Abstract
The global epidemic of type 2 diabetes mellitus (T2D) is one of the most challenging problems of the 21st century leading cause of and the fifth death worldwide. Substantial evidence suggests that T2D is a multifactorial disease with a strong genetic component. Recent genome-wide association studies (GWAS) have successfully identified and replicated nearly 75 susceptibility loci associated with T2D and related metabolic traits, mostly in Europeans, and some in African, and South Asian populations. The GWAS serve as a starting point for future genetic and functional studies since the mechanisms of action by which these associated loci influence disease is still unclear and it is difficult to predict potential implication of these findings in clinical settings. Despite extensive replication, no study has unequivocally demonstrated their clinical role in the disease management beyond progression to T2D from impaired glucose tolerance. However, these studies are revealing new molecular pathways underlying diabetes etiology, gene-environment interactions, epigenetic modifications, and gene function. This review highlights evolving progress made in the rapidly moving field of T2D genetics that is starting to unravel the pathophysiology of a complex phenotype and has potential to show clinical relevance in the near future.
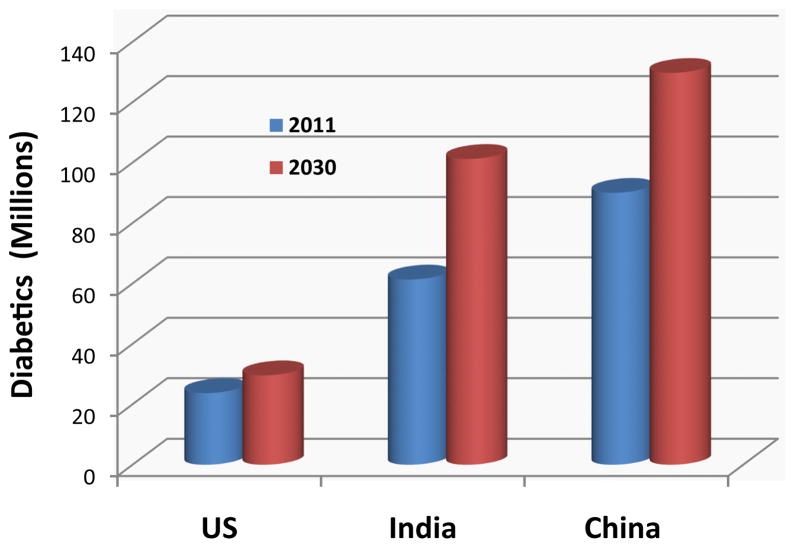
The global epidemic of type 2 diabetes (T2D) is a major public health problem of 21st century and the fifth leading cause of death worldwide [1]. The disease is also a leading cause of morbidity and contributes to development of premature coronary heart disease (CHD), stroke, peripheral vascular disease, renal failure, and amputation. According to latest statistics released by the International Diabetes Federation, the number of people living with diabetes is expected to rise from 366 million in 2011 to 552 million by 2030; 80% of these people with diabetes will live in developing countries (http://www.idf.org/diabetesatlas/5e/the-global-burden). According to these predictions, in three leading countries with diabetes populations USA, India, and China the approximate estimate of 23.7, 61.3 and 90 million people with diabetes in US, India, and China in 2011 will increase to 29.6, 101.2, 129.7 million by 2030 (Figure 1). The global health expenditure on diabetes is expected to increase from 376 billion dollars in 2010 to 490 billion in 2030 [2].
Figure 1.
It compares the prevalence of diabetes for the year 2011 to the projections for 2030 in USA, China, and India according to the latest projections by International Diabetes Federation (http://www.idf.org/diabetesatlas/5e/the-global-burden).
Substantial evidence suggests that T2D is a multifactorial disease with a strong genetic component. High concordance rate obtained in monozygotic twins (96%) supports a substantial contribution of genetic factors to T2D [3–7]. Furthermore, 40% of first-degree relatives of T2D patients develop diabetes as compared to 6% in the general population [8]. Segregation analysis also points to the polygenic nature of T2D, in addition to the existence of a major genetic component [9,10]. The general estimates of heritability (h2) of T2D is 0.49 and the relative recurrence risk for a sib of an affected person (λs) to develop T2D is 3.5 [11,12]. However, the findings and the results of recent genome-wide association studies (GWAS) have significantly underestimated these heritability estimates and presents a challenge for ongoing and future investigation.
Clinical Phenotype of T2D
Unlike simple characterization of type 1 diabetes (T1D) which is primarily due to autoimmune mediated destruction of pancreatic beta cells resulting in insulin deficiency, the pathogenesis of T2D is more complex and remains a matter of debate. Hyperglycemia in T2D is a consequence of complex interplay between insulin resistance (sensitivity) and abnormal insulin secretion [13]. It is initially characterized by compensatory insulin secretion associated with insulin resistance. However, the β-cell’s response is inadequate for the increased demand during progressive resistance to insulin-mediated glucose disposal, eventually resulting in β-cell failure and overt diabetes. There is also accumulating evidence that the β-cell is adversely influenced by influx of fatty acids [14] and cholesterol, which accumulates and exerts toxic effects when efflux by HDL is limited [15].
The progression of insulin resistance often associated with obesity and leading to mild glucose intolerance preceding an increase in glucose levels above defined thresholds for diabetes, has made it difficult to define the true phenotype, possibly because of early involvement of multiple tissues such as muscle, fat and pancreatic β-cells. Furthermore, the insulin resistant state, usually preceding diabetes by several years, is associated with a cluster of co-morbidities including obesity, dyslipidemias, and elevated blood pressure [16,17], and the presence of three or more traits is currently recognized as the metabolic syndrome which is also predictive of diabetes [18].
More than 80% of diabetic subjects are obese and these individuals typically have an android body type (upper body obesity) manifesting as an increased waist circumference [19]. Obesity, the metabolic syndrome, and T2D are becoming increasingly prevalent even in children and adolescents living in rapidly developing countries in different parts of world [20–24]. The prevalence of T2D in the US is higher among minorities and ethnic populations like African Americans, Native Americans, Hispanic Americans, Asian Americans, and Pacific Islanders than in the general population [25–28]. High prevalence is even seen in children belonging to ethnic minorities [29–32]. Although environmental factors play an important role in determining the risk of disease, overwhelming data support that genetic factors influence the disease susceptibility [33]. Lifestyle modifications (weight reduction and physical activity) delay or even prevent the development of T2D [34,35], such changes are exceedingly difficult to sustain outside the research setting and seem to have contributed little to control the diabetes epidemic [36].
T2D remains undiagnosed for many years because hyperglycemia is usually not severe enough to provoke noticeable symptoms, unlike T1D which often presents with keto-acidosis requiring admission to hospital for correction and initiation of treatment. But hyperglycemia can cause significant pathological and functional changes, which can cause organ damage before the diagnosis of T2D is made. The long term effects of diabetes include micro-vascular and macro-vascular complications and those who have genetic susceptibility are at greater risk [37]. Micro-vascular complications are progressive development of disease in fine capillaries supplying blood to the kidneys and retina of the eye that results in blindness. Macro-vascular complications include hypertension, coronary artery disease, peripheral vascular disease, cerebral vascular disease, and hyperlipidemia. Diabetic patients also have neuropathy, which may lead to foot ulcers, amputations, sexual dysfunction, and non-healing skin wounds. Certain infections such as staphylococcal sepsis is more common in diabetics and infections of the ear, nose, throat as well as reactivation of tuberculosis associated with high rate of mortality and morbidity are more likely with poor blood glucose control. Consequently, diabetes is a disproportionably expensive disease. The economic impact of diabetes in the US is enormous, and is expected to increase from $113 billion to $336 billion [38].
Linkage and Candidate Gene Association Studies
Progress in identifying the genetic basis of simple Mendelian (monogenic) diseases during the past decade has been substantial. More than 3000 monogenic disorders have been successfully mapped by linkage and family based studies (http://www.ncbi.nlm.nih.gov/omim). On the other hand, complex diseases like T2D do not segregate in simple Mendelian fashion but rather are affected by multiple genetic and environmental factors. Linkage and candidate-gene focused studies were successful in identifying some rare familial forms of T2D presenting at young ages called maturity onset diabetes of young (MODY), mitochondrial diabetes and neonatal diabetes. However, linkage and association studies on the common form of T2D provided inconsistent results and failed replication in multiple populations [39]. Only PPARG, KCNJ11 and TCF7L2 were identified as established genes associated with common forms of T2D [40]. Apparently, inconsistencies across populations were due to the heterogeneity of the disease itself or its pathogenesis, incorrect candidate selection because of incomplete knowledge of molecular mechanisms, variation in study design, sample size, population-specific linkage disequilibrium, choice, analytical methods, or over-interpretation of results [40–43].
Genome-Wide Association Studies (GWAS)
Completion of the Human Genome Project in 2003 [44] led to subsequent advances in biomedical research. Since 2007, a new technology in the form of ‘genome-wide chips’ has facilitated remarkable progress in T2D genetic research with the first publication of five large GWA scans within the span of four months, showing that more than 500,000 SNP markers distributed across the genome [45–49]. This approach has been successful in locating genes for other diseases besides T2D and obesity [40] namely, type 1 diabetes [50], prostate cancer [51], rheumatoid arthritis [52], Crohns disease [53,54], and cardiovascular disease [55] and is being applied to other complex disorders. Use of this ‘hypothesis-free’ approach involved in GWAS has opened new areas of biology to explore as discoveries of more than seventy entirely new T2D loci clearly suggest that associations are not limited to candidate genes and by applying GWAS and re-sequencing approaches, new genes involved in disease pathogenesis can be identified [56] (Table 1).
Table 1.
