Abstract
p300 is a transcriptional coactivator that participates in many important processes in the cell, including proliferation, differentiation and apoptosis. The viral oncoproteins, adenovirus (Ad) E1A and human papillomavirus (HPV) E7, have been implicated in binding to p300. The Ad-E1A/p300 interaction has been shown to result in an induction of cellular proliferation, epigenetic reprogramming, as well as cellular transformation and cancer. The HPV-E7/p300 interaction, on the other hand, is not well understood. p300 contains three zinc-binding domains, CH1-CH3, and studies have shown that Ad-E1A can bind to the p300 CH1 and CH3 domains, whereas E7 can bind to the CH1 domain and to a lesser extent to the CH2 and CH3 domains. Here we address how high risk HPV16-E7 and Ad5-E1A, which have different structures, can both bind the p300 CH1 domain. Using pull down, gel filtration, and analytical ultracentrifugation studies, we show that the N-terminus and CR1 domains of Ad5-E1A and that the CR1-CR2 domains of HPV16-E7 bind to the p300 CH1 domain competitively and with mid- nanomolar and low micromolar dissociation constants, respectively. We also show that Ad5-E1A can form a ternary complex with the p300 CH1 domain and the retinoblastoma pRb transcriptional repressor, whereas HPV16-E7 cannot. These studies suggest that the HPV16-E7 and Ad5-E1A viral oncoproteins bind to the same p300 CH1 domain to disrupt p300 function by distinct mechanisms.
Keywords: p300, adenovirus, human papillomavirus, zinc-binding domain
INTRODUCTION
p300 is a versatile transcriptional coactivator that participates in many important processes in the cell, including proliferation, differentiation and apoptosis (1–3). It promotes gene transcription by acting as a protein bridge, or scaffold, that connects different transcription factors to the basic transcriptional apparatus (3, 4). This allows for the assembly of multi-component transcription coactivator complexes (5, 6). It also has histone acetyltransferase (HAT) activity which allows it to influence chromatin structure by modulating nucleosomal histones (7). It has also been shown to acetylate tumor suppressor transcription factors, such as p53 and pRb, thereby regulating protein function (8, 9). Therefore, p300 is involved in multiple, signal-dependent transcription events and its deregulation is implicated in many types of diseases (1, 10, 11).
Interestingly, p300 was originally identified through its specific interaction with E1A from adenovirus (12). Ad-E1A has been shown to hijack the cellular transcription machinery by competing with essential transcription factors for binding to p300 as well as its paralog, CREB-binding protein (CBP), thereby inactivating a number of cellular and viral promoters and enhancers (13, 14). The Ad-E1A/p300 complex has been shown to stimulate cellular proliferation, whereas mutants of E1A that cannot bind p300 are defective in cellular transformation (15). Some of these effects are also mediated by the ability of Ad-E1A to interact with the retinoblastoma protein, pRb (16, 17). Ad-E1A/p300 interactions have also been implicated in epigenetic reprogramming, leading to cellular transformation (14). The formation of viral oncoprotein complexes with p300 has also been indicated to cause a loss of cell growth control and a block of cellular differentiation, leading to cancers (15). The E7 protein from high risk human papillomavirus 16 (HPV 16), which is functionally homologous to Ad-E1A, but structurally dissimilar, has also been shown to bind to p300 (18). However, the downstream implications of this interaction are not well understood. HPV 16 E6, the other viral oncoprotein from this virus, has also been shown to bind to and inhibit the functions of p300/CBP and inhibit p53-dependent transcription (19, 20). Furthermore, this has only been observed for high risk E6 proteins that are associated with cancerous lesions.
The viral oncoproteins E1A and E7 have been reported to bind to the transactivation zinc-finger (TAZ) domains of p300 (13, 18). p300 contains two TAZ domains, called TAZ1 and TAZ2, that contain two cysteine/histidine-rich regions, called CH1 and CH3, respectively and whose primary function is protein recognition (5). Currently, more than thirty transcription factors have been found to bind to the TAZ domains (21). The sequences of the CH1 and CH3 domains are structurally homologous, but bind different proteins (21, 22). Interestingly, HPV16-E7 has been suggested to bind most prominently to the CH1 domain and to a lesser extent to the CH2 (another putative p300 zinc-finger domain) and CH3 domains, whereas Ad5-E1A was suggested to bind to the CH1 and CH3 domains of p300 (13, 18, 23). The interaction between Ad5-E1A and the CH1 domain, however, is not well understood. Since these viral oncoproteins bind to regions of p300 that are known to bind many transcription factors, these interactions with E7 or E1A can have many negative downstream effects in the cell.
Even though HPV16-E7 and Ad5-E1A are functionally homologous, their sequences and structures differ greatly. Both of these viral oncoproteins contain highly conserved regions CR1, CR2 and CR3, however, Ad-E1A shares limited sequence homology with HPV-E7, mainly found within the strictly conserved LxCxE motif of their CR2 domains (24, 25). This motif has been shown to mediate high-affinity binding to pRb and the related pocket proteins p107 and p130 (26). The CR1 domains of these oncoproteins have some sequence similaritiy, however, the long stretches of amino acids flanking the CR1 domain of Ad-E1A have no sequence homology to HPV-E7. Interestingly, the CR1 domains appear to have different binding partners: whereas the CR1 domain of Ad-E1A can mediate binding to pRb, HPV-E7 cannot (27). The CR3 domains are zinc-binding domains, however, Ad-E1A is a monomer, while HPV-E7 forms an obligate zinc-homodimer (28, 29).
Since HPV16-E7 and Ad5-E1A are functionally homologous and structurally diverse, we were interested to biochemically compare their binding to the p300 CH1 domain. To do this, we mapped the domains of the viral oncoproteins that were required for p300CH1 binding and quantified their interactions using analytical ultracentrifugation sedimentation equilibrium studies. The dissociation constants of the Ad5-E1A/p300CH1 and HPV16-E7/p300CH1 were found to be in the mid-nanomolar and low micromolar range, respectively, indicative of strong interactions. Furthermore, we were able to show that these viral oncoproteins bind competitively to p300CH1, suggesting that they may bind to the same site on p300CH1. We also show that Ad5-E1A can form a ternary complex with the p300 CH1 domain and the retinoblastoma pRb transcriptional repressor, whereas HPV16-E7 cannot. These studies suggest that the HPV16-E7 and Ad5-E1A viral oncoproteins may bind to a common surface on p300CH1 to disrupt p300 function by distinct mechanisms.
