Abstract
The polyphenol quercetin (Quer) represses expression of the cardiovascular disease risk factor plasminogen activator inhibitor-1 (PAI-1) in cultured endothelial cells (ECs). Transfection of PAI-1 promoter-luciferase reporter deletion constructs identified a 251-bp fragment (nucleotides −800 to −549) responsive to Quer. Two E-box motifs (CACGTG), at map positions −691 (E-box1) and −575 (E-box2), are platforms for occupancy by several members of the c-MYC family of basic helix-loop-helix leucine zipper (bHLH-LZ) proteins. Promoter truncation and electrophoretic mobility shift/supershift analyses identified upstream stimulatory factor (USF)-1 and USF-2 as E-box1/E-box2 binding factors. ECs co-transfected with a 251 bp PAI-1 promoter fragment containing the two E-box motifs (p251/luc) and a USF-2 expression vector (pUSF-2/pcDNA) exhibited reduced luciferase activity versus p251/luc alone. Overexpression of USF-2 decreased, while transfection of a dominant-negative USF construct increased, EC growth consistent with the known anti-proliferative properties of USF proteins. Quer-induced decreases in PAI-1 expression and reduced cell proliferation may contribute, at least in part, to the cardioprotective benefit associated with daily intake of polyphenols.
Keywords: PAI-1, ENDOTHELIAL CELLS, TRANSFECTION, PROLIFERATION, TRANSCRIPTION FACTOR, USF
Polyphenols are abundant dietary micronutrients with a potential wide range of beneficial effects on human health. Epidemiologic data indicate, in fact, that the intake of polyphenols, such as quercetin (Quer), are inversely associated with mortality from cardiovascular disease (CVD) [Hertog et al., 1993; Knekt et al., 2001]. Quer is a potent anti-oxidant and metal ion chelator with anti-inflammatory, anti-neoplastic, and cardio-protective activities [Fresco et al., 2006]. This polyphenol inhibits the proliferation and migration of human aortic smooth muscle cells [Alcocer et al., 2002] likely through the transcriptional regulation of relevant genes [Abou-Agag et al., 2001; Pan et al., 2008] among the most prominent of which is plasminogen activator inhibitor-1 (PAI-1), the major regulator of pericellular plasmin generation [Pasten et al., 2007]. Elevated PAI-1 expression is an important pathologic contributor to the development and progression of CVD leading to neointimal disease, thrombosis formation, perivascular fibrosis, and eventual myocardial infarction [DeYoung et al., 2001; Kaikita et al., 2001; Pandolfi et al., 2001; Wu and Zhao, 2002; Chen et al., 2006; Westrick and Eitzman, 2007; Alessi and Juhan-Vague, 2008; Samarakoon and Higgins, 2008].
Among the PAI-1 transcriptional control elements implicated in CVD risk is the hexanucleotide E-box motif CANNTG that is a recognition platform for several members of the MYC family of the bHLH-LZ transcription factors including c-MYC, upstream stimulatory factors 1 and 2 (USF-1/2) as well as hypoxia-inducible factor-α (HIF-α) [Sawadogo and Roeder, 1985; Samoylenko et al., 2001]. The USF-1 and USF-2 genes encode the 43 and 44 kDa USF proteins 1 and 2, respectively, that function as both homo- and heterodimers to impact transcriptional outcomes [e.g., Corre and Galibert, 2005; Qi et al., 2006]. USF-1 and USF-2 have identical DNA binding specificities and, although their transcriptional activities are often gene-dependent, distinct cell-type ratios of USF homodimers and heterodimers are common which appears important in gene regulation [Sirito et al., 1994; Viollet et al., 1996; Massari and Murre, 2000; Dimova and Kietzmann, 2006]. USF-mediated PAI-1 transcriptional controls, however, may depend on cell-type rather than promoter context requiring confirmation of target gene activity in any specific cell system [e.g., Dimova and Kietzmann, 2006].
In this article we report that Quer stimulates binding of USF-2 to two distinct E-box sequences in the PAI-1 promoter resulting in down-regulation of PAI-1 expression in human coronary artery endothelial cells (HCAECs). Consistent with the anti-proliferative effects of USF proteins in other cell systems [Ismail et al., 1999; Qyang et al., 1999; Szentirmay et al., 2003; Corre and Galibert, 2005; Jung et al., 2007], transfection of human microvessel ECs with a dominant-negative (DN) USF construct stimulated, while USF-2 overexpression inhibited, cell growth. USF-2 clearly affects both the proliferative and proteolytic programs in human ECs and may, therefore, be an important contributor to the cardiovascular benefits of quercetin exposure.