Summary of type 2 diabetes genes discovered in GWA studies (in order of gene region).
| SNP | Gene | Gene Region | Population | Trait | Risk Allele Freq in Controls | OR/Beta [95% CI] | P-Value | First Author. Journal; Published Year |
|---|---|---|---|---|---|---|---|---|
| rs10923931 | NOTCH2, ADAM30 | 1p12 | European | T2D | T[0.11] | 1.13[1.08–1.17] | 4×10-8 | Zeggini et al. Nat Genet; 2008 |
| rs243021 | BCL11A | 2p16.1 | European | T2D | A[NR] | 1.08[1.06–1.10] | 3×10-15 | Voight et al. Nat Genet; 2010 |
| rs7578326 | IRS1 | 2q36.3 | European | T2D | A[NR] | 1.11[1.08–1.13] | 5×10-20 | Voight et al. Nat Genet; 2010 |
| rs2943641 | LOC64673, IRS1 | 2q36.3 | European | T2D/Other traits | C[0.63] | 1.19[1.13–1.25] | 9×10-12 | Rung et al. Nat Genet; 2009 |
| rs7593730 | RBMS1, ITGB6 | 2q24.2 | European | T2D | ?[0.78] | 1.11[1.08–1.16] | 4×10-8 | Qi et al. Hum Mol Genet; 2010 |
| rs7578597 | THADA | 2p21 | European | T2D | T[0.9] | 1.15[1.10–1.20] | 1×10-9 | Zeggini et al. Nat Genet; 2008 |
| rs4607103 | ADAMTS9 | 3p14.1 | European | T2D | C[0.76] | 1.09[1.06–1.12] | 1×10-8 | Zeggini et al. Nat Genet; 2008 |
| rs4402960 | IGF2BP2 | 3q27.2 | European | T2D | T[0.3] | 1.14[1.11–1.18] | 9×10-16 | Scott et al. Science; 2007 |
| rs358806 | NR | 3p14 | European | T2D | ?[0.8] | 1.16[1.03–1.33] | 3×10-6 | WTCC Consortium Nature; 2007 |
| rs1801282 | PPARG | 3p25.2 | European | T2D | C[0.82] | 1.14[1.08–1.20] | 2×10-6 | Scott et al. Science; 2007 |
| rs17036101 | SYN2, PPARG | 3p25.2 | European | T2D | G[0.93] | 1.15[1.10–1.21] | 2×10-7 | Zeggini et al. Nat Genet; 2008 |
| rs1801214 | WFS1 | 4p16.1 | European | T2D | T[NR] | 1.13[1.08–1.18] | 3×10-8 | Voight et al. Nat Genet; 2010 |
| rs4689388 | WFS1, PPP2R2C | 4p16.1 | European | T2D/Other traits | T[0.57] | 1.16[1.10–1.21] | 1×10-8 | Rung et al. Nat Genet; 2009 |
| rs12518099 | LOC72901, CETN3 | 5q14.3 | European | T2D/Other traits | C[0.23] | 1.16[1.10–1.22] | 7×10-7 | Rung et al. Nat Genet; 2009 |
| rs4457053 | ZBED3 | 5q13.3 | European | T2D | G[NR] | 1.08[1.06–1.11] | 3×10-12 | Voight et al. Nat Genet; 2010 |
| rs7754840 | CDKAL1 | 6p22.3 | European | T2D | C[0.36] | 1.12[1.08–1.16] | 4×10-11 | Scott et al. Science; 2007 |
| rs896854 | TP53INP1 | 6q22.1 | European | T2D | T[NR] | 1.06[1.04–1.09] | 1×10-9 | Voight et al. Nat Genet; 2010 |
| rs9472138 | VEGFA | 6p21.1 | European | T2D | T[0.28] | 1.06[1.04–1.09] | 4×10-6 | Zeggini et al. Nat Genet; 2008 |
| rs864745 | JAZF1 | 7p15.1 | European | T2D | T[0.5] | 1.1[1.07–1.13] | 5×10-14 | Zeggini et al. Nat Genet; 2008 |
| rs972283 | KLF14 | 7q32.3 | European | T2D | G[NR] | 1.07[1.05–1.10] | 2×10-10 | Voight et al. Nat Genet; 2010 |
| rs13266634 | SLC30A8 | 8q24.11 | European | T2D | C[0.61] | 1.12[1.07–1.16] | 5×10-8 | Scott et al. Science; 2007 |
| rs564398 | CDKN2A, CD-KN2B | 9p21.3 | European | T2D | T[0.56] | 1.13[1.08–1.19] | 1×10-6 | Zeggini et al. Science; 2007 |
| rs7018475 | CDKN2B | 9p21.3 | European | T2D | ?[NR] | 1.35[1.18–1.56] | 3×10-8 | Huang J et al. Eur J Hum Genet; 2012 |
| rs13292136 | CHCHD9 | 9q21.31 | European | T2D | C[NR] | 1.11[1.07–1.15] | 3×10-8 | Voight et al. Nat Genet; 2010 |
| rs12779790 | CDC123, CAMK1D | 10p13 | European | T2D | G[0.18] | 1.11[1.07–1.14] | 1×10-10 | Zeggini et al. Nat Genet; 2008 |
| rs1111875 | HHEX | 10q23.33 | European | T2D | C[0.52] | 1.13[1.09–1.17] | 6×10-10 | Scott et al. Science; 2007 |
| rs7903146 | TCF7L2 | 10q25.2 | European | T2D | T[0.26] | 1.38[1.31–1.46] | 2×10-31 | Saxena et al. Science; 2007 |
| rs1552224 | ARAP1 | 11q13.4 | European | T2D | A[NR] | 1.14[1.11–1.17] | 1×10-22 | Voight et al. Nat Genet; 2010 |
| rs5219 | KCNJ11 | 11p15.1 | European | T2D | T[0.46] | 1.14[1.10–1.19] | 7×10-11 | Scott et al. Science; 2007 |
| rs231362 | KCNQ1 | 11p15.5 | European | T2D | G[NR] | 1.08[1.06–1.10] | 3×10-13 | Voight et al. Nat Genet; 2010 |
| rs1387153 | MTNR1B | 11q14.3 | European | T2D | T[NR] | 1.09[1.06–1.11] | 8×10-15 | Voight et al. Nat Genet; 2010 |
| rs1153188 | DCD | 12q13.2 | European | T2D | A[0.73] | 1.08[1.05–1.11] | 2×10-7 | Zeggini et al. Nat Genet; 2008 |
| rs1531343 | HMGA2 | 12q14.3 | European | T2D | C[NR] | 1.1[1.07–1.14] | 4×10-9 | Voight et al. Nat Genet; 2010 |
| rs7957197 | HNF1A | 12q24.31 | European | T2D | T[NR] | 1.07[1.05–1.10] | 2×10-8 | Voight et al. Nat Genet; 2010 |
| rs7961581 | TSPAN8, LGR5 | 12q21.1 | European | T2D | C[0.27] | 1.09[1.06–1.12] | 1×10-9 | Zeggini et al. Nat Genet; 2008 |
| rs8042680 | PRC1 | 15q26.1 | European | T2D | A[NR] | 1.07[1.05–1.09] | 2×10-10 | Voight et al. Nat Genet; 2010 |
| rs11634397 | ZFAND6 | 15q25.1 | European | T2D | G[NR] | 1.06[1.04–1.08] | 2×10-9 | Voight et al. Nat Genet; 2010 |
| rs8050136 | FTO | 16q12.2 | European | T2D | A[0.4] | 1.23[1.18–1.32] | 7×10-14 | Zeggini et al. Science; 2007 |
| rs4430796 | HNF1B | 17q12 | European | T2D | G[NR] | 1.14[1.08–1.20] | 2×10-6 | Voight et al. Nat Genet; 2010 |
| rs5945326 | DUSP9 | Xq26 | European | T2D | G[NR] | 1.27[1.18–1.37] | 3×10-10 | Voight et al. Nat Genet; 2010 |
| rs7542900 | SLC44A3, F3 | 1p21.3 | African Amer. | T2D | C[0.56] | 1.16[1.09–1.25] | 6×10-6 | Palmer ND et al. PLoS One; 2012 |
| rs7560163 | RBM43, RND3 | 2q23.3 | African Amer. | T2D | C[0.86] | 1.33[1.19–1.49] | 7×10-9 | Palmer ND et al. PLoS One; 2012 |
| rs2722769 | GALNTL4, LOC729013 | 11p15.3 | African Amer. | T2D | C[0.53] | 1.35[1.19–1.54] | 2×10-6 | Palmer ND et al. PLoS One; 2012 |
| rs7107217 | TMEM45B, BARX2 | 11q24.3 | African Amer. | T2D | C[0.91] | 1.18[1.10–1.27] | 3×10-7 | Palmer ND et al. PLoS One; 2012 |
| rs17045328 | CR2 | 1q32.2 | Asian | T2D | G[0.3] | 1.38[1.20–1.59] | 7×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs12027542 | PCNXL2 | 1q42.2 | Asian | T2D | A[0.61] | 1.41[1.23–1.61] | 4×10-7 | Sim X et al. PLoS Genet; 2011 |
| rs7630877 | PEX5L | 3q26.33 | Asian | T2D | A[0.17] | 1.32[1.17–1.49] | 7×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs3773506 | PLS1 | 3q23 | Asian | T2D | C[0.06] | 1.81[1.39–2.35] | 9×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs831571 | PSMD6 | 3p14.1 | Asian | T2D | c[0.61] | 1.09[1.06–1.12] | 8×10-11 | Cho YS et al. Nat Genet; 2011 |
| rs2063640 | ZPLD1 | 3q12.3 | Asian | T2D | A[0.17] | 1.23[1.13–1.34] | 3×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs6815464 | MAEA | 4p16.3 | Asian | T2D | C[0.58] | 1.13[1.10–1.16] | 2×10-20 | Cho YS et al. Nat Genet; 2011 |
| rs1048886 | C6orf57 | 6q13 | Asian | T2D | G[0.18] | 1.54[1.32–1.80] | 3×10-8 | Sim X et al. PLoS Genet; 2011 |
| rs1535500 | KCNK16 | 6p21.2 | Asian | T2D | T[0.42] | 1.08[1.05–1.11] | 2×10-8 | Cho YS et al. Nat Genet; 2011 |
| rs9470794 | ZFAND3 | 6p21.2 | Asian | T2D | C[0.27] | 1.12[1.08–1.16] | 2×10-10 | Cho YS et al. Nat Genet; 2011 |
| rs7636 | ACHE | 7q22.1 | Asian | T2D | A[0.06] | 1.85[1.42–2.41] | 5×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs6467136 | GCC1, PAX4 | 7q32.1 | Asian | T2D | G[0.79] | 1.11[1.07–1.14] | 5×10-11 | Cho YS et al. Nat Genet; 2011 |
| rs7041847 | GLIS3 | 9p24.2 | Asian | T2D | A[0.41] | 1.1[1.07–1.13] | 2×10-14 | Cho YS et al. Nat Genet; 2011 |
| rs17584499 | PTPRD | 9p24.1 | Asian | T2D | T[0.06] | 1.57[1.36–1.82] | 9×10-10 | Tsai et al. PLoS Genet; 2010 |
| rs6583826 | KIF11 | 10q23.33 | Asian | T2D | G[0.26] | 1.18[1.10–1.27] | 7×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs10741243 | TCERG1L | 10q26.3 | Asian | T2D | G[0.93] | 1.75[1.38–2.23] | 5×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs1359790 | SPRY2 | 13q31.1 | Asian | T2D | G[0.71] | 1.15[1.10–1.20] | 6×10-9 | Shu et al. PLoS Genet; 2010 |
| rs1436955 | C2CD4B | 15q22.2 | Asian | T2D | C[0.73] | 1.13[1.08–1.19] | 7×10-7 | Shu et al. PLoS Genet; 2010 |
| rs16955379 | CMIP | 16q23.2 | Asian | T2D | C[0.8] | 1.08[1.05–1.12] | 3×10-7 | Cho YS et al. Nat Genet; 2011 |