EXPERIMENTAL PROCEDURES
Expression and Purification of Proteins
GST-tagged E7 and E1A proteins
The DNA encoding Ad5-E1A1–77, Ad5-E1A1–138, HPV16-E71–98 (full length), HPV16-E717–98, HPV16-E746–98, HPV16-E71–51, HPV16-E71–46, HPV16-E717–46, HPV16-E71–17, and HPV1A-E71–93 (full length) were cloned into the pGEX-4T-1 vector, containing an N-terminal GST tag. Proteins were expressed in E. coli BL21(DE3) cells overnight at 18 °C using 1mM IPTG. 100μM Zn(OAc)2 was also added at induction to constructs containing a zinc-binding domain (HPV16-E71–98, HPV16-E717–98, HPV16-E746–98, and HPV1A-E71–93). Cells were lysed by sonication in a buffer containing 1x PBS, 7.4, 100mM NaCl, 10mM BME and 1x PMSF. 10μM Zn(OAc)2 was added to the lysis buffer of constructs containing a zinc-binding domain. The cell lysate was centrifuged at 18,000 RPM for 30 minutes and the resulting supernatant was loaded onto GST superflow resin (CLONTECH) pre-equilibrated with 1x PBS, 7.4, 100mM NaCl, and 10mM BME. The column with loaded protein was then washed with the same buffer. The GST-fused protein was then eluted using 1x PBS, 7.4, 100mM NaCl, 10mM GSH, and 10mM BME. The eluent was then concentrated and further purified using a superdex analytical column (GE Healthcare Life Sciences) in a buffer containing 20mM Tris, 7.5, 150mM NaCl, and 10mM BME.
Untagged HPV-E7 proteins
To purify untagged HPV-E7 constructs, HPV-E7 was expressed and purified as a GST-fusion as described above. After loading on the GST superflow column and washing off the contaminants, the protein was subjected to thrombin (Enzyme Research) cleavage at 20°C for 1 hour. The cleaved protein and protease were washed off the column. Ion exchange was then performed to separate E7 from the thrombin using high-trap Q HP column (Fisher) with a salt gradient (buffer A: 20mM Tris, 7.5, 50mM NaCl, 10mM BME; buffer B: 20mM Tris, 7.5, 750mM NaCl, 10mM BME). Fractions containing HPV-E7 were pooled, concentrated and run on gel filtration using a superdex 200 analytical column (GE Healthcare Life Sciences) in a buffer containing 20mM Tris 7.5, 150mM NaCl, and 10mM BME.
His-tagged Ad5-E1A and HPV16-E7 proteins
The DNA for Ad5-E1A1–189, Ad5-E1A77–189, Ad5-E1A138–189, Ad5-E1A1–138, Ad5-E1A1–77, were cloned into a pET vector containing an N-terminal 6x-histidine tag. HPV16-E71–98 was cloned into the pRSET vector, also containing an N-terminal 6x-histidine tag. Ad5-E1A and HPV16-E7 were expressed in E. coli BL21(DE3) cells overnight at 18 °C and 25 °C, respectively, using 1mM IPTG. 100μM Zn(OAc)2 was also added at induction to constructs containing a zinc-binding domain (Ad5-E1A1–189, Ad5-E1A77–189, Ad5-E1A138–189, and HPV16-E71–98). Cells were lysed by sonication in a buffer containing 20mM Tris, 7.5, 500mM NaCl, 35mM imidazole, 10mM BME and 1x PMSF. 10μM Zn(OAc)2 was added to the zinc-binding domain containing constructs. The cell lysate was centrifuged at 18,000 RPM for 30 minutes and the resulting supernatant was loaded onto a Ni-NTA column (Fisher) pre-equilibrated with 20mM Tris, 7.5, 500mM NaCl, 35mM imidazole, and 10mM BME. The column was washed and the bound protein was eluted using an imidazole gradient from 35mM to 250mM. Ion exchange was then performed using high-trap Q HP column (Fisher) with a salt gradient (buffer A: 20mM Tris, 7.5, 50mM NaCl, 10mM BME; buffer B: 20mM Tris, 7.5, 750mM NaCl, 10mM BME). Fractions containing the desired protein were pooled, concentrated and run on gel filtration using a superdex 200 analytical column (GE Healthcare Life Sciences) in a buffer containing 20mM Tris 7.5, 150mM NaCl, and 10mM BME.
p300
The DNA encoding the CH1 domain of human p300 (residues 323–424) was cloned into the pGEX-4T-1 vector, containing an N-terminal GST tag. The protein was expressed, lysed, and purified, as described above for the GST-tagged HPV-E7 proteins with a zinc-binding domain. To generate untagged p300, the protein was subjected to on-column cleavage with TEV. The protease was removed by binding to Ni-NTA resin (Fisher). Ion exchange was then performed using high-trap SP HP column (Fisher) with a salt gradient (buffer A: 20mM Tris, 7.5, 50mM NaCl, 10mM BME; buffer B: 20mM Tris, 7.5, 750mM NaCl, 10mM BME). Fractions containing the desired protein were pooled, concentrated and run on gel filtration using a superdex 200 analytical column (GE Healthcare Life Sciences) in a buffer containing 20mM Tris 7.5, 150mM NaCl, and 10mM BME.
GST alone
In order to make GST protein as a control for pull down experiments, the pGEX-4T-1 vector was transformed into BL21 (DE3) cells and expressed and purified the same way as the GST-fused proteins.
pRb
The A and B domains of pRb used in pull down experiments was expressed and purified as described elsewhere (30).
Pull Downs
30 μg of GST protein (1μM – 4μM final concentration) was incubated with 25 μL GST superflow resin (Clontech) that was prewashed in binding buffer for 15 minutes at 4° C, and an equal molar amount of binding partner to be tested was added to the mixture with incubation for 1 hour with gentle rotation at 4° C. For ternary complex formation, an additional 1 hour incubation was used for the third protein. The buffer used in each pull down is given in the text and determined based on the ability to form a complex and the ability of the proteins to stay in solution. The final reaction volume was approximately 300–400μL. The beads were then collected by centrifugation at 600g, at 4° C, for 5 minutes and washed three times using 1mL of binding buffer. Samples were then subjected to SDS-PAGE analysis. For competition experiments, 12.5 μg of GST protein was incubated with 10 μL GST superflow resin (Clontech) in a buffer of 20mM Tris, pH 8.0, 100mM NaCl, 10mM BME, and 0.05% TWEEN20. The binding partner was then incubated with the GST-tagged protein for one hour prior to the addition of different concentrations of competitor protein. After another incubation of one hour, the beads were washed as described above, run on an SDS-PAGE gel, transferred to PVDF membrane, and then probed with anti-His (1:5000, Fisher), anti-HPV16-E7 (1:5000, Abcam), and anti-GST (1:5000, Millipore) antibodies, followed by anti-mouse conjugated to HRP (1:5000, Bio-RAD). Bands were visualized by chemiluminescence (Pierce) and exposed to film (Kodak).