MATERIALS AND METHODS
CELL CULTURE
Primary HCAECs (Cambrex, Walkersville, MD) were used between the 6th and 8th passage [Pasten et al., 2007]. Human microvessel endothelial (HMEC-1) cells were grown in DMEM/10% FBS/10 ng/ml EGF. Opti-MEM1 reduced serum medium was from BRL. Quer (Sigma) concentrations are indicated in the text.
pPAI-251/LUC, pE-BOX1/LUC, AND pE-BOX2/LUC CONSTRUCTS
The PAI-1 promoter reporter constructs, pPAI-800/luc, pPAI-549/luc, pPAI-308/luc, and pPAI-251/luc were described previously [Grenett et al., 1998a, 2001]. To generate the pE-box1/luc and pE-box2/luc constructs, complementary oligonucleotides containing three tandem E-box1 sequences (Forward: 5′-CGTCTGGACACGTGGGGGAGTGTCTGGACACGTGGGGGAGTGTCTGGACACGTGGGGGAGT-3′; Reverse: 5′-CTCGAGACTCCCCCACGTGTCCAGACACTCCCCCACGTGTCCAGACACTCCCCCACGTGTCCAGAGCT-3′) or three tandem E-box2 sequences (Forward: 5′-CGACAATCACGTGGCTGGCTAGACAATCACGTGGCTGGCTAGACAATCACGTGGCTGGCTAC-3′; Reverse: 5′-CTCGTAGCCAGCCACGTGATTGTCTAGCCAGCCACGTGATTGTCTAGCCAGCCACGTGATTGTCAGCT-3′) were synthesized with ScaI/XhoI restriction sites (underlined), annealed, and subcloned into the ScaI/XhoI site of the pGL3-control vector. Sequencing and restriction enzyme mapping verified all constructs. The pUSF-2/pcDNA3 expression vector was a kind gift of Dr. Benoit Viollet (Institut Cochin, University of Paris).
ENDOTHELIAL CELL TRANSFECTION
DNA-Lipofectin complexes were added to semi-confluent cultures in 12-well multiwell plates in Opti-MEM-1 for 5 h at 37°C [Grenett et al., 1998b, 2000]. After transfection, cells were rinsed, incubated in complete media for 18 h then stimulated with Quer (10 μmol/L) for 6 h at 37°C in serum-free medium. All constructs were co-transfected with a thymidine kinase promoter-driven Renilla luciferase reporter (pRL-TK) [Farr and Roman, 1992; Lorenz et al., 1996]. Firefly luciferase activities were normalized to corresponding Renilla luciferase signal to correct for variations in DNA uptake when using different plasmids.
ELECTROPHORETIC MOBILITY SHIFT ASSAY
Nuclear extracts [Grenett et al., 2001] were incubated with 32P-labeled E-box1 or E-box2 probes. Nonlabeled wild-type or mutant deoxyoligonucleotides were added at 100-fold molar excess prior to addition of the 32P-labeled target sequences (Table I). Supershift antibodies were to the N-terminus of USF-1 or USF-2 (Santa Cruz). For certain analyses, nuclear extracts were treated with lambda protein phosphatase. Dried gels were analyzed with a Molecular Dynamics PhosphoImager (Molecular Dynamics).
TABLE I.
E-box1 and E-box2 Probes Used in Electrophoretic Mobility Shift Assays
| Oligo E-box1 | 5′-GTCTGGACACGTGGGGGAGTC-3′ |
| Oligo mut E-box1 | 5′-GTCTGGACTTAATGGGGAGTC-3′ |
| Oligo E-box2 | 5′-AGACAATCACGTGGCTGGCTG-3′ |
| Oligo mut E-box2 | 5′-AGACAATCTTAATGCTGGCTG-3′ |
The wild-type E-box motifs are underlined and the mutant sequences underlined and highlighted in boldface. Only top strand illustrated.
PROLIFERATION ASSAY
Cells were cultured in 24-well plates at a density of 15 ×103/well, growth arrested in medium containing 0.5% calf serum for 24 h, incubated with or without Quer (10 μmol/L), or transfected with the pUSF-2/pcDNA [Fresco et al., 2006] construct or empty vector prior to maintenance for 0, 24, 48, and 72 h in media containing 5% calf serum. Cell proliferation was determined using the CyQUANT assay (Invitrogen). Briefly, CyQuant GR solution (200 μl) was added to each well, incubated for 5 min at room temperature, relative fluorescence (480-nm excitation, 520-nm emission) measured with a Synergy™ HT Multi-Detection Microplate Reader (BioTek, Winooski, VT) and standardized for cell number. HMEC-1 cells were transfected with a DN-USF (USF-A) construct [Allen et al., 2005], maintained in complete HMEC-1 growth medium and the fraction of Ki-67-expressing cells assessed immunocytochemically.
STATISTICAL ANALYSIS
All data were evaluated using the Student’s t-test; values of P < 0.05 were considered significant.
RESULTS
EFFECT OF QUER ON PAI-1 PROMOTER ACTIVITY
We demonstrated previously that Quer down-regulates PAI-1 expression in cultured HCAECs likely through responsive sequences in a 1.1 kb promoter region (pPAI-1100/luc construct) [Pasten et al., 2007]. To further define the molecular basis of PAI-1 gene regulation, ECs transiently transfected with 5′ deletion constructs of the pPAI-1100/luc were incubated in the absence or presence of 10 μmol/L Quer for 6 h. pPAI-800/luc reporter activity declined by ~55% in Quer-treated cells; pPAI-549/luc and pPAI-308/luc signals, in contrast, were unaffected (Fig. 1) suggesting that Quer-responsive promoter elements likely reside in the 251-bp promoter fragment upstream of bp −549 (i.e., bps −800 to −549) relative to the start site of PAI-1 transcription. This 251-bp region had significant independent functional potential since ECs transfected with the pPAI-251/luc reporter and stimulated with Quer exhibited the same ~50–60% reduction in luciferase activity compared with control as did the pPAI-800/luc construct (Fig. 2).