| rs17797882 | WWOX | 16q23.2 | Asian | T2D | T[0.32] | 1.08[1.05–1.12] | 9×10-7 | Cho YS et al. Nat Genet; 2011 |
| rs391300 | SRR | 17p13.3 | Asian | T2D | G[0.62] | 1.28[1.18–1.39] | 3×10-9 | Tsai et al. PLoS Genet; 2010 |
| rs10460009 | LPIN2 | 18p11.31 | Asian | T2D | C[0.6] | 1.35[1.18–1.54] | 9×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs472265 | PAPL | 19q13.2 | Asian | T2D | G[0.22] | 1.39[1.20–1.61] | 9×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs3786897 | PEPD | 19q13.11 | Asian | T2D | A[0.56] | 1.1[1.07–1.14] | 1×10-8 | Cho YS et al. Nat Genet; 2011 |
| rs2833610 | HUNK | 21q22.11 | Asian | T2D | A[0.57] | 1.17[1.09–1.24] | 4×10-6 | Sim X et al. PLoS Genet; 2011 |
| rs3923113 | GRB14 | 2q24.3 | South Asian | T2D | A[0.74] | 1.09[1.06–1.13] | 1×10-8 | Kooner JS et al. Nat Genet; 2011 |
| rs16861329 | ST6GAL1 | 3q27.3 | South Asian | T2D | G[0.75] | 1.09[1.06–1.12] | 3×10-8 | Kooner JS et al. Nat Genet; 2011 |
| rs1802295 | VPS26A | 10q22.1 | South Asian | T2D | A[0.26] | 1.08[1.05–1.12] | 4×10-8 | Kooner JS et al. Nat Genet; 2011 |
| rs2028299 | AP3S2 | 15q26.1 | South Asian | T2D | C[0.31] | 1.1[1.07–1.13] | 2×10-11 | Kooner JS et al. Nat Genet; 2011 |
| rs7178572 | HMG20A | 15q24.3 | South Asian | T2D | G[0.52] | 1.09[1.06–1.12] | 7×10-11 | Kooner JS et al. Nat Genet; 2011 |
| rs4812829 | HNF4A | 20q13.12 | South Asian | T2D | A[0.29] | 1.09[1.06–1.12] | 3×10-10 | Kooner JS et al. Nat Genet; 2011 |
| rs2237897 | KCNQ1 | 11p15.4 | Japanese | T2D | C[0.61] | 1.33[1.24–1.41] | 1×10-16 | Unoki et al. Nat Genet; 2008 |
| rs2237892 | KCNQ1 | 11p15.5 | Japanese | T2D | C[0.61] | 1.4[1.34–1.47] | 2×10-42 | Yasuda et al. Nat Genet; 2008 |
| rs7172432 | C2CD4A, C2CD4B | 15q22.2 | Japanese | T2D | ?[0.58] | 1.11[1.08–1.14] | 9×10-14 | Yamauchi et al. Nat Genet; 2010 |
| rs730570 | C14orf70 | 14q32.2 | Mexican | T2D | G[NR] | 1.14[1.08–1.21] | 8×10-6 | Parra EJ et al. Diabetologia; 2011 |
The number of risk alleles for complex diseases identified by GWAS since 2007 exceeded those identified in the entire preceding decade, and these studies can effectively detect multiple common variants with small effects with odds ratios (ORs) <1.2 [36,39,57] but offers limited power to detect rare variants with stronger effects [58]. However, GWAS have the best chance of detecting and identifying additional genes for T2D and are being pursued in different population groups, to provide a more realistic ‘genetic risk landscape’ of the disease [59], and account for variation in population-specific environmental interactions. Notably, most immediate advances from GWA studies are not disease prediction or prevention, rather these studies are providing a better understanding of the disease pathophysiology since further studies can lead to defining the role of the newly identified genes in key metabolic pathways involved in pathogenesis. Also GWAS, together with targeted resequencing of the identified loci allows recovery of a more complete inventory of the sequence variation leading to identification of novel metabolic pathways and proteins as potential targets for interventions [57]. Since substantial numbers, maybe thousands, of genes with small effects are operative in human disease and can be identified by this approach, their combined effect may be substantial [56]. Furthermore, gene-gene interaction may not only be additive but effects of a mutation can be more than doubled when compounding mutations are present. The phenomenon known as epistasis was first shown to occur in a rodent knockout model in which the insulin receptor when combined with IRS-1 knockout resulted in 40% increase in diabetes onset compared to isolated knock-outs [60]. The phenomenon could contribute to effect size and in part account for the low heritability accounted for by classic GWAS. It follows that multi locus association testing could be effective in the presence of epistasis. With this in mind the Welcome Trust Case Control Consortium used two-locus tests of association and detected epistatic signaling of association at 79 SNP pairs [61]. However the large number of possible associations in GWAS requires filtration strategies based on knowledge of the biochemistry and known interacting pathways [62]. However, several models for analysis have been proposed and consensus on the optimal methodology has not been reached [63].
The common strategy used for elucidating the inherited components of complex disease is based on common disease-common variant hypothesis [64,65]. The hypothesis has held true in the case of putative causal variants in APOE, including APOEε4, association with Alzheimer’s disease, interleukin 23 receptor association with Crohns disease (the at-risk allele has a frequency 93% in the general population), and PPARG2 association with T2D (at-risk (pro) allele frequency ~85% in general population). The assumption with the use of common variants is that either they will identify the association with the common disease, or a quantitative trait (QT) directly, or indirectly being in linkage disequilibrium (LD) with a functional variant. Most of the T2D loci identified using GWAS approach are common variants with small effects. The first GWAS in T2D was published by a French group of investigators who identified a zinc transporter and member of solute carrier family SLC30A8 and HHEX along with confirming the association of TCF7L2 and KCNJ11 with T2D [49]. Three additional GWAS in their jointly published findings confirmed TCF7L2, KCNJ11, PPARG, SLC30A8, and HHEX. They also discovered novel T2D loci called CD-KAL1 and a variant near CDKN2A-B [47,48,66]. These findings were also simultaneously confirmed by GWAS performed on an Icelandic sample by the deCODE researchers [67]. Association of FTO with T2D was also first discovered by GWAS for obesity [OR for T2D being 1.27, p < 10−8] [68] and was later confirmed in replication studies [46].
Additionally, a joint meta-analysis of three previously conducted T2D GWA scans by the Diabetes Genetics Replication And Meta-analysis (DIAGRAM) consortium [69] has detected an additional six novel loci with strong evidence of association; JAZF1, CDC123, TSPAN8, THADA, ADAMTS9, and NOTCH2 [69] in 10,128 individuals using 2.2 million SNPs. Also, another European study reported a strong association of an intronic SNP (rs560887) in G6PC2 with fasting plasma glucose levels by screening 392,935 SNPs in 654 non-T2D participants using GWAS [70]. Association of G6PC2 with fasting plasma glucose levels was replicated in the FUSION study [71]. Further, a variant near MTNR1B has been identified to increase FBG levels in two separate GWA studies [72,73]. Association of MTNR1B variants (rs10830963 and rs1387153) with FBG or T2D in our Punjabi cohort from India has not been confirmed. Instead a less common variant on 5′ UTR (rs1374645) of MTNR1B revealed a strong association with low FBG levels in normoglycemic individuals with low BMI (<25 kg/m2). Furthermore, our data showed strong interaction of this variant with BMI with respect to FBG levels [74]. Most recently, a meta-analysis was performed on 21 GWAS comprising 46,186 participants to identify loci influencing glycemic traits. They performed follow-up studies on 25 loci in up to 76,558 additional subjects and identified nine loci associated with fasting glucose and HOMA-B (ADCY5, MADD, ADRA2A, CRY2, FADS1, GLIS3, SLC2A2, PROX1 and C2CD4B) and one influencing fasting insulin and HOMA-IR (IGF1) in European populations [75]. Another study analyzed combined GWAS data from 8,130 individuals with T2D and 38,987 controls of European descent, and in follow-up meta-analysis found signals in a further 34,412 cases and 59,925 controls and identified 12 new T2D associations including new signals at KCNQ1, KLF14, DUSP9, CENTD2, HMGA2, HNF1A and PRC1 [76].