Size Exclusion Chromatography
To determine if the viral oncoproteins co-elute with p300CH1, equimolar amounts of each protein were added to a final volume of approximately 1mL buffer (as indicated in the text). The proteins were incubated for at least one hour prior to running on either a pre-equilibrated superdex 200 analytical column (Fisher Scientific) for the samples with Ad-E1A proteins or a pre-equilibrated superdex 75 preparatory column (Amersham Biosciences) for the samples with HPV-E7 proteins.
Equilibrium Sedimentation
Analytical ultracentrifugation of the Ad5-E1A/p300 and HPV16-E7/p300 complexes, as well as individual proteins, were performed at 4 °C with absorbance optics using a Beckman Optima XL-I analytical ultracentrifuge, using a 4-hole rotor. The partial specific volume and viscosity were estimated by using Sednterp (31). Analysis was performed by using six-channel centerpieces with quartz windows. To ensure equilibrium was reached, scans were performed every 4 hours and compared. Protein samples were analyzed at three different protein and protein complex concentrations; the optical densities used were 0.3, 0.5, and 0.7. A global fit of the data was performed for each sample using the program HeteroAnalysis. The quality of fit was assessed from the RMSD value.
RESULTS
p300CH1 Binds Strongly to Ad5-E1A, HPV16-E7 and HPV1A-E7 but with Differing Affinities
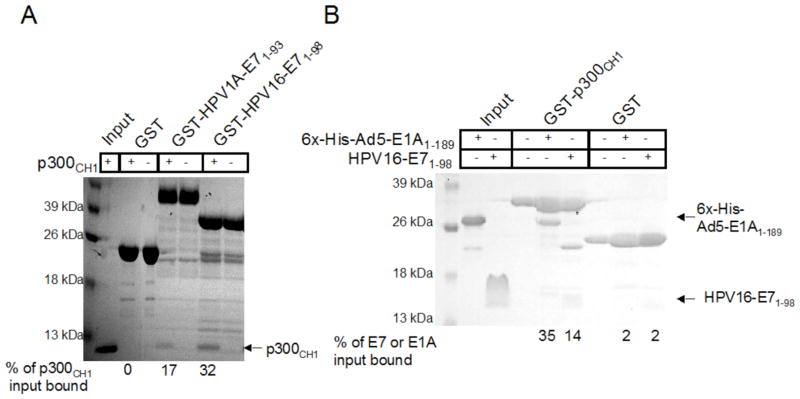
Ad5-E1A and HPV-E7 have been implicated to interact with the CH1 domain of p300 (1, 18, 23), therefore, we wanted to compare the binding of these different viral oncoproteins to p300CH1. We first wanted to determine whether full length high risk HPV16-E71–98 or low risk HPV1A-E71–93 proteins bind to p300CH1 and if so whether they bind with similar or differing affinities. To test this, full length E7 protein from each form of HPV was expressed with an N-terminal GST tag and tested at a final concentration of 2.6 μM. Binding was assessed with in vitro pull down assays on GST beads with equal molar amounts of untagged p300CH1. The buffer used for these studies was 20mM Tris, pH 8.0, 100mM NaCl, 10mM BME, and 0.05% TWEEN20 and the samples were analyzed by SDS-PAGE. As can be seen in Figure 1A, a very faint band is observed in the HPV1A-E7 lane with a darker band in the HPV16-E7 lane, suggesting that high risk HPV16-E7 binds more strongly than low risk HPV1A-E7 to p300CH1. p300CH1 did not bind to GST alone, indicating that the binding to E7 was specific and not mediated by p300CH1 non-specific aggregation.
Figure 1. Pull downs between p300 and the viral oncoproteins.
A. Comparison of the binding of HPV1A-E7 and HPV16-E7 to the CH1 domain of p300. An SDS-PAGE gel is shown for pull downs done between equal molar amounts of p300 CH1 and GST-tagged full length HPV- E7 proteins each tested at a final concentration of 2.6 μM. 3.7 μM GST was used as a control to ensure that p300 CH1 does not bind to the tag. B. Comparison of the binding of 6xHis-Ad5-E1A and untagged HPV16-E7 to p300 CH1. An SDS-PAGE gel is shown for pull downs done between GST-tagged p300CH1 and untagged HPV16-E7 or 6xHis-Ad5-E1A each tested at a final concentration of 2.6 μM. Thrombin that has not been completely removed from the HPV16-E7 purification cleaved a small amount of the GST-p300CH1 protein, resulting in an additional band, shown in lane 6. 3.7 μM GST was used as a control to ensure that HPV16-E7 and Ad5-E1A does not bind to the tag.
Since high risk HPV16-E7 bound to p300CH1 with a greater apparent affinity than low risk HPV1A-E7, we next wanted to compare the binding of full length HPV16-E7 and the CR1–3 domains of Ad5-E1A to p300CH1. For these studies, p300CH1 was fused to a GST-tag, and HPV16-E71–98 and Ad5-E1A1–189 were expressed with N-terminal 6x-histidine-tags. Similarly, pull down assays were done on GST beads with the viral oncoproteins and GST- p300CH1 at protein concentrations of 2.6 μM and 3.7 μM, respectively, with the same buffer as before and binding was assessed by running the samples on an SDS-PAGE gel. As can be seen in Figure 1B, the retained band for 6xHis-Ad5-E1A1–189 was much darker than the corresponding band for 6xHis-HPV16-E71–98 suggesting that Ad5-E1A1–189 binds more tightly to p300CH1 than HPV16-E71–98. 6xHis-Ad5-E1A1–189 and 6xHis-HPV16-E71–98, did not bind to the control GST only sample, indicating that the interactions of p300CH1 with Ad5-E1A1–189 and HPV16-E71–98 were specific and that they were not forming aggregates.