Fig. 1.
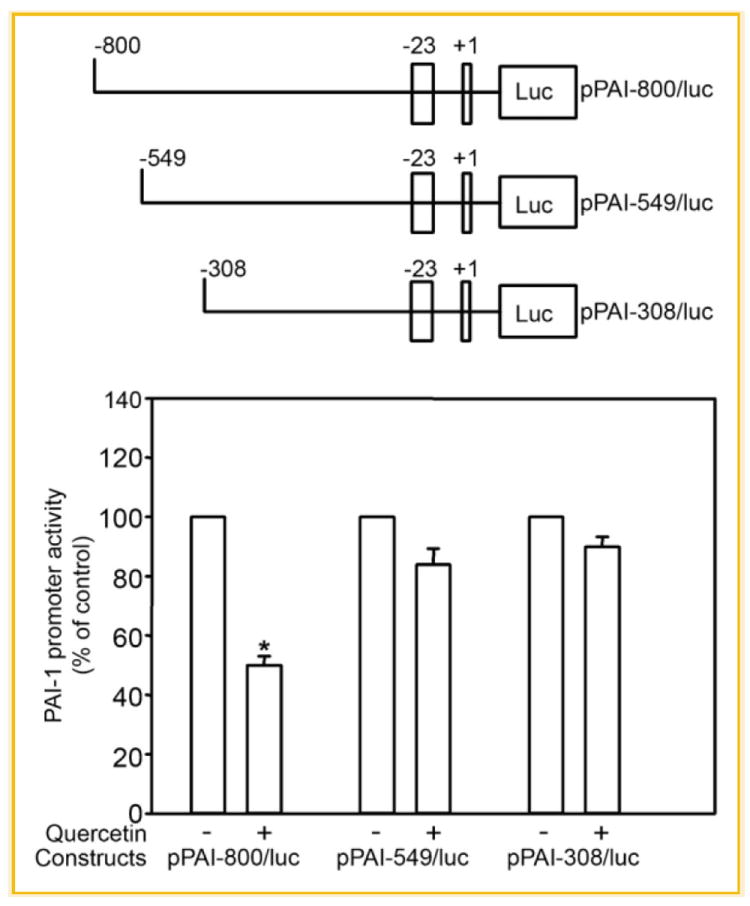
Identification of a quercetin responsive region between nt −800 and −549 in the PAI-1 promoter. A: Schematic representation of the luciferase reporter constructs containing the pPAI-800/luc, pPAI-549/luc, and pPAI-308/luc) of human PAI-1 promoter. B: ECs transiently transfected with pPAI-800/luc, pPAI-549/luc, and pPAI-308/luc were treated with quercetin or vehicle for 6 h. The columns and vertical bar represent means ± SE of five separated replicates; *P ≤ 0.05.
Fig. 2.
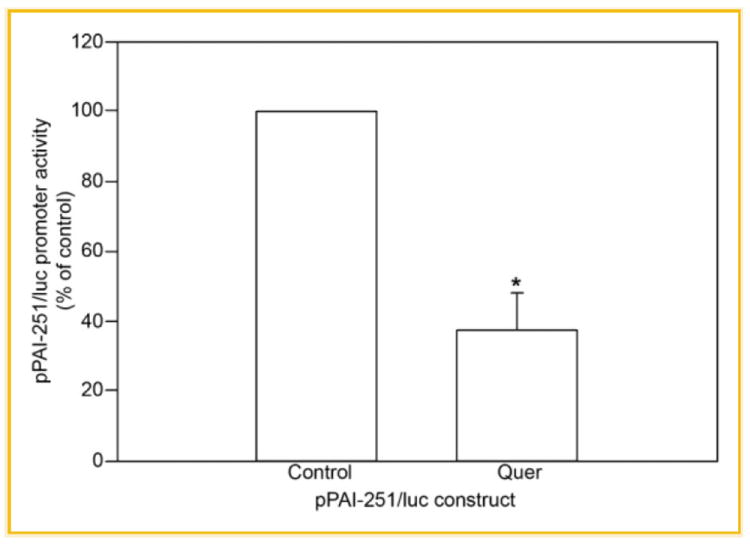
Functional analysis of the 251-bp promoter fragment of the PAI-1 gene. ECs were transfected with the pPAI-251/luc reporter construct and treated or not with 10 μM quercetin. The relative luciferase activity of unstimulated HCAEC was set as 100%. Values were expressed as mean ± SE of five individual experiments; *P ≤ 0.05.
FUNCTIONAL ANALYSIS OF THE PAI-1 E-BOX1 AND E-BOX2 SITES
The 251-bp PAI-1 promoter fragment contains two E-box motifs (CACGTG) that map to positions −691 (E-box1) and −575 (E-box2) (Fig. 3). These hexanucleotide sequences are recognized binding platforms for bHLH-LZ proteins of the c-MYC family. E-box2, moreover, is a typical classic “B” class sequence as the CACGTG motif is preceded by a T residue (Table I). EC cells transfected with luciferase reporters driven by tandomly generated triplicate sequences representing either E-box1 or E-box2 were incubated in the absence or presence of Quer (10 μmol/L) to determine the independent functionality of E-box1 and E-box2. Quer-treated transfectants (both E-box1 and E-box2) exhibited the typical 40–50% reduction in normalized luciferase activity compared to untreated controls (Fig. 4).