Genetic Diversity among Populations
The majority of GWAS so far (96%) have been predominantly performed in Caucasians [77,78]. Given the existence of a strong genetic diversity among the world communities due to biological traits, cultural histories, languages, caste system, physical appearance, food habits etc., the information from GWA findings on Caucasians should not be transferred and used to predict risk in other populations [79,80]. Even the largest and best executed GWAS is unlikely to provide a complete assessment of “the genetic risk landscape” of the disease [59]. These loci neither account for all cases of T2D, nor do they sufficiently account for the variation in quantitative sub-phenotypes of T2D. Extending the use of new methodologies such as GWA to study non-European ancestries with different mutational spectra and demographic and cultural histories is important for identifying the population-specific patterns for allele frequency and LD relationship of the susceptibility loci, and population-specific environmental factors for disease risk or protection [57]. Several GWAS in other ethnic groups are currently ongoing and will be capable of revealing novel susceptibility loci. GWAS have proven more successful in small homogenous populations like Finland, Iceland and Costa Rica, and have potential to detect regionally common variants which may be missed in outbred populations [78]. It is of interest to note that many of the common loci originally associated with diabetes in European populations have not been replicated in other non-European populations [49,67,77,81]. For example, loci associated with Crohns disease in Caucasian GWAS were not associated in Japanese [81], also the association of CDKAL1 locus with T2D was not seen in African Americans [67]. As discussed earlier, the common intron 1 FTO variant (rs9939609) was initially associated with diabetes in Europeans, and phenotypic interactions appear to be diabetogenic, but the associations were due to obesity [68,69]. However, the question has been explored in independent association and meta-analyses of South Asian populations in whom BMI and waist association with FTO is similar to that seen in Europeans, but a strong association with diabetes is only partly accounted for by BMI [82,83]. A recent large scale meta-analysis conducted on 96,551 individuals from East and South Asia confirmed that the association of rs9939609 with T2D was independent of obesity [84].
There is now a strong consensus that a single population is not sufficient to uncover all the variants underlying complex diseases [77]. Recent GWAS studies in non-European populations have yielded intriguing new variants, for instance, the KCNQ1 signal (first noticed in Japanese) provided insight into novel etiological pathways leading to insulin resistance and T2D. Joint meta-analysis of Caucasian GWA studies later discovered a second independent signal at the KCNQ1 locus (rs231362) and an overlap between loci implicated in monogenic and multifactorial forms of diabetes at HNF1A [76]. The new Caucasian signal at KCNQ1 was later confirmed in our study on Asian Indians [85]. These findings provide new insights into the pathophysiology of this extremely complex disease by discovering previously unknown variants and often replicating the findings but not always. Association of UBE2E2 with T2D was first described in East Asians [86]. Recent larger meta-analysis using East Asian GWAS cohorts yielded eight new loci for T2D in (GLIS3, PEPD, FITM2-R3HDML-HNF4A, KCNK16, MAEA, GCC1-PAX4, PSMD6 and ZFAND3) [87]. Six novel loci for T2D (GRB14, ST6GAL1, VPS26A, HMG20A, AP3S2 and HNF4A) were first discovered in our South Asian population GWAS by Kooner et al. [88]. To date, GWAS and meta-analyses in T2D and related quatitative traits have together identified approximately 75 susceptibility variants which together explain about 10% of the observed familial aggregation (Table 1). Although these effect sizes detected so far do not reflect biological or clinical significance of these variants, they highlight a particular genomic region likely to be associated with the trait. Of all the complex traits studied by GWAS, Crohns disease, has so far had the strongest association with genetic components of risk; up to 20% being explained by variants detected by GWAS [89]. Furthermore, GWAS provided important biological insight with direct clinical relevance adding to significant understanding of the disease compared to what was known about Crohns disease five years back.
Hidden Heritability and Rare Variant Hypothesis
Despite these successes, results of multiple GWAS and meta-analysis studies (based on the common disease-common variant hypothesis [64,65]) show that common variants discovered in these studies explain only a small fraction with odds ratios (ORs) ranging from 1.1–1.2 (p values <10−8) of the heritable component of T2D risk, and these studies have not pinpointed with certainty the causal variants at the associated loci [90]. Importantly, commercial arrays used in GWAS have captured much less than 60% of common SNP information in the majority of the GWAS conducted to date [91]. One approach could be to expand the sample sizes to detect more variants of moderate effects, however, the GWAS approach is usually underpowered to locate those variants with a frequency of <5% [57], and is considered unlikely to yield more information on the heritable control of disease than what has already been revealed by GWAS [92]. Moreover, association studies, based on the common disease-common variant hypothesis would have less relevance if the disease is caused by less common mutations at multiple sites and each arising on a different ancestral haplotype [93].
Many studies have suggested the large role of rare variants in complex diseases [93,94], and that the genes carrying common variants with small effects also are likely to carry rare variants with large effects [69,90] which can only be detected by targeted resequencing [57,95]. Also, several independently and dominantly acting rare variants from different genes, together can confer a detectable relative risk for a given “common disease-rare variant” hypothesis [96]. These low frequency variants with stronger penetrance would be expected if trait alleles have undergone purifying selection [94]. Resequencing of certain candidate genes have shown that certain rare variants can be identified in individuals with extreme phenotypes [97]. These rare variants when clustered together can produce a significant genetic effect in a complex disease [56]. For instance, rare functional recessive mutations contribute to HDL and triglyceride levels [98,99], suggesting the possibility of discovering such variants in a common disease such as T2D [100]. Fearhead and colleagues [101] detected several rare functional variants in five genes among 124 subjects with multiple adenomatous polyps showing combined ORs of 2.2; p = 0.0001. Furthermore, 25% of these individuals had a single rare variant and none of them had two or more. Their findings suggested that mainly missense and possibly also nonsense, promoter, and splicing variants can collectively explain a significant portion of inherited susceptibility to colorectal adenomas. It is realistic to expect similar findings in other chronic diseases [94,96] including diabetes. It is also important to note that rare variants are often founder variants caused by genetic drift and can be population-specific and less cosmopolitan. For instance, the rare variants discovered in the APC gene for colorectal adenomas in Ashkenazi Jews were not present in Koreans [102]. These findings therefore suggest that the search for genetic variation in diabetes-susceptible populations should be carried out in multiple ethnicities in carefully chosen candidate genes, and in clearly phenotyped disease groups. Further use of whole genome rare variants from the 1000 Genomes Project (www.1000genomes.org) and T2D-GENES Consortium with data from multiple ethnic groups will substantially improve power of these studies and capture heritability associated with rare variants from specific regions or even from the other regions of the genome, especially those (deletions/insertions, duplications, inversions) that have escaped detection in GWAS [103]. Additionally, imputation from 1000 Genomes Project will allow discovery of new association signals from the missing “dark matter”. It will also assist in post-GWAS fine mapping and functional characterization of known associations and enhance our ability to fine map signals of local adaptation as has been successfully achieved for refining the locus for smoking quantity at 15q25 (P = 9.4 × 10−19) using imputed data from 1000 Genomes which increased marker density to five-fold compared to HapMap2 [104].
In addition to major gene mutations and SNPs, changes in the normal genome sequence can result in significant miscoding with disease association [105]. Submicroscopic deletions, duplications, insertions, inversions and translocations can occur in a small DNA segment and are known as copy number variations (CNV) [106]. Since methodology for CNV detection is relatively new [107], few studies have looked for CNV associations with T2D. Since copy number variation in the leptin receptor gene (LEPR) is associated with T2D [108], it is possible that genome-wide detection of CNV could improve association with T2D as shown for insulin resistance in the HyperGEN study, in which joint tests of association for SNPs and CNVs were performed resulting in detection of association in the T-cell receptor gene (TCRVB) in African Americans [109]. Furthermore, generation of CNV maps [110] supports potential for studying CNV for missing variation in T2D and possibly accounting for the inter-ethnic variation found among diabetes-prone populations [111]; however, common CNVs may be tagged by SNPs and have been indirectly explored in previous SNP-based studies [112]. Further, the event resulting in a CNV may also disrupt regional genes and their regulatory elements accounting for disease association [106].
Role of Epigenetics in Missing Heritability
The assumption of SNP-based GWAS has been that genomic sequence variations account for disease phenotypes, however recent developments in the field of epigenetics has shown altered expression of genes that could lead to enhanced prediction of obesity and diabetes [113]. The modifications include DNA methylation and histone modifications such as ubiquitination, adenosine diphosphate ribosylation, phosphorylation, acetylation and methylation. Since the modifications are often observed during cellular growth and differentiation, it follows that the changes may influence embryological development of cells involved in carbohydrate homeostasis such as the β-cells, adipocytes and myocytes. For example, DNA methylation occurs in the leptin gene promotor and in GLUT4 in 3T3-L1 cells [114]. The epigenetic mechanism mediate the influence of the environment on gene expression [115]. For instance, leptin plays an important role in programming of obesity-related traits [116,117], change in expression could be involved in the pathogenesis of T2D, in part supporting epidemiological findings that fetal growth and early nutrition in childhood influence adult disease incl, HNF1B (rs 4430796 uding T2D [118]. For example, increased methylation of PDX-1 results in decreased expression in pancreatic islets from patients with T2D suggesting that modification of PDX-1 plays a role in T2D pathogenesis [119]. Besides, epigenetic modifications are sensitive to change by nutrients, hormones and toxins that can operate during developmental windows according to tissue-specific and sex-specific mechanisms [120], which potentially can be reversed with treatment and prevention [121,122].