The CR1 Domain of Ad5-E1A is Necessary for Interaction with p300CH1
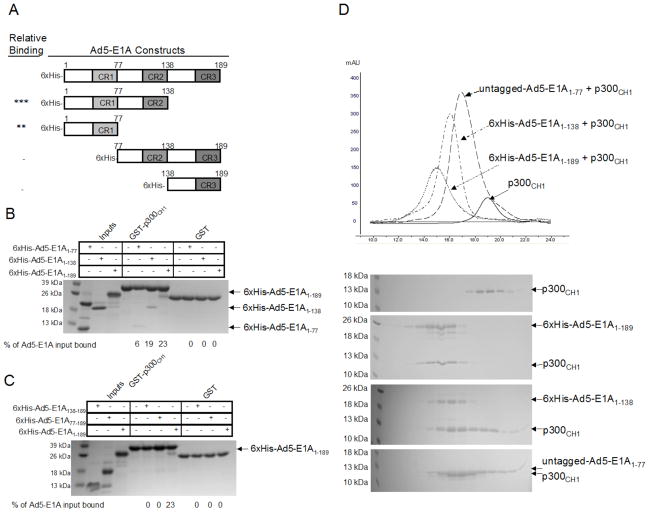
Since Ad5-E1A1–189 was able to bind to p300CH1, we next wanted to map the site of interaction on Ad5-E1A1–189. To do this, different deletion constructs of 6xHis-Ad5-E1A1–189 were made: 6xHis-Ad5-E1A1–138, 6xHis-Ad5-E1A1–77, 6xHis-Ad5-E1A77–189, and 6xHis-Ad5-E1A138–189 (Figure 2A), and tested for their ability to bind to GST- p300CH1 at protein concentrations of 2.6 μM using pull downs in a buffer of 20mM Tris, pH 8.0, 100mM NaCl, 10mM BME, and 0.05% TWEEN20. As can be seen in Figure 2B, 6xHis-Ad5-E1A1–189 and 6xHis-Ad5-E1A1–138 appeared to bind with similar affinities, suggesting that the CR3 domain of Ad5-E1A is not necessary for binding to p300CH1. This was further confirmed by the fact that 6xHis-Ad5-E1A138–189 (containing regions C-terminal to the CR2 domain) did not show detectable binding to p300CH1 (Figure 2C). 6xHis-Ad5-E1A1–77 (containing the N-terminus and CR1 domain) also showed significant binding, although the binding appeared reduced (Figure 2B). To determine if residues 77–138 were important for p300CH1 binding, 6xHis-Ad5-E1A77–189 was tested and shown not to interact with p300CH1, suggesting that Ad5-E1A1–77 is necessary and possibly sufficient for interaction with p300CH1 (Figure 2C).
Figure 2. Mapping the interaction between Ad5-E1A and the p300CH1.
A. Constructs of 6xHis-Ad5-E1A that were tested in pull down experiments. The asterisks (*) in the left column indicate the level of binding as compared to 6xHis-Ad5-E1A1–189, with more asterisks suggesting a stronger interaction to p300CH1. A minus sign (−) indicates that no interaction was observed. B. Pull downs with C-terminal deletions of 6xHis-Ad5-E1A. An SDS-PAGE gel is shown for each pull down done between the p300CH1 and the different constructs of Ad5-E1A each tested at a final concentration of 2.6 μM. The stronger upper band in the input lane for E1A1–77 in Figure 2B is a contaminant that was obtained from the expression of this construct in E. coli. and that could not be separated during the purification process. C. Pull downs with N-terminal deletions of 6xHis-Ad5-E1A. An SDS-PAGE gel is shown for each pull down experiment done between p300CH1 and the different constructs of 6xHis-Ad5-E1A each tested at a final concentration of 2.6 μM. D. Gel Filtration between 6xHis-Ad5-E1A or untagged Ad5-E1A and p300CH1. The chromatograms from size exclusion chromatography for the different complexes (final protein concentrations tested range from 40 μM to 80 μM), as well as for p300CH1 alone, are shown with the elution profiles using SDS-PAGE gels. The peaks in the gels are aligned with the corresponding peaks in the chromatograms.
Since an interaction could be seen between Ad5-E1A and p300CH1 using pull down experiments, we next wanted to determine if stable complexes of these two proteins could be isolated for more quantitative analysis. To test this, the different constructs of 6xHis-Ad5-E1A that bind to p300CH1 were mixed at a concentration of approximately 40 μM to 80 μM with the same concentration of p300CH1 and incubated for one hour before running the complex on a superdex 200 analytical column in a buffer of 20mM Tris pH 8.0, 100mM NaCl, and 5 mM BME. Since 6xHis-Ad5-E1A1–77 had many impurities, an untagged construct was made by expressing it as a GST-fusion. Consistent with results from the pull down experiments, 6xHis-Ad5-E1A1–189, 6xHis-Ad5-E1A1–138 and untagged Ad5-E1A1–77 were able to co-elute with p300CH1, as indicated by SDS-PAGE gels and by the shift in the gel filtration peak when compared to p300CH1 alone (Figure 2D). These results suggested that Ad5-E1A constructs containing residues 1–77 could form stable complexes with p300CH1.
The CR1–2 Domains of HPV16-E7 are Necessary for the Interaction with p300CH1
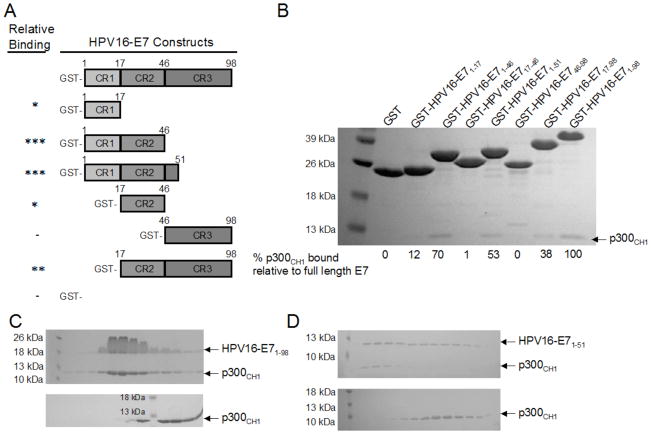
Since E1A and E7 are functionally homologous and we were able to demonstrate that residues 1–77 of Ad5-E1A, corresponding to its N-terminus and CR1 domain, are necessary for binding to p300CH1, we wanted to determine which domains of HPV16-E7 are needed for complex formation. This was especially of interest since the N-terminus of Ad5-E1A has no sequence homology to HPV16-E7. To map regions of HPV16-E7 that were required for p300CH1 interaction, we prepared different deletion constructs of GST-tagged HPV16-E7: GST-HPV16-E717–98, GST-HPV16-E746–98, GST-HPV16-E71–51, GST-HPV16-E71–46, GST-HPV16-E717–46, GST-HPV16-E71–17, and GST-HPV16-E71–98 (full length) (Figure 3A) to compare their abilities to bind to p300CH1 using pull downs in a buffer of 20mM Tris, pH 8.0, 50mM NaCl, 10mM BME, and 0.05% TWEEN20 (Figure 3B). Proteins were tested at a final concentration of 3 μM. As can be seen in Figure 3B, all of the constructs were able to bind to p300CH1 except for the GST-HPV16-E746–98 (containing only the CR3 domain). Of note, HPV16-E71–98, HPV16-E71–51, HPV16-E71–46 bound to p300CH1 most strongly, HPV16-E717–98 bound more weakly, and HPV16-E71–17 and HPV16-E717–46 bound the weakest. These results suggest that the CR2 domain, consisting of residues 17–46, was necessary but not sufficient for the interaction with the p300CH1. In fact both the CR1 and CR2 domains of HPV16-E7, consisting of residues 1–46, appeared to be sufficient for p300CH1 interaction. As was the case for Ad5-E1A, the CR3 domain of HPV16-E7 (residues 46–98) did not appear to interact at all with p300CH1 (Figure 3B).
Figure 3. Mapping the interaction between HPV16-E7 and p300CH1.