Fig. 3.
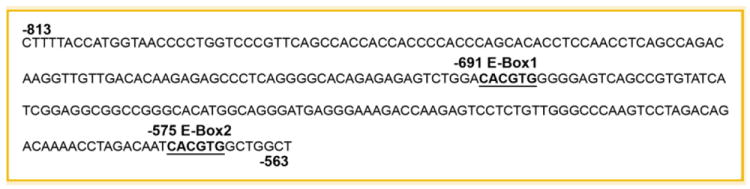
The human PAI-1 gene promoter region spanning nucleotides −813 to −563. Position of the E-box1 (−691) and E-box2 (−575) are represented in boldface.
Fig. 4.
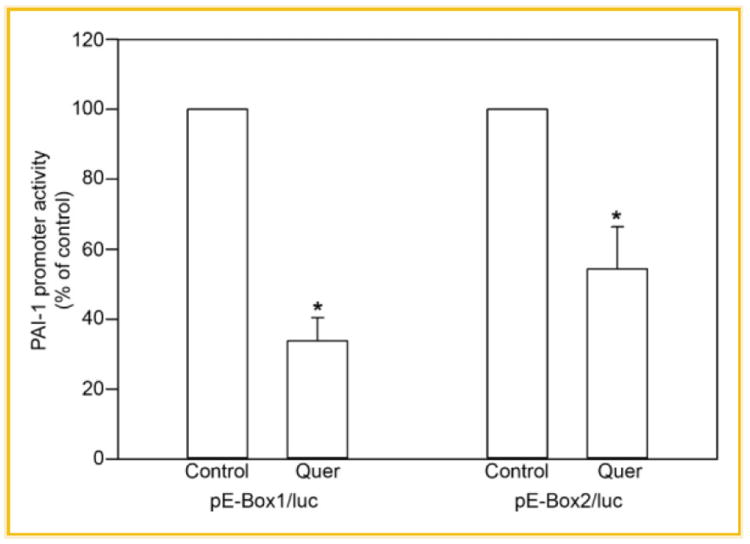
Functional analysis of the PAI-1 E-box1 and E-box2. ECs were transfected with reporter constructs bearing 3 tandem E-box1 or E-box2 sequences (see the Materials and Methods Section) cloned into the pGL-3 control vector. After transfection, cells were left untreated (control) or incubated with 10 μM quercetin. The relative luciferase activity of unstimulated HCAEC was set as 100%. Values were expressed as means ± SE of five individual experiments; *P ≤ 0.05.
Since E-box motifs are functionally significant in the response to Quer, nuclear extracts from Quer-treated or -untreated ECs were incubated with 32P-labeled double-stranded deoxyoligonucleotides spanning E-box1 and E-box2 and their respective flanking sequences to identify binding elements (Table I). Extracts from Quer-treated cells had enhanced binding to both E-box target probes compared to untreated controls (Fig. 5A,B). Binding specificity was confirmed by addition of a 100× molar excess of unlabeled competitor wild-type sequences or the appropriate mutants. As a first approximation to identify the potential binding factors, antibodies against USF-1, USF-2, or c-MYC were included in the EMSA reaction. Addition of either USF-1 or USF-2 (but not c-MYC) antibodies supershifted the DNA–protein complexes (Fig. 5) while pre-incubation of Quer-treated nuclear extracts with lambda phosphatase prior to addition of labeled probe effectively inhibited the formation of the typical band shift pattern, consistent with the phosphorylation requirements of USF proteins for probe recognition.
Fig. 5.
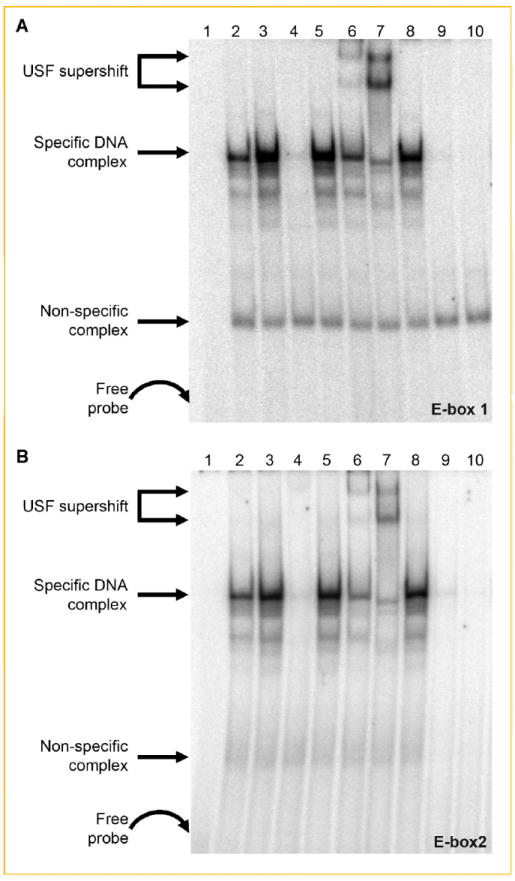
Identification of USF-1 and USF-2 as E-box-binding proteins. Nuclear proteins from untreated and Quercetin-treated cells were incubated with 32P-labeled double-stranded oligonucleotides to E-box1 (A) or E-box2 (B). Lane 1, probe only; lane 2, untreated NE; lane 3, quercetin-treated NE; lane 4, quercetin-treated NE and 100-fold excess of unlabeled probe; lane 5, quercetin-treated NE and 100-fold excess of mutated E-box1 (A) or E-box2 (B); lane 6, quercetin-treated NE and USF-1 antibody; lane 7, quercetin-treated NE and USF-2 antibody; lane 8, quercetin-treated NE and c-Myc anibody; lane 9, unstimulated NE treated with lambda protein phosphatase; and lane 10, quercetin-stimulated NE treated with phosphatase. The two supershifted bands in (A) and (B) lane 7 and, to a lesser extent, lane 6 likely are due to the relative abundance of different complexes (i.e., USF-2 homodimers vs. USF-1/2 heterodimers) that bind to E-box1 and E-box2 as a function of stimulus conditions [e.g., Allen et al., 2005; Qi et al., 2006].