Gene-Environment Interaction
Evidence from the epidemiology of T2D overwhelmingly supports a strong environmental influence interacting with genetic predisposition in a synergistic fashion as has been recently reviewed [123], however current state-of-the-art methods for measuring environmental effects lack precision and can result in changes in statistical power to detect interaction [123,124]. Since lifestyle factors are important in preventing diabetes [125,126], interaction of gene variants with measures of dietary intake and exercise have been selected for studies on gene-environment interaction. For example, HNF1B (rs 4430796) was shown to interact with exercise; low levels of activity enhanced the risk of T2D in association with absence of the risk allele, but there was no protective effect of exercise when the allele was present. It follows that subgrouping by genotype may serve to enhance risk prediction while considering gene-environment interaction as has been done for exercise [127]. Also lifestyle including exercise modified the effect of a CDKN2A/B variant on 2-hour glucose levels in the Diabetes Prevention Program [128] but was not confirmed in the HERITAGE study using different measurements and phenotypes involving insulin sensitivity and β-cell function [129]. The pro12ala PPARG variant also interacts with physical activity for effect on 2-hour glucose levels [130], which was confirmed in the smaller HERITAGE study [129]. In addition, a relationship of dietary fat intake with plasma insulin and BMI differs by the pro12ala PPARG genotype [131].
It is possible that there are genes that because of their known metabolic involvement are likely to interact with specific nutrients. For example, SLC30A8 which encodes a zinc transporter localized in secretory granules, interacted with dietary zinc to effect fasting insulin levels [132]. However, the majority of GWAS variants have not shown interaction with environmental factors for effect on diabetes or related traits. Therefore, it is likely that prospective future studies will utilize improved assessment methods to increase power and avoid false interpretation [133,134]. This could be enhanced by prioritizing variants that are most likely to have effects [135] or selective sampling according to extremes of the environmental factor could reduce the requirement for sample size [136]. These and other strategies such as meta-analysis, nested case control and genotype-based studies have been recently reviewed [123,133] and the difficulties in measuring environmental exposures have been emphasized, including the application of analyses based on logistic regression [124] and problems with instruments such as physical activity questionnaires [137]. Validated food frequency questionnaires are popular instruments for evaluation diabetes risk and are often used in conjunction with food analysis software [138,139]. Similar methodology has been adapted to assess two predominant food consumption patterns by Prudent and Western [140], and demonstrated synergistic interaction with genotype and a less healthy Western dietary pattern in determining male risk for T2D by showing that the gene-diet interaction was higher in men with a high genetic risk score determined by a gene counting method [141]. Also the effects of diet may predominate at specific developmental periods [142] suggesting that age and associated physiological changes are important as well as differences between genders. It has also been observed that homogeneity of an environmental factor such as physical activity in an Asian Indian study, may reduce ability to detect interaction, but could be solved by subgrouping by the level of activity [143], but increased recruitment would be needed to maintain power.
It is also speculated that several rapidly developing communities lived through centuries of famine and starvation and over the years have adapted to survival under conditions of unreliable food supply because they have “thrifty genes” which enable efficient storage of energy during periods of food deprivation [144]. The genes that once represented a survival advantage, now, in the modern environment with continuous low physical activity and dietary excess, result in obesity and T2D [145,146]. In addition, early protein deprivation, as indicated by low birth weight is considered as an important contributor to the high prevalence of T2D especially in people living in the under-developed world [147–150] possibly when followed by rapid catch-up growth in infancy and early childhood [151]. The “thrifty phenotype” hypothesis was based on data showing a positive association of low birth weight with cardiovascular disease and diabetes in adulthood [152], but the metabolic basis of gestational programming of the fetus is poorly understood. In Europeans, the risk of T2D increases with increase in birth weight whereas in Pima Indians and in South Asians T2D rates increase with both low and high birth weight [153] suggesting that the effective birth weight distribution is bimodal. Poor intra-uterine nutrition also may be associated with abnormal β-cell development and differentiation [154–156]. Therefore, it is imperative to develop a deeper understanding of genetic, epigenetic, environmental, cultural, and social factors that may interact to cause progression to T2D, and are responsible for the declining health of the fastest developing nations of the third world.
Prospects of Translation of Genetic Findings into Clinical Practice
As discussed in more detail in previous sections, investigations on genetic determinants of T2D have progressed substantially following increased characterization of the genome and use of GWAS, however a large portion of the heritability is unaccounted for, and many of the genes commonly found in GWAS have small effects [157–160]. Thus, state-of-the-art clinical use of genotyping is currently restricted to investigation. Also the use SNPs to predict disease onset or response to treatment is limited but can be done with some monogenic forms of diabetes [160]. Furthermore, metabolic pathways that have been responsive to treatment with pharmaceutical agents or exercise, or have been associated with disease have rarely contained genes that have been identified by large scale studies such as GWAS.
However, pharmacogenetic studies on association with the effects of anti-diabetes drugs have provided a significant clinical link with epidemiological findings [161], and frequently used agents in the treatment of T2D such as metformin, sulfonylureas, and thiozolidinediones have been shown to have gene associations. The principal metformin-associated pathway involves adenosine-monophosphate-activated kinase (AMPK), a regulator of glucose levels [162], and therefore has potential as a drug target [163] for the control of T2D by metformin and possibly other agents. It serves as a sensor of cellular energy status via its phosphorylation and activation. Liver AMPK controls hepatic glucose production by inhibiting expression of phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase) and this action is down-regulated by the drug, metformin. Candidate genes encoding enzymes, cofactors and transcription factors involved in AMPK-associated pathways have potential for studies on therapeutic responses. Since gene variants coding for steps in metabolic pathways involving metformin’s action may lead to defining treatment responses, investigators in the United Kingdom conducted a meta-analysis using the glycemic response to metformin as the phenotype. A variant at a locus containing ATM, the ataxia telangiectasia mutated gene (rs11212617) was associated with the response [164]. The importance of the variant was emphasized by showing that specific inhibition of ATM attenuated the phosphorylation and activation of AMPK in response to metformin in a rat hepatoma cell line [164]. Although the effect attributed to the variant was small, the study identified an important upstream AMPK regulator adding information on how metformin works, and in addition identified a possible mechanistic link with DNA repair and cancer prevention. Shu et al. have investigated polymorphisms in SLC22A1 which encodes organic cation transporter 1(OCT-1) which functions to facilitate absorption of metformin into hepatocytes and identified association with reduced responsiveness to metformin’s effect on glucose levels during an oral glucose tolerance test [165]. Also association with HbA1c was shown with a variant in SL-C47A1 encoding the multidrug and toxin extrusion protein 1 involved in the excretion of metformin into bile and urine [166]. Furthermore, the two genes both acting to increase intracellular metformin, have an interactive effect [167]. The roles of both genes were supported by the Diabetes Prevention Program investigators [168] who confifrmed association of the glucose-lowering response to metformin and variants in SLC47A1, and interactions with SLC22A1 and the AMPK genes STK11, PRKAA1, and PRKAA2.
Genes encoding the receptor and pathways for PPARγ agonists constituting the thiozolidinediones, are often required for glucose control because of their role in promoting insulin sensitivity. Of 133 PPARG variants tested in the TRIPOD study, eight showed evidence for association with response to troglitazone therapy defined as change in tolerance to intravenous glucose (IVGTT) [169] but no association with fasting glucose suggesting that the response shown in the IVGTT represents insulin resistance whereas fasting glucose is more likely to be a measure of β-cell failure.
In studies where overt T2D has been the phenotype the majority of associated polymorphisms have encoded proteins known to be involved in β-cell metabolism; for example TCF7L2, KCNJ11 and HHEX have shown robust association [170,171]. This suggests that these genes could prove useful in predicting β-cell preservation during the course of T2D. The glucokinase gene (GCK) coding for the initial glucose-sensing step in the β-cell can have activating mutations causing hypoglycemia that might provide structural and functional models leading to drug targets for treating T2D [172]. In the GoDARTs study, investigators examined the medication response of metformin and sulphonylurea based on the TCF7L2 variants mainly affecting the β-cell. The carriers of the at risk ‘T’ allele responded less well to sulphonylurea therapy than metformin [173]. Also it is of significant public health interest that in the Diabetes Prevention Program, lifestyle modifications were shown to reduce the risk of diabetes conferred by risk variants of TCF7L2 at rs7093146, and in placebo participants who carried the homozygous risk genotype (TT), there was 80% higher risk for developing diabetes compared to the lifestyle intervention group carrying the same risk genotypes [35]. These findings could herald significant future progress in the field of T2D pharmacogenomics, possibly leading to the development and use of agents tailored on the basis of genotype.
Predictive Power of Genetics beyond Traditional T2D Risk Factors
Traditional clinical assessment includes a complete history of the presenting traits including family history of the traits, and followed by laboratory studies that present either as continuous variables or dichotomous values according to a cut-point. From this basic clinical information it has been possible to derive risk scores that predict T2D such as the Finnish Diabetes Risk Score and the Diabetes Risk Calculator [174,175], which have been used to assess predictive ability using receiver operating characteristic curves or the area under the curve (AUC) as an index of discriminative accuracy. Using these scores prediction has only been considered to be modest [176], leading to the question whether genetic risk prediction might be superior. Studies have been based on genetic variants alone and in combination with environmental factors, but studies have differed in the number and choice of genes and environmental factors. However, disappointingly the genetic risk models had lower prediction capacity than the clinical environment-based models [176]. Differences in populations, BMI, age and duration of follow-up may offer some explanation [176,177], but improving genetic methodologies may be the main issue needing to be addressed.