A. Constructs of HPV16-E7 that were tested in pull down experiments. The asterisks (*) in the left column indicate the level of binding as compared to the full length GST-HPV16-E71–98, with more asterisks suggesting a stronger interaction to p300CH1. A minus sign (−) indicates that no interaction was observed. B. Pull downs with the different constructs of HPV16-E7, tagged to GST. An SDS-PAGE gel is shown for each pull down done between p300CH1 and the different GST-tagged constructs of HPV16-E7, each tested at a final concentration of 3 μM. GST was used as a control to ensure that p300CH1 does not bind to the tag. C and D. Gel Filtration between HPV16-E7 and p300CH1. The elution profiles from size exclusion chromatography are shown using SDS-PAGE gels. Full length HPV16-E71–98 (C) in complex with p300CH1 (final protein concentrations tested range from 40 μM to 80 μM) is compared to the elution profile of p300CH1 alone. Similarly, residues 1–51 of HPV16-E7 (D) in complex with p300CH1 (final protein concentrations tested range from 40 μM to 80 μM) is compared to the elution profile of p300CH1 alone.
Since interactions could be seen between HPV16-E7 and p300CH1 using pull down experiments, complexes formed between the different HPV16-E7 constructs and p300CH1 were run on a superdex 75 preparatory column to assess if stable complexes could form. Untagged HPV16-E7 constructs, at a concentration of approximately 100 μM to 140 μM were used for these experiments, and they were combined with p300CH1 at the same final concentration. Size exclusion chromatography was carried out with a buffer of 20mM Tris, 50mM NaCl, and 10mM BME. The buffer was adjusted to pH 8.0 for HPV16-E71–98 and HPV16-E717–98 and to pH 7.0 for HPV16-E71–51. These proteins were also run individually for comparison against any complexes that formed. Consistent with results from the pull down experiments, only HPV16-E71–98 and HPV16-E71–51, revealed co-elution with p300CH1, suggesting that a stable complex was formed between the two proteins (Figure 3C, Figure 3D). Interestingly, HPV16-E71–98 and HPV16-E71–51 did not co-elute with p300CH1 when using a buffer with higher salt: 20mM Tris, 100mM NaCl, and 10mM BME, suggesting that E7 binds with a lower affinity to p300 than E1A, consistent with our previous results (Figure 1B). HPV16-E717–98, whose binding to p300CH1 was weaker, did not co-elute with p300CH1 (data not shown). As indicated from the SDS-PAGE gels, adding HPV16-E7 to p300CH1 shifted the elution pattern to the left, when compared to p300CH1 alone, indicative of complex formation (Figure 3C, Figure 3D).
HPV16-E7 and Ad5-E1A bind competitively to p300CH1
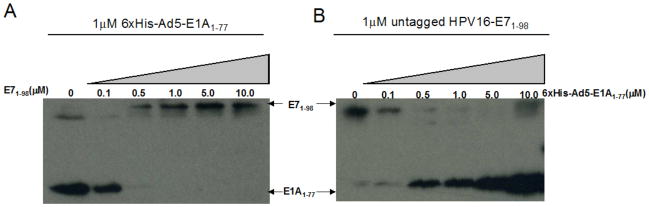
Our data indicate that the N-terminus and CR1 domain of Ad5-E1A and the CR1 and CR2 domains of HPV16-E7, respectively, are necessary and sufficient for interaction with p300CH1. These regions of the viral oncoproteins have very limited sequence homology, suggesting that they might bind to different regions of p300CH1. To test this, pull downs were performed in which either 1 μM 6xHis-Ad5E1A1–77 or untagged HPV16-E71–98 was pre-incubated with GST-tagged p300CH1 on GST-beads and then titrated with different concentrations of either untagged HPV16-E71–98 or 6xHis-Ad5E1A1–77, respectively, in a buffer of 20mM Tris pH 8.0, 100mM NaCl, and 10mM BME, and 0.05% TWEEN20. The amount of viral oncoprotein remaining bound was determined by western blotting. Surprisingly, as shown in Figure 4A and 4B, each viral oncoprotein was able to compete the other for binding to p300CH1. This suggests that these two viral oncoproteins bind competitively to p300CH1, and possibly bind to the same region of the CH1 domain of p300.
Figure 4. Competition between HPV16-E7 and Ad5-E1A for binding to p300CH1.
A.Titration of HPV16-E7 into 6xHis-Ad5-E1A1–77/p300CH1 complexes. Different concentrations of HPV16-E71–98 were added to preformed 6xHis-Ad5-E1A1–77/p300CH1 complexes and the amount of 6xHis-Ad5-E1A1–77 remaining bound to GST-p300CH1 was probed using an anti-His antibody (left panel). The amount of HPV16-E71–98 added was probed using anti-HPV16-E7 antibody (left panel). The higher molecular weight band in lane 1 corresponds to 6xHis-Ad5-E1A1–77, due to its high concentration in that sample. B. Titration of 6xHis-Ad5-E1A1–77 into HPV16-E71–98/p300CH1 complexes. Different concentrations of 6xHis-Ad5-E1A1–77 were added to preformed HPV16-E71–98/p300CH1 complexes and the amount of HPV16-E71–98 remaining bound to GST-p300CH1 was probed using an anti-HPV16-E7 antibody (right panel). The amount of 6xHis-Ad5-E1A1–77 added was probed using anti-His antibody (right panel). The higher molecular weight band in lane 6 corresponds to 6xHis-Ad5-E1A1–77, due to its high concentration in that sample.
p300CH1 Binds with a 1:1 Stochiometry to the Viral Oncoproteins
Because stable complexes could be formed between the viral oncoproteins and p300CH1, their binding stoichiometry and dissociation constants were determined. To do this, analytical ultracentrifugation (AUC) sedimentation equilibrium experiments were performed on the different complexes, as well as on the individual proteins to determine their oligomerization states. In each case, AUC sedimentation equilibrium experiments were performed at 4 °C using protein optical densities 0.7, 0.5, and 0.3 and three different speeds and a global fit of the data was performed. A summary of the constructs tested, the protein concentrations used, the speeds and models employed, as well as the results obtained, is given in Table 1. The data with residuals is provided in Figure S1 for two of the complexes. The residuals were randomly distributed above and below the baseline, indicative of good fits. To ensure that the proteins and their respective complexes stayed in solution, experiments with HPV16-E7 were done in a buffer of 20mM Tris, 50mM NaCl, and 1mM TCEP, where the pH was adjusted to 8.0 for full length HPV16-E7 and to 7.0 for HPV16-E71–51. Experiments with Ad5-E1A were done in a buffer of 20mM Tris pH 8.0, 100mM NaCl, and 1mM TCEP. Complexes were pre-run on gel filtration prior to their analysis using analytical ultracentrifugation sedimentation equilibrium experiments.
Table 1.
