family (review) Int. J. Mol. Med. 2009;23:135–140. [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K. A planarian orthopedia homolog is specifically expressed in the branch region of both the mature and regenerating brain. Dev. Growth Differ. 1997;39:723–727. doi: 10.1046/j.1440-169x.1997.t01-5-00008.x. [DOI] [PubMed] [Google Scholar]
- van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Krishnamurthy K, Umapathy NS, Verin AD, Bieberich E. The carboxyl-terminal domain of atypical protein kinase Cζ binds to ceramide and regulates junction formation in epithelial cells. J Biol Chem. 2009a;284:14469–14475. doi: 10.1074/jbc.M808909200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Elghazi L, Parker SE, Kizilocak H, Asano M, Sussel L, Sosa-Pineda B. The concerted activities of Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell differentiation. Dev. Biol. 2004;266:178–189. doi: 10.1016/j.ydbio.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Wang YC, Gallego-Arteche E, Iezza G, Yuan X, Matli MR, Choo SP, Zuraek MB, Gogia R, Lynn FC, German MS, et al. Homeodomain transcription factor NKX2.2 functions in immature cells to control enteroendocrine differentiation and is expressed in gastrointestinal neuroendocrine tumors. Endocr. Relat. Cancer. 2009b;16:267–279. doi: 10.1677/ERC-08-0127. [DOI] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev. Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willier BH, Hyman LH, Rifenburgh SA. A histochemical study of intracellular digestion in triclad flatworms. J. Morph. Phys. 1925;40:299–340. [Google Scholar]
- Worgall TS. Sphingolipid synthetic pathways are major regulators of lipid homeostasis. Adv. Exp. Med. Biol. 2011;721:139–148. doi: 10.1007/978-1-4614-0650-1_9. [DOI] [PubMed] [Google Scholar]
- Xu F, Yang CC, Gomillion C, Burg KJ. Effect of ceramide on mesenchymal stem cell differentiation toward adipocytes. Appl. Biochem. Biotechnol. 2010;160:197–212. doi: 10.1007/s12010-008-8505-8. [DOI] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zattara EE, Bely AE. Evolution of a novel developmental trajectory: fission is distinct from regeneration in the annelid Pristina leidyi. Evol. Dev. 2011;13:80–95. doi: 10.1111/j.1525-142X.2010.00458.x. [DOI] [PubMed] [Google Scholar]
- Zayas RM, Cebrià F, Guo T, Feng J, Newmark PA. The use of lectins as markers for differentiated secretory cells in planarians. Dev. Dyn. 2010;239:2888–2897. doi: 10.1002/dvdy.22427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. USA. 2005;102:18491–18496. doi: 10.1073/pnas.0509507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.