Conclusions
The escalating current and predicted world-wide prevalence of T2D underscores the need to develop effective strategy for early detection, prevention and targeted treatments. In this regard the field of genetic epidemiology has potential to progress to effective translation to patient care possibly within the next decade [178]. Current state-of-the-art approaches that may lead to this goal include defining the pathogenesis of T2D and the phasic nature of the pre-diabetes phenotype beginning in utero and progressing to overt T2D associated with β-cell failure. Thus far, genes identified by GWAS using diagnosed T2D as the phenotype have mainly been associated with β-cell failure but have only accounted for less than 5% of the heritability. Although the greater than 60% heritability appears clinically obvious, environmental interactions, biochemical DNA modifications (epigenetic), and the combined effect of rare variants and other genes present potential challenges in defining risk association. However, the contribution of GWAS over the past decade should not be minimized because of low contribution to heritability, since the genes showing association have been pointers to key metabolic pathways involved in pathogenesis and in the action of drugs used for T2D treatment. Future functional studies on model organisms or cell lines will enable functional characterization of identified risk loci and will provide better understanding of the biological mechanisms of disease development. Improved SNP- and expression-array technology, sequencing, and targeted re-sequencing are likely to enhance current understanding and progress. In addition, inclusion of “at risk” populations in the developing world is likely to improve understanding of gene-environment interaction and is more likely to lead to detection of rare variants and novel variants that associate with unique phenotypes and disease associations.
Acknowledgments
This work was partly supported by NIH grants -R01DK082766 funded by the National Institute of Diabetes and Digestive and Kidney Diseases and NOT-HG-11-009 funded by National Genome Research Institute, USA. Superb technical assistance provided by Latonya Been in manuscript preparation is duly acknowledged.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Zhang P, Zhang X, Brown J, Vistisen D, Sicree R, et al. Global health-care expenditure on diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:293–301. doi: 10.1016/j.diabres.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Barnett AH, Eff C, Leslie RD, Pyke DA. Diabetes in identical twins. A study of 200 pairs. Diabetologia. 1981;20:87–93. doi: 10.1007/BF00262007. [DOI] [PubMed] [Google Scholar]
- 4.Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, et al. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763–768. doi: 10.1007/BF00275741. [DOI] [PubMed] [Google Scholar]
- 5.Kaprio J, Tuomilehto J, Koskenvuo M, Romanov K, Reunanen A, et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992;35:1060–1067. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 6.Lo SS, Tun RY, Hawa M, Leslie RD. Studies of diabetic twins. Diabetes Metab Rev. 1991;7:223–238. doi: 10.1002/dmr.5610070403. [DOI] [PubMed] [Google Scholar]
- 7.Medici F, Hawa M, Ianari A, Pyke DA, Leslie RD. Concordance rate for type II diabetes mellitus in monozygotic twins: actuarial analysis. Diabetologia. 1999;42:146–150. doi: 10.1007/s001250051132. [DOI] [PubMed] [Google Scholar]
- 8.Kobberling JTH. Emperical risk figures of first degree relatives of non-insulin dependent diabetes. London: Academic Press; 1982. [Google Scholar]
- 9.Ferrell RE, Iyengar Sudha. Molecular studies of the genetics of non-insulin-dependent diabetes mellitus. Am J Hum Biol. 1993;5:415–424. doi: 10.1002/ajhb.1310050406. [DOI] [PubMed] [Google Scholar]
- 10.Stern MP, Mitchell BD, Blangero J, Reinhart L, Krammerer CM, et al. Evidence for a major gene for type II diabetes and linkage analyses with selected candidate genes in Mexican-Americans. Diabetes. 1996;45:563–568. doi: 10.2337/diab.45.5.563. [DOI] [PubMed] [Google Scholar]
- 11.Risch N. Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am J Hum Genet. 1990;46:242–253. [PMC free article] [PubMed] [Google Scholar]
- 12.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 13.DeFronzo RA. Pathogenesis of type 2 diabetes mellitus. Med Clin North Am. 2004;88:787–835. doi: 10.1016/j.mcna.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Newsholme P, Keane D, Welters HJ, Morgan NG. Life and death decisions of the pancreatic beta-cell: the role of fatty acids. Clin Sci (Lond) 2007;112:27–42. doi: 10.1042/CS20060115. [DOI] [PubMed] [Google Scholar]
- 15.Kruit JK, Brunham LR, Verchere CB, Hayden MR. HDL and LDL cholesterol significantly influence beta-cell function in type 2 diabetes mellitus. Curr opin lipidol. 2010;21:178–185. doi: 10.1097/MOL.0b013e328339387b. [DOI] [PubMed] [Google Scholar]
- 16.Haffner SM, Stern MP, Hazuda HP, Mitchell BD, Patterson JK. Cardiovascular risk factors in confirmed prediabetic individuals. Does the clock for coronary heart disease start ticking before the onset of clinical diabetes? JAMA. 1990;263:2893–2898. doi: 10.1001/jama.263.21.2893. [DOI] [PubMed] [Google Scholar]
- 17.Ford ES, Li C, Sattar N. Metabolic syndrome and incident diabetes: current state of the evidence. Diabetes Care. 2008;31:1898–1904. doi: 10.2337/dc08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ford ES, Giles WH, Mokdad AH. Increasing prevalence of the metabolic syndrome among u.s. Adults. Diabetes Care. 2004;27:2444–2449. doi: 10.2337/diacare.27.10.2444. [DOI] [PubMed] [Google Scholar]
- 19.Nyamdorj R, Qiao Q, Lam TH, Tuomilehto J, Ho SY, et al. BMI compared with central obesity indicators in relation to diabetes and hypertension in Asians. Obesity (Silver Spring) 2008;16:1622–1635. doi: 10.1038/oby.2008.73. [DOI] [PubMed] [Google Scholar]
- 20.Danaraj TJ, Acker MS, Danaraj W, Wong HO, Tan BY. Ethnic group differences in coronary heart disease in Singapore: an analysis of necropsy records. Am Heart J. 1959;58:516–526. doi: 10.1016/0002-8703(59)90085-7. [DOI] [PubMed] [Google Scholar]
- 21.Enas EA, Garg A, Davidson MA, Nair VM, Huet BA, et al. Coronary heart disease and its risk factors in first-generation immigrant Asian Indians to the United States of America. Indian Heart J. 1996;48:343–353. [PubMed] [Google Scholar]
- 22.McKeigue PM, Marmot MG, Syndercombe Court YD, Cottier DE, Rahman S, et al. Diabetes, hyperinsulinaemia, and coronary risk factors in Bangladeshis in east London. Br Heart J. 1988;60:390–396. doi: 10.1136/hrt.60.5.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller GJ, Beckles GL, Alexis SD, Byam NT, Price SG. Serum lipoproteins and susceptibility of men of Indian descent to coronary heart disease. The St James Survey, Trinidad. Lancet. 1982;2:200–203. doi: 10.1016/s0140-6736(82)91041-8. [DOI] [PubMed] [Google Scholar]
- 24.Tuomilehto J, Ram P, Eseroma R, Taylor R, Zimmet P. Cardiovascular diseases and diabetes mellitus in Fiji: analysis of mortality, morbidity and risk factors. Bull World Health Organ. 1984;62:133–143. [PMC free article] [PubMed] [Google Scholar]
- 25.Sundquist J, Winkleby MA, Pudaric S. Cardiovascular disease risk factors among older black, Mexican-American, and white women and men: an analysis of NHANES III, 1988–1994. Third National Health and Nutrition Examination Survey. J Am Geriatr Soc. 2001;49:109–116. doi: 10.1046/j.1532-5415.2001.49030.x. [DOI] [PubMed] [Google Scholar]
- 26.International Diabetes Federation: Diabetes Prevalence. 2001 http://www.idf.org/idf/burlp/buefx_art_a_forecast.htm.