Summary of results from AUC sedimentation equilibrium studies.
| Concentrations Used (μM)a | Speeds Usedb (RPM) | Model Used for Fitting | Stoichiometry/ Oligomerization State | Monomer Mw (kDa) | Monomer Mw(app) (kDa) | KDf (μM) | RMS Dg (μM) | |
|---|---|---|---|---|---|---|---|---|
| p300CH1 | 55, 90, 125 | 24K, 30K, 42.5K | Single Species | Monomerd | 11.6 | 12.1d | NAh | 0.017 |
| 6xHis-Ad5- E1A1–189 | 30, 50, 65 | 20.5K, 26K, 36K | Single Species | Monomerd | 24.2 | 25.9d | NAh | 0.029 |
| 6xHis-Ad5- E1A1–138 | 65, 110, 160 | 23K, 28K, 40K | Single Species | Monomerd | 18.3 | 19.6d | NAh | 0.029 |
| Ad5-E1A1–77 | NCi | NCi | NCi | NCi | 8.5 | NCi | NAh | NCi |
| 6xHis-Ad5- E1A1–189/p300CH1 | 30, 50, 65 | 16K, 20.5K, 30K | A + B ↔ ABc | 1:1e | NAh | NAh | 0.32 | 0.026 |
| 6xHis-Ad5- E1A1–138/p300CH1 | 50, 80, 115 | 19K, 23K, 34K | A + B ↔ ABc | 1:1e | NAh | NAh | 0.10 | 0.025 |
| Ad5-E1A1–77 /p300CH1 | 100, 170, 240 | 20K, 24K, 35.5K | A + B ↔ ABc | 1:1e | NAh | NAh | 0.22 | .030 |
| HPV16-E71–98 | 50, 85, 120 | 22K, 25.8K, 36.5K | Monomer ↔ n-merc, n=2 | Dimerd | 11.0 | 11.4d | 0.53 | 0.006 |
| HPV16-E71–51 | 65, 110, 160 | 26K, 35K, 46K | Single Species | Monomerd, k | 5.8 | 5.8d | NAh | 0.018 |
| HPV16- E71–98/p300CH1 | 50, 85, 120 | 15.5K, 19K, 25.8K | nA ↔ Anc, An + B ↔ AnBj n=2 | 2:2e | NAh | NAh | 3.5 | 0.019 |
| HPV16- E71–51/p300CH1 | 90, 150, 210 | 19K, 26K, 35K | A + B ↔ ABc | 1:1e | NAh | NAh | 0.88 | 0.008 |
Concentrations were chosen so that three different optical densities would be obtained (0.3, 0.5, and 0.7) for each sample. Consequently, proteins with lower extinction coefficients were used at a higher concentration so that visible curves could be obtained.
Speeds were chosen such that the ratio of the squares of the highest to lowest speed was greater than 3, and the ratio of the squares of the middle and lowest speed was greater than 1.4.
The model chosen for the complexes was determined by the molecular weight as determined using the single species model.
The oligomerization state of the individual proteins and its apparent monomeric weight-average molecular mass was determined by using a single species model.
The binding stoichiometry of the complexes was determined by using the single species model followed by the indicated model, where the molecular weight for each monomeric protein was used for A and B.
A global KD was determined by analyzing three different concentrations of the protein complex at the different speeds simultaneously.
Root-mean-square deviation of the residuals is given.
NA denotes that the information is not applicable.
NC denotes data that was not not collected. This was due to the poorly behaved nature of this construct when alone in solution.
A molecular weight corresponding to two molecules of p300CH1 was used for B.
The KD for dimerization of E7 is comparable to results from elsewhere (28).
The individual proteins, p300CH1, 6xHis-Ad5-E1A1–189, 6xHis-Ad5-E1A1–138, and HPV16-E71–51 were fit to a single species model and their apparent molecular weights were generally within 10% of their actual molecular weights, suggesting they were monomers when alone in solution (Table 1). Untagged Ad5-E1A1–77, precipitated when alone in solution and could therefore not be tested using this technique. HPV16-E71–98, which is known to dimerize, was fit to a monomer ↔ dimer equilibrium and its monomeric molecular weight was also found to be within 10% of its actual molecular weight. Furthermore, its KD was found to be 0.53 μM, similar to what is reported elsewhere (28).
For the complexes between the viral oncoproteins and p300CH1, the data was first fit to a single species model to determine the molecular weight of the complex and the binding stoichiometry. In all cases except for HPV16-E71–98/p300CH1, the molecular weight of the complex corresponded to a 1:1 p300CH1:viral oncoprotein complex (Table 1). Therefore, in these cases, the model of A + B ↔ AB was used for fitting, where the molecular weights of each protein were used for A and B. In the case of HPV16-E71–98/p300CH1, the molecular weight of the complex appeared to be approximately 40 kDa, and so the 2A ↔ A2, A2 + B ↔ A2B model was used for fitting the data, where twice the molecular weight of p300CH1 was used for B. The model fit well, indicating that the HPV16-E71–98/p300CH1 complex binding stoichiometry is 2:2.
The dissociation constants for p300CH1 binding to 6xHis-Ad5-E1A1–189, 6xHis-Ad5-E1A1–138, and 6xHis-Ad5-E1A1–77, were calculated to be 320 nM, 100 nM, and 220 nM, respectively, suggesting a strong interaction between Ad5-E1A and p300CH1. HPV16-E7 was found to bind more weakly to p300CH1, consistent with our results using pull-downs. The dissociation constants of HPV16-E71–98 and HPV16-E71–51 binding to p300CH1 were found to be 3.5 μM and 0.88 μM, respectively.