The most prominent supershift was generated using USF-2 antibodies (Fig. 5A,B) and whereas the transcriptional activities of USF family members often gene-specific [Dimova and Kietzmann, 2006], it was important to determine if USF-2 represses luciferase activity of the p251/luc construct. Cells were co-transfected with p251/luc and a plasmid encoding the USF-2 transcription factor (pUSF-2/pcDNA3) and luceriferase reporter signal assessed as described in Figure 1. Co-transfectants had a ~70–80% reduction in luciferase activity compared with ECs transfected with the p251/luc alone (Fig. 6).
Fig. 6.

Effect of USF-2 on pPAI-251/luc transcription. pPAI-251/luc and pUSF-2/pcDNA were introduced simultaneously into HCAECs. Overexpression of USF-2 significantly decreased the relative luciferase activity of the pPAI-251/luc reporter. Signal from ECs transfected with pPAI-251 alone was set as 100%. Values were expressed as mean ± SE of five individual experiments; *P ≤ 30.05.
EFFECT OF QUER AND USF-2 ON ENDOTHELIAL CELL PROLIFERATION
Quer inhibits proliferation in both human vascular smooth muscle cells and microvascular ECs [Alcocer et al., 2002; Fan et al., 2003]. PAI-1 is differentially associated with both cellular proliferation as well as the onset of replicative senescence [e.g., Kortlever et al., 2006; Qi et al., 2006] likely as a consequence of the relative level of expression of this SERPIN in specific cell types. USF family members also regulate an anti-proliferative program by initiating a cell-cycle stage-specific arrest [Jung et al., 2007]. Since Quer represses PAI-1 expression through USF-2-binding E-box sites and overexpression of USF-2 suppresses activation of a PAI-1 promoter-dependent reporter (Fig. 6), we examined the effect of Quer and USF-2 on cell proliferation as well as the effect of inhibition of USF function on HMEC-1 growth. ECs exposed to Quer, or transfected with pUSF-2/pcDNA, showed a time-dependent proliferative arrest compared with untreated cells or empty vector transfectants (Fig. 7A). Conversely, expression of the DN-USF construct (USF-A) markedly increased the number of Ki-67-positve HMEC-1 cells (Fig. 7B) indicating that manipulation of the level or function of these c-MYC family members has profound consequences on EC growth (Fig. 7B).
Fig. 7.
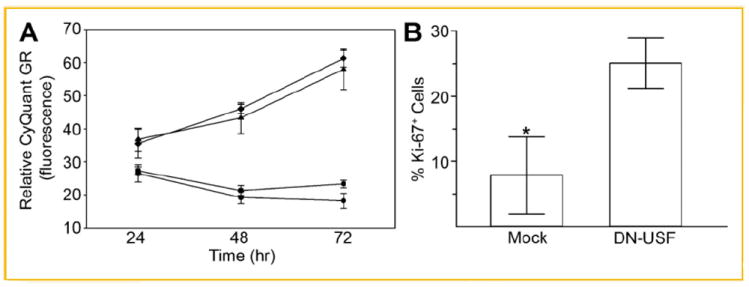
Effects of Quer and USF-2 on cell proliferation. A: ECs were incubated with Quer (10 μM) or transfected with the pUSF-2/pcDNA construct for 24, 48, and 72 h. Quercetin and overexpression of USF-2 significantly decreased proliferation. ◆, control media; ▼, pcDNA vector; ■, quercetin; ●, pUSF-2/pcDNA. Values were expressed as mean ± SE of five individual experiments; *P ≤ 0.05. B: ECs were transfected with a DN-USF construct and the fraction of cells expressing Ki-67, a marker for cell proliferation, assessed immunocytochemically. Data plotted is the mean ± SE for three arbitrary-selected sets of 100 cells each demonstrating nuclear Ki-67 immunofluorescence. *P ≤ 0.05. It was unlikely that apoptosis was a consideration in either the USF-2-dependent proliferative restriction or the growth response to DN-USF since DAPI staining (as a nuclear marker) did not indicate any significant change in the number of apoptotic cells.