- 27.Centers for Disease Control and Prevention. Atlanta, Georgia: Centers for Disease Control and Prevention; 2001. CDC at a Glance Diabetes: A Serious Public Health Problem. [Google Scholar]
- 28.Mokdad AH, Bowman BA, Ford ES, Vinicor F, Marks JS, et al. The continuing epidemics of obesity and diabetes in the United States. JAMA. 2001;286:1195–1200. doi: 10.1001/jama.286.10.1195. [DOI] [PubMed] [Google Scholar]
- 29.Cockram CJC. Diabetes threat on the rise among US children Chron Dis Notes Reports. Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Centers for Disease Control and Prevention; Atlanta, Georgia: 1999. pp. 10–12. [Google Scholar]
- 30.Mokdad AH, Serdula MK, Dietz WH, Bowman BA, Marks JS, et al. The spread of the obesity epidemic in the United States, 1991–1998. JAMA. 1999;282:1519–1522. doi: 10.1001/jama.282.16.1519. [DOI] [PubMed] [Google Scholar]
- 31.Clinical Guidelines on the Identification, Evaluation and Treatment of Overweight and Obesity in Adults--The Evidence Report. National Institutes of Health. Obes Res. 1998;2:51S–209S. [PubMed] [Google Scholar]
- 32.Fagot-Campagna A, Pettitt DJ, Engelgau MM, Burrows NR, Geiss LS, et al. Type 2 diabetes among North American. children and adolescents: an epidemiologic review and a public health perspective. J Pediatr. 2000;136:664–672. doi: 10.1067/mpd.2000.105141. [DOI] [PubMed] [Google Scholar]
- 33.Zimmet P. Type 2 (non-insulin-dependent) diabetes--an epidemiological overview. Diabetologia. 1982;22:399–411. doi: 10.1007/BF00282581. [DOI] [PubMed] [Google Scholar]
- 34.Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hämäläinen H, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 35.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy MI. Progress in defining the molecular basis of type 2 diabetes mellitus through susceptibility-gene identification. Hum Mol Genet. 2004;13:R33–R41. doi: 10.1093/hmg/ddh057. [DOI] [PubMed] [Google Scholar]
- 37.Doria A. Genetics of diabetes complications. Curr Diab Rep. 2010;10:467–475. doi: 10.1007/s11892-010-0147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang ES, Basu A, O’Grady M, Capretta JC. Projecting the future diabetes population size and related costs for the U.S. Diabetes Care. 2009;32:2225–2229. doi: 10.2337/dc09-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barroso I. Genetics of Type 2 diabetes. Diabet Med. 2005;22:517–535. doi: 10.1111/j.1464-5491.2005.01550.x. [DOI] [PubMed] [Google Scholar]
- 40.Zeggini E. A new era for Type 2 diabetes genetics. Diabet Med. 2007;24:1181–1186. doi: 10.1111/j.1464-5491.2007.02274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barroso I, Luan J, Middelberg RP, Harding AH, Franks PW, et al. Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol. 2003;1:E20. doi: 10.1371/journal.pbio.0000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh S, Schork NJ. Genetic analysis of NIDDM. The study of quantitative traits. Diabetes. 1996;45:1–14. doi: 10.2337/diab.45.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Elbein SC. The genetics of human noninsulin-dependent (type 2) diabetes mellitus. J Nutr. 1997;127:1891S–1896S. doi: 10.1093/jn/127.9.1891S. [DOI] [PubMed] [Google Scholar]
- 44.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 45.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 46.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 48.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 50.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 52.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petretto E, Liu ET, Aitman TJ. A gene harvest revealing the archeology and complexity of human disease. Nat Genet. 2007;39:1299–1301. doi: 10.1038/ng1107-1299. [DOI] [PubMed] [Google Scholar]
- 57.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 58.McCarthy MI. Genomics, type 2 diabetes, and obesity. N Engl J Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 59.Genomics of common diseases. Nature genetics. 2007;39:569. [Google Scholar]
- 60.Bruning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, et al. Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell. 1997;88:561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 61.Bell JT, Timpson NJ, Rayner NW, Zeggini E, Frayling TM, et al. Genome-wide association scan allowing for epistasis in type 2 diabetes. Ann Hum Genet. 2011;75:10–19. doi: 10.1111/j.1469-1809.2010.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann Hum Genet. 2011;75:172–182. doi: 10.1111/j.1469-1809.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKinney BA, Pajewski NM. Six Degrees of Epistasis: Statistical Network Models for GWAS. Front Genet. 2011;2:109. doi: 10.3389/fgene.2011.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 65.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 66.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 68.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- 71.Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118:2620–2628. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparsø T, Holmkvist J, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 73.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet. 2009;41:82–88. doi: 10.1038/ng.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Been LF, Hatfield JL, Shankar A, Aston CE, Ralhan S, et al. A low frequency variant within the GWAS locus of MTNR1B affects fasting glucose concentrations: Genetic risk is modulated by obesity. Nutr Metab Cardiovasc Dis. 2011 doi: 10.1016/j.numecd.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, et al. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475:163–165. doi: 10.1038/475163a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, et al. Deep common ancestry of indian and western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331–1334. doi: 10.1016/s0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- 80.RoyChoudhury A. The genetics of Indian populations. Science and Culture. 1979;45:87–91. [Google Scholar]
- 81.Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, et al. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 82.Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, et al. Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO risk. BMC variants confer a significant. Med Genet. 2008;9:59. doi: 10.1186/1471-2350-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rees SD, Islam M, Hydrie MZ, Chaudhary B, Bellary S, et al. An FTO variant is associated with Type 2 diabetes in South Asian populations after accounting for body mass index and waist circumference. Diabetic Med. 2011;28:673–680. doi: 10.1111/j.1464-5491.2011.03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Kilpelainen TO, Liu C, Zhu J, Liu Y, et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55:981–995. doi: 10.1007/s00125-011-2370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Been LF, Ralhan S, Wander GS, Mehra NK, Singh J, et al. Variants in KCNQ1 increase type II diabetes susceptibility in South Asians: a study of 3,310 subjects from India and the US. BMC Med Genet. 2011;12:18. doi: 10.1186/1471-2350-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 87.Cho YS, Chen CH, Hu C, Long J, Ong RT, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44:67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McCarthy MI, Hirschhorn JN. Genome-wide association studies: past, present and future. Hum Mol Genet. 2008;17:R100–R101. doi: 10.1093/hmg/ddn298. [DOI] [PubMed] [Google Scholar]
- 90.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17:R156–R165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature. 1000;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 93.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant … or not? Hum Mol Genet. 2002;11:2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 94.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bodmer W. Familial adenomatous polyposis (FAP) and its gene, APC. Cytogenet Cell Genet. 1999;86:99–104. doi: 10.1159/000015360. [DOI] [PubMed] [Google Scholar]
- 97.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 99.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fearnhead NS, Winney B, Bodmer WF. Rare variant hypothesis for multifactorial inheritance: susceptibility to colorectal adenomas as a model. Cell Cycle. 2005;4:521–525. doi: 10.4161/cc.4.4.1591. [DOI] [PubMed] [Google Scholar]
- 102.Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, et al. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci U S A. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 104.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, et al. Meta-analysis and imputation refines. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Almal SH, Padh H. Implications of gene copy-number variation in health and diseases. J Hum Genet. 2012;57:6–13. doi: 10.1038/jhg.2011.108. [DOI] [PubMed] [Google Scholar]
- 106.Lupski JR. Structural variation in the human genome. N Engl J Med. 2007;356:1169–1171. doi: 10.1056/NEJMcibr067658. [DOI] [PubMed] [Google Scholar]
- 107.Dhawan D, Padh H. Pharmacogenetics: technologies to detect copy number variations. Curr Opin Mol Ther. 2009;11:670–680. [PubMed] [Google Scholar]
- 108.Jeon JP, Shim SM, Nam HY, Ryu GM, Hong EJ, et al. Copy number variation at leptin receptor gene locus associated with metabolic traits and the risk of type 2 diabetes mellitus. BMC Genomics. 2010;11:426. doi: 10.1186/1471-2164-11-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Irvin MR, Wineinger NE, Rice TK, Pajewski NM, Kabagambe EK, et al. Genome-wide detection of allele specific copy number variation associated with insulin resistance in African Americans from the HyperGEN study. PLoS One. 2011;6:e24052. doi: 10.1371/journal.pone.0024052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 111.Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet. 2010;77:201–213. doi: 10.1111/j.1399-0004.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 112.Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713–720. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Slomko H, Heo HJ, Einstein FH. Minireview: Epigenetics of obesity and diabetes in humans. Endocrinology. 2012;153:1025–1030. doi: 10.1210/en.2011-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yokomori N, Tawata M, Onaya T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. Diabetes. 1999;48:685–690. doi: 10.2337/diabetes.48.4.685. [DOI] [PubMed] [Google Scholar]
- 115.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cottrell EC, Ozanne SE. Early life programming of obesity and metabolic disease. Physiol Behav. 2008;94:17–28. doi: 10.1016/j.physbeh.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 117.Cottrell EC, Ozanne SE. Developmental programming of energy balance and the metabolic syndrome. Proc Nutr Soc. 2007;66:198–206. doi: 10.1017/S0029665107005447. [DOI] [PubMed] [Google Scholar]