Ad5-E1A, but not HPV16-E7, can form a ternary complex with p300 and pRb
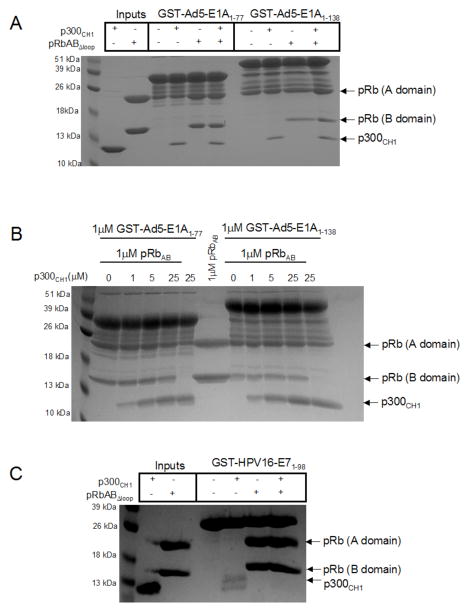
Given that the CR2 domain of HPV16-E7 and the CR1 and CR2 domains of Ad5-E1A bind to the pocket domain of pRb (26, 27, 32, 33), and that we were able to show that these same domains bind to p300CH1, we were interested to determine whether p300CH1 and the pRb pocket domain bind competitively or form a ternary complex with the viral oncoproteins. For these studies, we employed a pRb pocket domain that is missing the loop between the A and B cyclin folds (pRbABLΔloop) and therefore forms a globular heterodimer but forms 2 bands on SDS-PAGE (see Figure 5, lane 3). We first carried out pull down studies with GST-Ad5-E1A1–77 and GST-Ad5-E1A1–138, which were immobilized on GST-resin with the addition of p300CH1, pRbABΔloop or both proteins. Proteins were tested at a final concentration of 1 μM. The proteins bound to Ad5-E1A were identified by running the samples on SDS-PAGE gels. These experiments revealed that both p300CH1 and pRbABΔloop alone and in combination formed a complex with both forms of Ad5-E1A (Figure 5A). To ensure that both p300CH1 and pRbABΔloop could bind simultaneously to Ad5-E1A, the experiment was repeated where increasing amounts of p300CH1, to 25-fold excess over pRb and Ad5-E1A, were added to pre-formed complexes of pRbABΔloop/GST-Ad5-E1A. p300CH1 did not appear to compete off pRbABΔloop from either GST-Ad5-E1A1–77 or GST-Ad5-E1A1–138, suggesting that a ternary complex can form between these three proteins (Figure 5B). Interestingly, analogous pull down studies with GST-HPV16-E71–98 did not reveal the presence of an analogous ternary complex between p300CH1 and pRbABΔloop (Figure 5C), suggesting that there are molecular differences for how Ad-E1A and HPV-E7 encage p300CH1 for disruption of p300 activity.
Figure 5. Ternary complex viral oncoprotein formation with p300CH1 and pRbABΔloop.
A. Pull downs between GST-Ad5-E1A1–77, or GST-Ad5-E1A1–138, and p300CH1, and pRbABΔloop. An SDS-PAGE gel is shown in which equimolar amounts of p300CH1 alone, pRbABLloop alone, or both p300CH1 and pRbABLloop were added to the indicated construct of GST-Ad5-E1A on GST beads (final concentration of 1 μM). B. Titration of p300CH1 into GST-Ad5-E1A/pRbAB, complexes. 5-fold dilutions of p300CH1, starting at 25μM, was added to preformed GST-Ad5-E1A/pRbABLloop or GST-Ad5-E1A/pRbABLloop complex on GST-beads to see if p300CH1 could compete pRbABLloop off. The maximum amount of p300CH1 that could bind to GST-Ad5-E1A is shown in the lanes in which pRbABLloop was not added. C. Pull downs between GST-HPV16-E71–98, p300CH1, and pRbABLloop (final concentration of 2.6 μM). An SDS-PAGE gel is shown in which equimolar amounts of p300CH1 alone, pRbABLloop alone, or both p300CH1 and pRbABΔloop were added to GST-HPV16-E71–98 on GST-beads. The amount of proteins remaining bound is shown using an SDS-PAGE gel.
DISCUSSION
Our gel filtration and analytical ultracentrifugation sedimentation equilibrium data show that the interactions between the viral oncoproteins and p300CH1 are significant. The proteins were shown to bind with a 1:1 stochiometry and with dissociation constants for the Ad5-E1A/p300CH1 and HPV16-E7/p300CH1 complexes in the mid-nanomolar and low micromolar range, respectively. The poorer dissociation constants (0.88 μM–3.5 μM) for the HPV16-E7/p300CH1 complex could be due to the fact that either additional regions of p300 are necessary for the interaction, or additional proteins may be needed to further stabilize this complex. The 100–320 nM KD values that were obtained for the Ad5-E1A/p300CH1 complex, on the other hand, suggest that this interaction is very stable. Interestingly, the interaction between low risk HPV1A-E7, which is generally associated with benign lesions, and p300CH1 was much weaker than that of high risk HPV16-E7, which is known to cause a number of cancers, and p300CH1 (34–36). Consistent with our results, others have also shown that there is a stronger interaction between high risk HPV16-E7 and p300 than a different low risk E7 protein, from HPV11, and p300 (18). This suggests that the high risk HPV16-E7/p300CH1 interaction could contribute to cellular transformation and cancer. This is also consistent with results showing that Ad5-E1A can cause cellular transformation by its interaction with p300 (14–17). The ~10-fold difference in the dissociation constant between HPV16-E7 versus Ad5-E1A binding to p300 suggests that they may induce p300-mediated cellular transformation by different mechanisms.
Taking together the fact that the dissociation constants determined by analytical ultracentrifugation sedimentation equilibrium were similar (within 4-fold) for several different truncation constructs of each viral oncoprotein to p300CH1, and that these proteins exhibited similar binding to p300CH1 using pull downs and gel filtration, we were able to map regions of the viral oncoproteins necessary for the interaction. Whereas the unstructured CR1 and CR2 domains of HPV16-E7 (residues 1–46) were needed for binding to p300CH1, the N-terminus and CR1 domains of Ad5-E1A (residues 1–77) were required for binding. It is possible that p300CH1 is recruited by the the CR1-CR2 domains of HPV16-E7 and the N-terminus and CR1 domains of Ad5-E1A, after which p300 histone acetyltransferase (HAT) activity is inhibited by the other domains of the viral oncoproteins. This has already been demonstrated for E1A (37) and since Ad-E1A and HPV-E7 are functionally homologous, HPV-E7 may work the same way.
While others were able to show ternary complex formation between the CR1 domain of E1A, the CH3 domain of p300, and pRb (13), we were also able to show that a ternary complex can form with the CH1 domain of p300 as well. On the other hand, we could not detect a complex of HPV16-E7 with p300CH1 and pRb. Ternary complex formation between Ad5-E1A, pRb and p300 could result in a deregulation of pRb acetylation by p300 and therefore a change in pRb phosphorylation by cyclin dependent kinases (8). This can then result in an aberration of cell cycle control since pRb is a regulator of the G1 to S phase transition of the cell cycle (38–40). The fact that we were unable to detect an analogous ternary complex with HPV16-E7 supports the hypothesis that HPV16-E7 may induce cellular transformation by a different mechanism. It is also possible that the interaction between each viral oncoprotein and p300 may result in a modulation of other acetylated targets, such as histones, thereby providing another route through which Ad-E1A and HPV-E7 may influence gene regulation and result in cellular transformation (7).