DISCUSSION
The polyphenols catechin and quercetin transcriptionally repress PAI-1 gene expression in cultured HCAEC [Pasten et al., 2007]; the 251-bp fragment that maps to nt −800 and −549 in the PAI-1 promoter likely mediates Quer-initiated PAI-1 repression. Two resident E-box motifs (CACGTG), at positions −691 (E-box1) and position −575 (E-box2), are binding platforms for bHLH-LZ transcription factors including members of the MYC family (e.g., USF-1/2, c-Myc/Max). Functional analysis of this pPAI-251/luc fragment confirmed that Quer treatment effectively suppressed p251luc reporter activity. pE-box1/luc or pE-box2/luc constructs, moreover, independently exhibited Quer-dependent transcriptional inhibition implicating both E-box motifs as functional sites in Quer-mediated PAI-1 repression in human ECs.
Gel shift/competition analyses using labeled E-Box1 or E-Box2 oligonucleotide probes further indicated that intact E-box consensus motifs are necessary for site occupancy by USF-1/2 dimers. It appears, however, that the two USF species differ in affinity for, or relative abundance on, the two PAI-1 E box sites reflecting, perhaps, chromatin context, particular flanking DNA sequences or the low abundance of USF-1 in ECs but also cooperative or indirect binding with other accessory co-repressors. Although these PAI-1 E-Box motifs can, theoretically, bind both USF and c-MYC, c-MYC does not occupy the PAI-1 promoter in Quer-treated ECs likely because B-class E boxes are, at least in certain genes, preferential USF targets. Overexpression of wild-type or DN forms of USF or c-MYC revealed selectivity in gene activation by USF or c-MYC, highlighting the importance role of promoter context in determining whether a given E-box is selectively regulated by USF and c-MYC [Bruno et al., 2004]. Nuclear extracts treated with phosphatase before the binding assays, moreover, confirmed that USF phosphorylation is required for PAI-1 E-box motif occupancy in ECs as is the case in human keratinocytes [e.g., Qi et al., 2006], consistent with the phosphorylation requirement of certain bHLH-LZ factors for E-Box motif recognition [Cheung et al., 1999; Providence et al., 2002; Ma et al., 2007]. Finally, the role of USF-2 in PAI-1 repression of PAI-1 was confirmed by co-transfection of ECs with p251/luc and USF-2/pcDNA constructs, illustrating that USF-2-dependent expression controls may reflect both cell type [e.g., Qi et al., 2006; this article] and Quer- or growth factor-related recruitment of co-repressors or co-activators, respectively.
Given the central role of USF-2 in growth control, it was necessary to determine whether increased expression of USF-2 affected vascular cell proliferation. Cell proliferation was significantly decreased in cells treated with Quer or transfected with USF-2 and stimulated almost threefold in ECs expressing a DN-USF construct. USF overexpression inhibits cell proliferation in papillary thyroid cancer cells line and rat embryo fibroblasts among others [Aperlo et al., 1996; Luo and Sawadogo, 1996; Qyang et al., 1999; Jung et al., 2007] and loss of USF transcriptional activity is a frequent occurrence in breast cancer [Ismail et al., 1999]. The anti-proliferative effect of USF, however, may not be common to most cells types. In Saos-2 osteosarcoma, for example, USF did not inhibit proliferation [Qyang et al., 1999]. Thus, the transcriptional activity of USF proteins appear to be context dependent and may indicate that modulation of USF action by transcriptional co-activators and post-translational modification may be cell-type specific. While the exact mechanism responsible for suppression of cellular proliferation is not clear, USF might exert an inhibitory effect through the activation of cell-cycle regulatory proteins [Corre and Galibert, 2005; Jung et al., 2007]. Collectively, the present findings suggest that USF-2 is an important regulator of both PAI-1 expression and proliferation by Quer in ECs. These results may provide both a rationale for the potential use of quercetin in the prophylaxis of CVDs and insights as to the molecular basis for such novel therapies.
Acknowledgments
This work was supported by National Institutes of Heath grants PO1 HL70610, Project 2 (H.E.G.) and GM57242 (P.J.H.).
Grant sponsor: National Institutes of Heath grants; Grant numbers: PO1 HL70610, GM57242.