- 118.Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- 119.Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, et al. Increased DNA Methylation and Decreased Expression of PDX-1 in Pancreatic Islets from Patients with Type 2 Diabetes. Mol Endocrinol. 2012 doi: 10.1210/me.2012-1004. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Attig L, Gabory A, Junien C. Early nutrition and epigenetic programming: chasing shadows. Curr Opin Clin Nutr Metab Care. 2010;13:284–293. doi: 10.1097/MCO.0b013e328338aa61. [DOI] [PubMed] [Google Scholar]
- 121.Szyf M, McGowan P, Meaney MJ. The social environment and the epigenome. Environ Mol Mutagen. 2008;49:46–60. doi: 10.1002/em.20357. [DOI] [PubMed] [Google Scholar]
- 122.Szyf M. The early-life social environment and DNA methylation. Clin Genet. 2012;81:341–349. doi: 10.1111/j.1399-0004.2012.01843.x. [DOI] [PubMed] [Google Scholar]
- 123.Cornelis MC, Hu FB. Gene-Environment Interactions in the Development of Type 2 Diabetes: Recent Progress and Continuing Challenges. Annu Rev Nutr. 2012 doi: 10.1146/annurev-nutr-071811-150648. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 124.Cornelis MC, Tchetgen EJ, Liang L, Qi L, Chatterjee N, et al. Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. Am J Epidemiol. 2012;175:191–202. doi: 10.1093/aje/kwr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Knowler WC, Narayan KM. Prevention of non-insulin-dependent diabetes mellitus. Prev Med. 1994;23:701–703. doi: 10.1006/pmed.1994.1118. [DOI] [PubMed] [Google Scholar]
- 126.Knowler WC, Fowler SE, Hamman RF, Christophi CA, Hoffman HJ, et al. 10-year follow-up of diabetes incidence and weight loss in the Diabetes Prevention Program Outcomes Study. Lancet. 2009;374:1677–1686. doi: 10.1016/S0140-6736(09)61457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brito EC, Lyssenko V, Renstrom F, Berglund G, Nilsson PM, et al. Previously associated type 2 diabetes variants may interact with physical activity to modify the risk of impaired glucose regulation and type 2 diabetes: a study of 16,003 Swedish adults. Diabetes. 2009;58:1411–1418. doi: 10.2337/db08-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, et al. Extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes. 2008;57:2503–2510. doi: 10.2337/db08-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ruchat SM, Rankinen T, Weisnagel SJ, Rice T, Rao DC, et al. Improvements in glucose homeostasis in response to regular exercise are influenced by the PPARG Pro12Ala variant: results from the HERITAGE Family Study. Diabetologia. 2010;53:679–689. doi: 10.1007/s00125-009-1630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lindi VI, Uusitupa MI, Lindstrom J, Louheranta A, Eriksson JG, et al. Association of the Pro12Ala polymorphism in the PPAR-gamma2 gene with 3-year incidence of type 2 diabetes and body weight change in the Finnish Diabetes Prevention Study. Diabetes. 2002;51:2581–2586. doi: 10.2337/diabetes.51.8.2581. [DOI] [PubMed] [Google Scholar]
- 131.Memisoglu A, Hu FB, Hankinson SE, Manson JE, De Vivo I, et al. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12:2923–2929. doi: 10.1093/hmg/ddg318. [DOI] [PubMed] [Google Scholar]
- 132.Kanoni S, Nettleton JA, Hivert MF, Ye Z, van Rooij FJ, et al. Total zinc intake may modify the glucose-raising effect of a zinc transporter (SLC30A8) variant: a 14-cohort meta-analysis. Diabetes. 2011;60:2407–2416. doi: 10.2337/db11-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Franks PW. Gene x environment interactions in type 2 diabetes. Curr Diab Rep. 2011;11:552–561. doi: 10.1007/s11892-011-0224-9. [DOI] [PubMed] [Google Scholar]
- 134.Franks PW, Nettleton JA. Invited commentary: Gene X lifestyle interactions and complex disease traits--inferring cause and effect from observational data, sine qua non. Am J Epidemiol. 2010;172:992–997. doi: 10.1093/aje/kwq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pare G, Cook NR, Ridker PM, Chasman DI. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women’s Genome Health Study. PLoS Genet. 2010;6:e1000981. doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boks MP, Schipper M, Schubart CD, Sommer IE, Kahn RS, et al. Investigating gene environment interaction in complex diseases: increasing power by selective sampling for environmental exposure. Int J Epidemiol. 2007;36:1363–1369. doi: 10.1093/ije/dym215. [DOI] [PubMed] [Google Scholar]
- 137.Vanhees L, Lefevre J, Philippaerts R, Martens M, Huygens W, et al. How to assess physical activity? How to assess physical fitness? Eur J Cardiovasc Prev Rehabil. 2005;12:102–114. doi: 10.1097/01.hjr.0000161551.73095.9c. [DOI] [PubMed] [Google Scholar]
- 138.de Koning L, Fung TT, Liao X, Chiuve SE, Rimm EB, et al. Low-carbohydrate diet scores and risk of type 2 diabetes in men. Am J Clin Nutr. 2011;93:844–850. doi: 10.3945/ajcn.110.004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pan A, Sun Q, Bernstein AM, Schulze MB, Manson JE, et al. Red meat consumption and risk of type 2 diabetes: 3 cohorts of US adults and an updated meta-analysis. Am J Clin Nutr. 2011;94:1088–1096. doi: 10.3945/ajcn.111.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hu FB, Rimm EB, Stampfer MJ, Ascherio A, Spiegelman D, et al. Prospective study of major dietary patterns and risk of coronary heart disease in men. Am J Clin Nutr. 2000;72:912–921. doi: 10.1093/ajcn/72.4.912. [DOI] [PubMed] [Google Scholar]
- 141.Qi L, Cornelis MC, Zhang C, van Dam RM, Hu FB. Genetic predisposition, Western dietary pattern, and the risk of type 2 diabetes in men. Am J Clin Nutr. 2009;89:1453–1458. doi: 10.3945/ajcn.2008.27249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Moore SC, Gunter MJ, Daniel CR, Reddy KS, George PS, et al. Common Genetic Variants and Central Adiposity Among Asian-Indians. Obesity (Silver Spring) 2011 doi: 10.1038/oby.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 145.McKeigue PM, Shah B, Marmot MG. Relation of central obesity and insulin resistance with high diabetes prevalence and cardiovascular risk in South Asians. Lancet. 1991;337:382–386. doi: 10.1016/0140-6736(91)91164-p. [DOI] [PubMed] [Google Scholar]
- 146.Joeff B, Zimmet P. The thrifty genotype in type 2 diabetes: an unfinished symphony moving to its finale? Endocrine. 1998;9:139–141. doi: 10.1385/ENDO:9:2:139. [DOI] [PubMed] [Google Scholar]
- 147.Simmons D. Ethnic comparisons in diabetes and insulin levels. Asia Pacific J Clin Nutr. 1995;4:346–348. [PubMed] [Google Scholar]
- 148.Simmons D, Williams DR, Powell MJ. Prevalence of diabetes in a predominantly Asian community: preliminary findings of the Coventry diabetes study. BMJ. 1989;298:18–21. doi: 10.1136/bmj.298.6665.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, et al. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36:62–67. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- 150.Yajnik CS, Fall CH, Vaidya U, Pandit AN, Bavdekar A, et al. Fetal growth and glucose and insulin metabolism in four-year-old Indian children. Diabet Med. 1995;12:330–336. doi: 10.1111/j.1464-5491.1995.tb00487.x. [DOI] [PubMed] [Google Scholar]
- 151.Ibanez L, Ong K, Dunger DB, de Zegher F. Early development of adiposity and insulin resistance after catch-up weight gain in small-for-gestational-age children. J Clin Endocrinol Metab. 2006;91:2153–2158. doi: 10.1210/jc.2005-2778. [DOI] [PubMed] [Google Scholar]
- 152.Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 153.Pettitt DJ, Jovanovic L. Birth weight as a predictor of type 2 diabetes mellitus: the U-shaped curve. Curr Diab Rep. 2001;1:78–81. doi: 10.1007/s11892-001-0014-x. [DOI] [PubMed] [Google Scholar]
- 154.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 156.Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 158.Sookoian S, Pirola CJ. Metabolic syndrome: from the genetics to the pathophysiology. Curr Hypertens Rep. 2011;13:149–157. doi: 10.1007/s11906-010-0164-9. [DOI] [PubMed] [Google Scholar]
- 159.Florez JC. Genetic susceptibility to type 2 diabetes and implications for therapy. J Diabetes Sci Technol. 2009;3:690–696. doi: 10.1177/193229680900300413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008;8:186–200. doi: 10.1016/j.cmet.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Distefano JK, Watanabe RM. Pharmacogenetics of Anti-Diabetes Drugs. Pharmaceuticals (Basel) 2010;3:2610–2646. doi: 10.3390/ph3082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008;4:S7–12. doi: 10.1038/ijo.2008.116. [DOI] [PubMed] [Google Scholar]
- 163.Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–416. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 164.Zhou K, Bellenguez C, Spencer CC, Bennett AJ, Coleman RL, et al. Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet. 2011;43:117–120. doi: 10.1038/ng.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shu Y, Sheardown SA, Brown C, Owen RP, Zhang S, et al. Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J Clin Invest. 2007;117:1422–1431. doi: 10.1172/JCI30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, et al. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose-lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes. 2009;58:745–749. doi: 10.2337/db08-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Becker ML, Visser LE, van Schaik RH, Hofman A, Uitterlinden AG, et al. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet Genomics. 2010;20:38–44. doi: 10.1097/FPC.0b013e328333bb11. [DOI] [PubMed] [Google Scholar]
- 168.Jablonski KA, McAteer JB, de Bakker PI, Franks PW, Pollin TI, et al. Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes. 2010;59:2672–2681. doi: 10.2337/db10-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wolford JK, Yeatts KA, Dhanjal SK, Black MH, Xiang AH, et al. Sequence variation in PPARG may underlie differential response to troglitazone. Diabetes. 2005;54:3319–3325. doi: 10.2337/diabetes.54.11.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 171.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 172.Gloyn AL, Noordam K, Willemsen MA, Ellard S, Lam WW, et al. Insights into the biochemical and genetic basis of glucokinase activation from naturally occurring hypoglycemia mutations. Diabetes. 2003;52:2433–2440. doi: 10.2337/diabetes.52.9.2433. [DOI] [PubMed] [Google Scholar]
- 173.Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes. 2007;56:2178–2182. doi: 10.2337/db07-0440. [DOI] [PubMed] [Google Scholar]
- 174.Saaristo T, Peltonen M, Lindstrom J, Saarikoski L, Sundvall J, et al. Cross-sectional evaluation of the Finnish Diabetes Risk Score: a tool to identify undetected type 2 diabetes, abnormal glucose tolerance and metabolic syndrome. Diab Vasc Dis Res. 2005;2:67–72. doi: 10.3132/dvdr.2005.011. [DOI] [PubMed] [Google Scholar]
- 175.Heikes KE, Eddy DM, Arondekar B, Schlessinger L. Diabetes Risk Calculator: a simple tool for detecting undiagnosed diabetes and pre-diabetes. Diabetes Care. 2008;31:1040–1045. doi: 10.2337/dc07-1150. [DOI] [PubMed] [Google Scholar]
- 176.Willems SM, Mihaescu R, Sijbrands EJ, van Duijn CM, Janssens AC. A methodological perspective on genetic risk prediction studies in type 2 diabetes: recommendations for future research. Curr Diab Rep. 2011;11:511–518. doi: 10.1007/s11892-011-0235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359:2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 178.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]