Since both Ad5-E1A and HPV16-E7 were shown to bind competitively to p300CH1, this suggests that they may bind to the same surface of p300. Interestingly, the CH1 domain of p300 can bind many transcription factors using different parts of its surface. For example, the NMR structures of the transcription factor HIF-1α and the CITED2 transactivator were determined in complex the CH1 (TAZ1) domain of CBP, which is very similar to the corresponding domain of p300CH1, and shown to bind the TAZ1 domain in different ways (22, 41). It is also possible that the two viral oncoproteins compete with each other by some allosteric mechanism that involves binding to distinct sites on p300CH1. This would be consistent with the sequence divergance between the regions of Ad-E1A and HPV-E7 that mediate p300CH1 interaction. Either way, HPV-E7 and Ad-E1A may interfere with a certain subset of transcription factors to either activate or repress transcription. Therefore, the viral oncoproteins can cause cellular transformation by disrupting important protein complexes in addition to modulating HAT activity of p300. Notably, p53, which is an important transcription regulator and modulator of protein function, has also been suggested to bind to the CH1 domain of p300 (42, 43). Therefore, it is possible that one way in which HPV-E7 and Ad-E1A can lead to cellular transformation is by interfering with the normal activities between p300 and p53. Alternatively, these viral oncoproteins may also interfere with the binding of p300 and MDM2, a negative regulator of p53, also shown to bind to the CH1 domain of p300 (44). Although additional studies are clearly required to delineate the molecular details of p300 regulation by the HPV-E7 and Ad-E1A viral oncoproteins, our studies represent the first important demonstration that these viral oncoproteins use non-homologous regions to bind to the same p300 CH1 domain to disrupt p300 function by distinct mechanisms. This may be analogous to how HPV-E7 and Ad-E1A both disrupt pRb function by binding to the same pRb pocket domain but through distinct molecular mechanisms (26, 27, 29). Indeed, it appears that DNA tumor viruses have evolved several different strategies for disrupting normal cellular functions.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA094165 and GM060293 and a Hiliary Koprowski, M.D. Professorship awarded to R.M. D.F was supported by NIH training grant GM071339. We would like to acknowledge support of the Protein Expression and Libraries and Proteomics core facilities at the Wistar Institute (supported by NIH grant CA010815) for the studies presented here.
Footnotes
Analytical ultracentrifugation sedimentation equilibrium curves of two different p300CH1/viral oncoprotein complexes are provided along with their corresponding residuals curves. Supplemental materials may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Janknecht R. The versatile functions of the transcriptional coactivators p300 and CBP and their roles in disease. Histol Histopathol. 2002;17:657–668. doi: 10.14670/HH-17.657. [DOI] [PubMed] [Google Scholar]
- 2.Giordano A, Avantaggiati ML. p300 and CBP: partners for life and death. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 4.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell. 1999;4:365–376. doi: 10.1016/s1097-2765(00)80338-x. [DOI] [PubMed] [Google Scholar]
- 5.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 6.Blobel GA. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood. 2000;95:745–755. [PubMed] [Google Scholar]
- 7.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 8.Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 9.Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem. 2001;268:2773–2778. doi: 10.1046/j.1432-1327.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 10.Giles RH, Peters DJ, Breuning MH. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 1998;14:178–183. doi: 10.1016/s0168-9525(98)01438-3. [DOI] [PubMed] [Google Scholar]
- 11.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM, Delhanty JD, Ponder BA, Kouzarides T, Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 12.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc Natl Acad Sci U S A. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. Epigenetic reprogramming by adenovirus e1a. Science. 2008;321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnell AS, Mymryk JS. Roles for the coactivators CBP and p300 and the APC/C E3 ubiquitin ligase in E1A-dependent cell transformation. Br J Cancer. 2006;95:555–560. doi: 10.1038/sj.bjc.6603304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HG, Moran E, Yaciuk P. E1A promotes association between p300 and pRB in multimeric complexes required for normal biological activity. J Virol. 1995;69:7917–7924. doi: 10.1128/jvi.69.12.7917-7924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ait-Si-Ali S, Polesskaya A, Filleur S, Ferreira R, Duquet A, Robin P, Vervish A, Trouche D, Cabon F, Harel-Bellan A. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene. 2000;19:2430–2437. doi: 10.1038/sj.onc.1203562. [DOI] [PubMed] [Google Scholar]
- 18.Bernat A, Avvakumov N, Mymryk JS, Banks L. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene. 2003;22:7871–7881. doi: 10.1038/sj.onc.1206896. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann H, Degenkolbe R, Bernard HU, O’Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. Embo J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Guzman RN, Wojciak JM, Martinez-Yamout MA, Dyson HJ, Wright PE. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry. 2005;44:490–497. doi: 10.1021/bi048161t. [DOI] [PubMed] [Google Scholar]
- 22.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurokawa R, Kalafus D, Ogliastro MH, Kioussi C, Xu L, Torchia J, Rosenfeld MG, Glass CK. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 24.Barbosa MS, Edmonds C, Fisher C, Schiller JT, Lowy DR, Vousden KH. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and Sv40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation. Embo J. 1990;9:153–160. doi: 10.1002/j.1460-2075.1990.tb08091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JO, Russo AA, Pavletich NP. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Marmorstein R. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor. Genes & Development. 2007;21:2711–2716. doi: 10.1101/gad.1590607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clements A, Johnston K, Mazzarelli JM, Ricciardi RP, Marmorstein R. Oligomerization properties of the viral oncoproteins adenovirus E1A and human papillomavirus E7 and their complexes with the retinoblastoma protein. Biochemistry. 2000;39:16033–16045. doi: 10.1021/bi002111g. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Clements A, Zhao K, Marmorstein R. Structure of the Human Papillomavirus Oncoprotein and Its Mechanism for Inactivation of the Retinoblastoma Tumor Suppressor. Journal of Biological Chemistry. 2006;281:578– 586. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- 30.Xiao B, Spencer J, Clements A, Ali-Khan N, Mittnacht S, Broceno C, Burghammer M, Perrakis A, Marmorstein R, Gamblin SJ. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2363–2368. doi: 10.1073/pnas.0436813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lave TM, SB, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; London, United Kingdom: 1992. [Google Scholar]
- 32.Fattaey AR, Harlow E, Helin K. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol Cell Biol. 1993;13:7267–7277. doi: 10.1128/mcb.13.12.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda MA, Nevins JR. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol Cell Biol. 1993;13:7029–7035. doi: 10.1128/mcb.13.11.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burd EM. Human papillomavirus and cervical cancer. Clinical Microbiology Reviews. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 36.zur Hausen H. Papillomavirus infections--a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 37.Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, Evans RM. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 38.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 39.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes & Development. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 40.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 41.Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, Eck MJ. Structural basis for negative regulation of hypoxia-inducible factor–1alpha by CITED2. Nat Struct Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 42.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci U S A. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Kim HJ, Lim SC, Kim SH, Kim TY. Induction of apoptosis and expression of cell cycle regulatory proteins in response to a phytosphingosine derivative in HaCaT human keratinocyte cells. Mol Cells. 2003;16:331–337. [PubMed] [Google Scholar]
- 44.Kobet E, Zeng X, Zhu Y, Keller D, Lu H. MDM2 inhibits p300-mediated p53 acetylation and activation by forming a ternary complex with the two proteins. Proc Natl Acad Sci U S A. 2000;97:12547–12552. doi: 10.1073/pnas.97.23.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.