References
- Abou-Agag LH, Aikens ML, Tabengwa EM, Benza RL, Shows SR, Grenett HE, Booyse FM. Polyphyenolics increase t-PA and u-PA gene transcription in cultured human endothelial cells. Alcohol Clin Exp Res. 2001;25:155–162. [PubMed] [Google Scholar]
- Alcocer F, Whitley D, Salazar-Gonzalez JF, Jordan WD, Sellers MT, Eckhoff DE, Suzuki K, Macrae C, Bland KI. Quercetin inhibits human vascular smooth muscle cell proliferation and migration. Surgery. 2002;131:198–204. doi: 10.1067/msy.2002.119190. [DOI] [PubMed] [Google Scholar]
- Alessi MC, Juhan-Vague I. Metabolic syndrome, haemostasis and thrombosis. Thromb Haemost. 2008;99:995–1000. doi: 10.1160/TH07-11-0682. [DOI] [PubMed] [Google Scholar]
- Allen RR, Qi L, Higgins PJ. Upstream stimulatory factor regulates E Box-dependent PAI-1 transcription in human epidermal keratinocytes. J Cell Physiol. 2005;203:156–165. doi: 10.1002/jcp.20211. [DOI] [PubMed] [Google Scholar]
- Aperlo C, Boulukos KE, Pognonec P. The basic region/helix-loop-helix/leucine repeat transcription factor USF interferes with Ras transformation. Eur J Biochem. 1996;241:249–253. doi: 10.1111/j.1432-1033.1996.0249t.x. [DOI] [PubMed] [Google Scholar]
- Bruno ME, West RB, Schneeman TA, Bresnick EH, Kaetzel CS. Upstream stimulatory factor but not c-Myc enhances transcription of the human polymeric immunoglobulin receptor gene. Mol Immunol. 2004;40:695–708. doi: 10.1016/j.molimm.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Chen Y, Budd RC, Kelm RJ, Jr, Sobel BE, Schneider DJ. Augmentation of proliferation of vascular smooth muscle cells by plasminogen activator inhibitor type 1. Arterioscler Thromb Vasc Biol. 2006;26:1777–1783. doi: 10.1161/01.ATV.0000227514.50065.2a. [DOI] [PubMed] [Google Scholar]
- Cheung E, Mayr P, Coda-Zabetta F, Woodman PG, Boam DS. DNA-binding activity of the transcription factor upstream stimulatory factor 1 (USF-1) is regulated by cyclin-dependent phosphorylation. Biochem J. 1999;344:145–152. [PMC free article] [PubMed] [Google Scholar]
- Corre S, Galibert MD. Upstream stimulating factors: Highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- DeYoung MB, Tom C, Dichek DA. Plasminogen activator inhibitor type 1 increases neointima formation in balloon-injured rat carotid arteries. Circulation. 2001;104:1972–1977. doi: 10.1161/hc4101.097110. [DOI] [PubMed] [Google Scholar]
- Dimova EY, Kietzmann T. Cell type-dependent regulation of the hypoxia-responsive plasminogen activator inhibitor-1 gene by upstream stimulatory factor-2. J Biol Chem. 2006;281:2999–3005. doi: 10.1074/jbc.M512078200. [DOI] [PubMed] [Google Scholar]
- Fan PS, Gu ZL, Sheng R, Liang ZQ, Wang XX, Zhu Y. Inhibitory effect of quercetin on proliferation of human microvascular endothelial cells in vitro. Acta Pharmacol Sin. 2003;24:1231–1234. [PubMed] [Google Scholar]
- Farr A, Roman A. A pitfall of using a second plasmid to determine transfection efficiency. Nucleic Acids Res. 1992;20:920–922. doi: 10.1093/nar/20.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco P, Borges F, Diniz C, Marques MP. New insights on the anti-cancer properties of dietary polyphenols. Med Res Rev. 2006;26:747–766. doi: 10.1002/med.20060. [DOI] [PubMed] [Google Scholar]
- Grenett HE, Benza RL, Fless GM, Li XN, Davis GC, Booyse FM. Genotype-specific transcriptional regulation of PAI-1 gene expression by insulin, hypertriglyceridemic VLDL, Lp(a) in transfected cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 1998a;18:1803–1809. doi: 10.1161/01.atv.18.11.1803. [DOI] [PubMed] [Google Scholar]
- Grenett HE, Aikens ML, Torres JA, Demissie S, Tabengwa EM, Davis GC, Booyse FM. Ethanol transcriptionally upregulates t-PA and u-PA gene expression in cultured human endothelial cells. Alcohol Clin Exp Res. 1998b;22:849–853. [PubMed] [Google Scholar]
- Grenett HE, Aikens ML, Tabengwa EM, Davis GC, Booyse FM. Ethanol downregulates transcription of the PAI-1 gene in cultured human endothelial cells. Thromb Res. 2000;97:247–255. doi: 10.1016/s0049-3848(99)00172-3. [DOI] [PubMed] [Google Scholar]
- Grenett HE, Wolkowicz PE, Benza RL, Tresnak JK, Wheeler CG, Booyse FM. Identification of a 251-bp fragment of the PAI-1 gene promoter that mediates the ethanol-induced suppression of PAI-1 expression. Alcohol Clin Exp Res. 2001;25:629–636. [PubMed] [Google Scholar]
- Hertog MG, Feskens EJ, Hollman PC, Katan MB, Kromhout D. Dietary antioxidant flavonoids and risk of coronary heart disease: The Zutphen Elderly Study. Lancet. 1993;342:1007–1011. doi: 10.1016/0140-6736(93)92876-u. [DOI] [PubMed] [Google Scholar]
- Ismail PM, Lu T, Sawadogo M. Loss of USF transcriptional activity in breast cancer cell lines. Oncogene. 1999;18:5582–5591. doi: 10.1038/sj.onc.1202932. [DOI] [PubMed] [Google Scholar]
- Jung HS, Kim KS, Chung YJ, Chung HK, Min YK, Lee MS, Lee MK, Kim KW, Chung JH. USF inhibits cell proliferation through delay in G2/M phase in FRTL-5 cells. Endocr J. 2007;54:275–285. doi: 10.1507/endocrj.k06-120. [DOI] [PubMed] [Google Scholar]
- Kaikita K, Fogo AB, Ma L, Schoenhard JA, Brown NJ, Vaughan DE. Plasminogen activator inhibitor-1 deficiency prevents hypertension and vascular fibrosis in response to long-term nitric oxide synthase inhibition. Circulation. 2001;104:839–844. doi: 10.1161/hc3301.092803. [DOI] [PubMed] [Google Scholar]
- Knekt P, Ritva J, Antti R, Jouni M. Flavoid intake and coronary mortality in Finland: A Cohort Study. Br Med J. 2001;312:478–481. doi: 10.1136/bmj.312.7029.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescnce. Nature Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz WW, Cormier MJ, O’Kane DJ, Hua D, Escher AA, Szalay AA. Expression of the Renilla reniformis luciferase gene in mammalian cells. J Biolumin Chemilumin. 1996;11:31–37. doi: 10.1002/(SICI)1099-1271(199601)11:1<31::AID-BIO398>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Luo X, Sawadogo M. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc Natl Acad Sci USA. 1996;93:1308–1313. doi: 10.1073/pnas.93.3.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Jhun B, Oh CK. Upstream stimulating factor-1 mediates the E-box-dependent transcriptional repression of the plasminogen activator inhibitor-1 gene in human mast cells. FEBS Lett. 2007;581:4485–4490. doi: 10.1016/j.febslet.2007.08.034. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Chang MJ, Booyse FM, Grenett HE, Bradley KM, Wolkowicz PE, Shang Q, Tabengwa EM. Quercetin induced tissue-type plasminogen activator expression is mediated through Sp1 and p38 mitogen-activated protein kinase in human endothelial cells. J Thromb Haemost. 2008;6:976–985. doi: 10.1111/j.1538-7836.2008.02977.x. [DOI] [PubMed] [Google Scholar]
- Pandolfi A, Cetrullo D, Polishuck R, Alberta MM, Calafiore A, Pellegrini G, Vitacolonna E, Capani F, Consoli A. Plasminogen activator inhibitor type 1 is increased in the arterial wall of type II diabetic subjects. Arterioscler Thromb Vasc Biol. 2001;21:1378–1382. doi: 10.1161/hq0801.093667. [DOI] [PubMed] [Google Scholar]
- Pasten C, Olave NC, Zhou L, Tabengwa EM, Wolkowicz PE, Grenett HE. Polyphenols downregulate PAI-1 gene expression in cultured human coronary artery endothelial cells: Molecular contributor to cardiovascular protection. Thromb Res. 2007;121:59–65. doi: 10.1016/j.thromres.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Providence KM, White LA, Tang J, Gonclaves J, Staiano-Coico L, Higgins PJ. Epithelial monolayer wounding stimulates binding of USF-1 to an E-box motif in the plasminogen activator inhibitor type 1 gene. J Cell Sci. 2002;115:3767–3777. doi: 10.1242/jcs.00051. [DOI] [PubMed] [Google Scholar]
- Qi L, Allen RR, Lu Q, Higgins CE, Garone R, Staiano-Coico L, Higgins PJ. PAI-1 transcriptional regulation during the G0→G1 transition in human epidermal keratinocytes. J Cell Biochem. 2006;99:495–507. doi: 10.1002/jcb.20885. [DOI] [PubMed] [Google Scholar]
- Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M. Cell-type-dependent activity of the ubiquitous transcription factor USF in cellular proliferation and transcriptional activation. Mol Cell Biol. 1999;19:1508–1517. doi: 10.1128/mcb.19.2.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon R, Higgins PJ. Integration of non-SMAD and SMAD signaling in TGF-β1-induced plasminogen activator inhibitor type-1 gene expression in vascular smooth muscle cells. Thromb Haemost. 2008;100:976–983. [PMC free article] [PubMed] [Google Scholar]
- Samoylenko A, Roth U, Jungermann K, Kietzmann T. The upstream stimulatory factor-2a inhibits plasminogen activator inhibitor-1 gene expression by binding to a promoter element adjacent to the hypoxia-inducible factor-1 binding site. Blood. 2001;97:2657–2666. doi: 10.1182/blood.v97.9.2657. [DOI] [PubMed] [Google Scholar]
- Sawadogo M, Roeder RG. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous expression of the 43-and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentirmay MN, Yang HX, Pawar SA, Vinson C, Sawadogo M. The IGF2 receptor is a USF2-specific target in nontumorigenic mammary epithelial cells but not in breast cancer cells. J Biol Chem. 2003;278:37231–37240. doi: 10.1074/jbc.M305791200. [DOI] [PubMed] [Google Scholar]
- Viollet B, Lefrancois-Martinez AM, Henrion A, Kahn A, Raymondjean M, Martinez A. Immunochemical characterization and transacting properties of upstream stimulatory factor isoforms. J Biol Chem. 1996;271:1405–1415. doi: 10.1074/jbc.271.3.1405. [DOI] [PubMed] [Google Scholar]
- Westrick RJ, Eitzman DT. Plasminogen activator inhibitor-1 in vascular thrombosis. Curr Drug Targets. 2007;8:966–1002. doi: 10.2174/138945007781662328. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhao Z. Inhibition of PAI-1: A new anti-thrombotic approach. Curr Drug Targets Cardiovasc Haematol Disord. 2002;2:27–42. doi: 10.2174/1568006023337727. [DOI] [PubMed] [Google Scholar]


